Comparative Assessment of Bacteriophage and Antibiotic Activity against Multidrug-Resistant Staphylococcus aureus Biofilms
Abstract
:1. Introduction
2. Results
2.1. MIC Determination
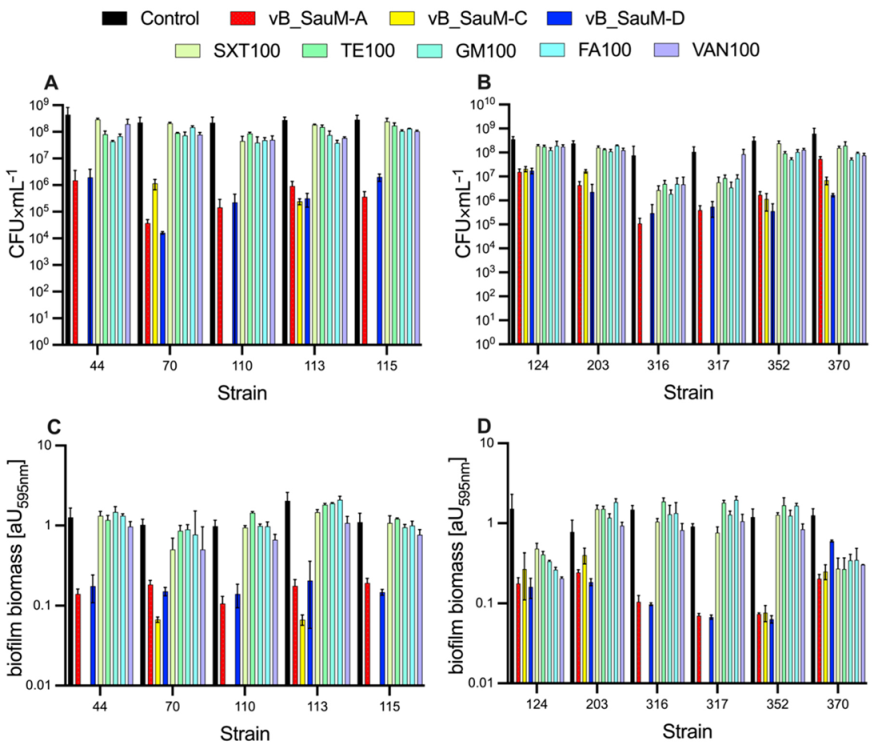
2.2. Biofilm Eradication by Phages
2.3. Biofilm Eradication by Antibiotics
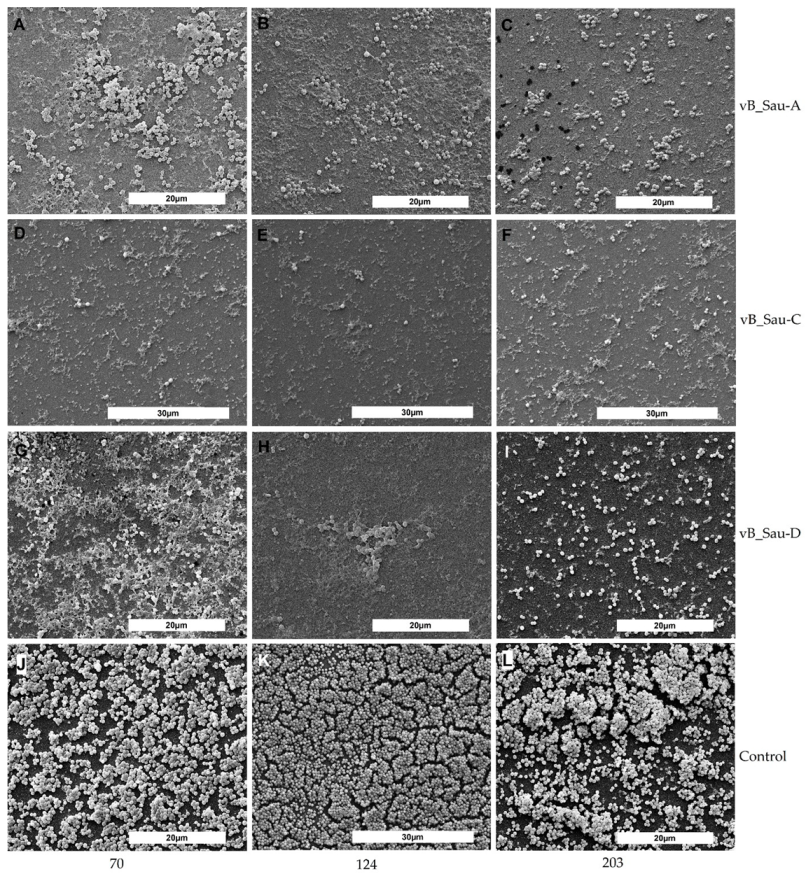
2.4. SEM Analysis of Biofilm
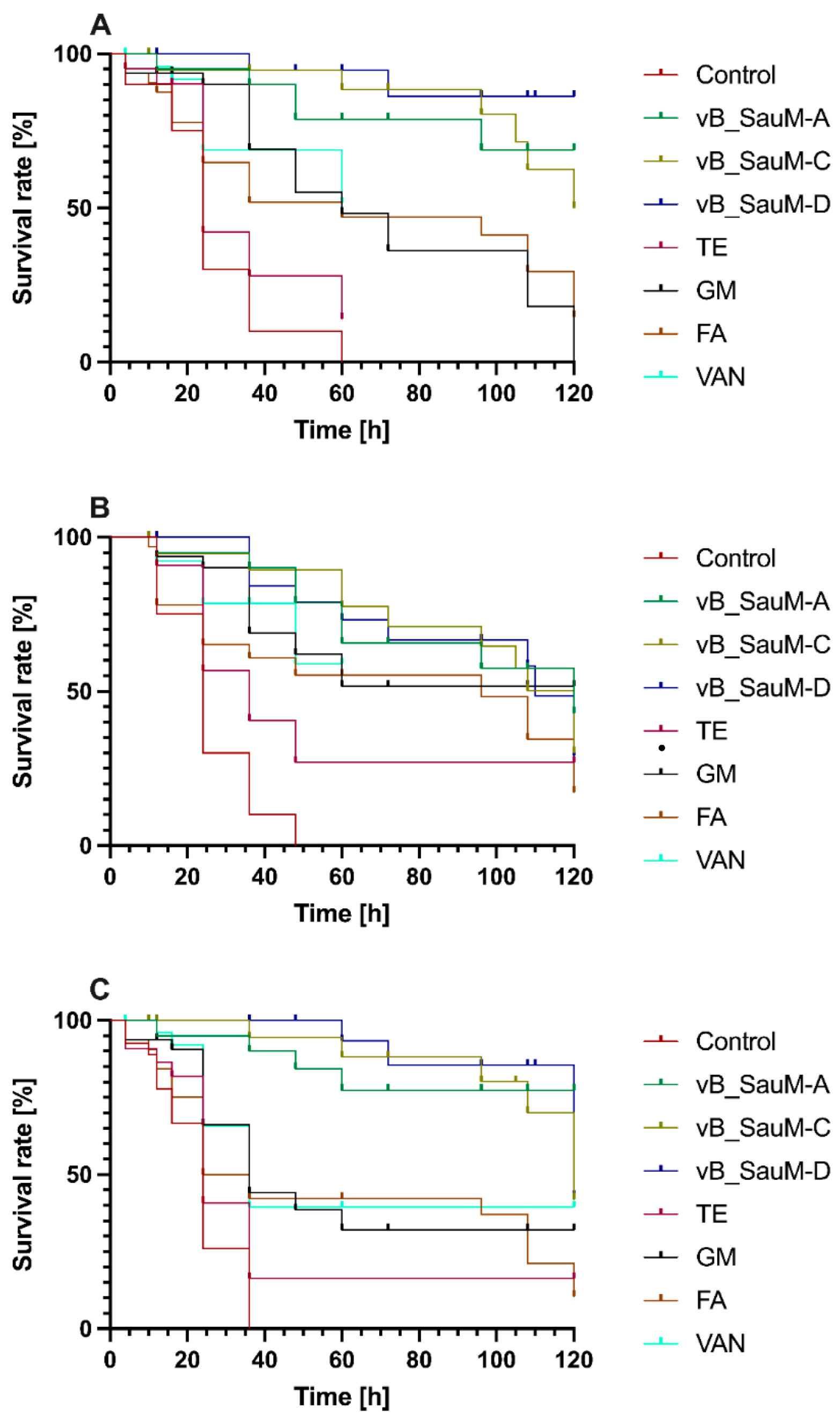
2.5. Galleria mellonella Survival Assay
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains
4.2. Phages
4.3. MIC Determination
4.4. Biofilm Eradication
4.5. Microscopic Examination of Biofilms
4.6. Phages and Antibiotics Effectiveness in an In Vivo Model—Galleria mellonella
4.7. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Karaca, B.; Akcelik, N.; Akcelik, M. Effects of P22 bacteriophage on Salmonella enterica subsp. enyerica serovar Typhimurium DMC4 strain biofilm formation and eradication. Arch. Biol. Sci. 2015, 67, 1361–1367. [Google Scholar] [CrossRef]
- Høiby, N.; Ciofu, O.; Johansen, H.K.; Song, Z.; Moser, C.; Jensen, P.Ø.; Molin, S.; Givskov, M.; Tolker-Nielsen, T.; Bjarnsholt, T. The clinical impact of bacterial biofilms. Int. J. Oral Sci. 2011, 3, 55–65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumaran, D.; Taha, M.; Yi, Q.L.; Ramirez-Arcos, S.; Diallo, J.S.; Carli, A.; Abdelbary, H. Does treatment order matter? Investigating the ability of bacteriophage to augment antibiotic activity against staphylococcus aureus biofilms. Front. Microbiol. 2018, 9, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaźmierczak, N.; Grygorcewicz, B.; Piechowicz, L. Biofilm Formation and Prevalence of Biofilm-Related Genes among Clinical Strains of Multidrug-Resistant Staphylococcus aureus. Microb. Drug Resist. 2021, 27, 956–964. [Google Scholar] [CrossRef] [PubMed]
- Drilling, A.; Hons, B.B.; Morales, S.; Sc, M.; Jardeleza, C.; Vreugde, S.; Speck, P.; Wormald, P.-J. Bacteriophage reduces biofilm of Staphylococcus aureus ex vivo isolates from chronic rhinosinusitis patients. Am. J. Rhinol. Allergy 2014, 28, 3–11. [Google Scholar] [CrossRef]
- Kropinski, A.M. Bacteriophage research–What we have learnt and what still needs to be addressed. Res. Microbiol. 2018, 169, 481–487. [Google Scholar] [CrossRef]
- Grygorcewicz, B.; Roszak, M.; Golec, P.; Śleboda-Taront, D.; Łubowska, N.; Górska, M.; Jursa-Kulesza, J.; Rakoczy, R.; Wojciuk, B.; Dołęgowska, B. Antibiotics act with vb_abap_agc01 phage against acinetobacter baumannii in human heat-inactivated plasma blood and galleria mellonella models. Int. J. Mol. Sci. 2020, 21, 4390. [Google Scholar] [CrossRef]
- Melo, L.D.R.; Brandão, A.; Akturk, E.; Santos, S.B.; Azeredo, J. Characterization of a new Staphylococcus aureus Kayvirus harboring a lysin active against biofilms. Viruses 2018, 10, 182. [Google Scholar] [CrossRef] [Green Version]
- Jensen, K.C.; Hair, B.B.; Wienclaw, T.M.; Murdock, M.H.; Hatch, J.B.; Trent, A.T.; White, T.D.; Haskell, K.J.; Berges, B.K. Isolation and Host Range of Bacteriophage with Lytic Activity against Methicillin-Resistant Staphylococcus aureus and Potential Use as a Fomite Decontaminant. PLoS ONE 2015, 10, e0131714. [Google Scholar] [CrossRef]
- Abatángelo, V.; Peressutti Bacci, N.; Boncompain, C.A.; Amadio, A.A.; Carrasco, S.; Suárez, C.A.; Morbidoni, H.R. Broad-range lytic bacteriophages that kill Staphylococcus aureus local field strains. PLoS ONE 2017, 12, e0181671. [Google Scholar] [CrossRef]
- Fernandez, L.; Gutierrez, D.; Garcia, P.; Rodriguez, A. The Perfect Bacteriophage for Therapeutic. Antibiotics 2019, 8, 126. [Google Scholar] [CrossRef] [Green Version]
- Łubowska, N.; Grygorcewicz, B.; Kosznik-Kwaśnicka, K.; Zauszkiewicz-Pawlak, A.; Węgrzyn, A.; Dołęgowska, B.; Piechowicz, L. Characterization of the three new kayviruses and their lytic activity against multidrug-resistant Staphylococcus aureus. Microorganisms 2019, 7, 471. [Google Scholar] [CrossRef] [Green Version]
- Mah, T.F. Biofilm-specific antibiotic resistance. Future Microbiol. 2012, 7, 1061–1072. [Google Scholar] [CrossRef] [Green Version]
- Fu, W.; Forster, T.; Mayer, O.; Curtin, J.J.; Lehman, S.M.; Donlan, R.M. Bacteriophage cocktail for the prevention of biofilm formation by Pseudomonas aeruginosa on catheters in an in vitro model system. Antimicrob. Agents Chemother. 2010, 54, 397–404. [Google Scholar] [CrossRef] [Green Version]
- Kasprzyk, J.; Piechowicz, L.; Wiśniewska, K.; Dziewit, Ł.; Bronk, M.; Świeć, K. Zrónicowanie typów spa i kaset SCCmec u klinicznych szczepów Staphylococcus aureus opornych na metycylinę izolowanych w ośrodkach regionu gdańskiego. Med. Dosw. Mikrobiol. 2015, 67, 79–88. [Google Scholar]
- Andersson, D.I.; Hughes, D. Microbiological effects of sublethal levels of antibiotics. Nat. Rev. Microbiol. 2014, 12, 465–478. [Google Scholar] [CrossRef]
- Drulis-Kawa, Z.; Majkowska-Skrobek, G.; Maciejewska, B. Bacteriophages and phage-derived proteins—Application approaches. Curr. Med. Chem. 2015, 22, 1757–1773. [Google Scholar] [CrossRef]
- Bertozzi Silva, J.; Sauvageau, D. Bacteriophages as antimicrobial agents against bacterial contaminants in yeast fermentation processes. Biotechnol. Biofuels 2014, 7. [Google Scholar] [CrossRef]
- Chan, B.K.; Abedon, S.T. Bacteriophages and their enzymes in biofilm control. Curr. Pharm. Des. 2015, 21, 85–99. [Google Scholar] [CrossRef]
- Thiel, K. Old dogma, new tricks—21st Century phage therapy. Nat. Biotechnol. 2004, 22, 31–36. [Google Scholar] [CrossRef]
- Harper, D.; Parracho, H.; Walker, J.; Sharp, R.; Hughes, G.; Werthén, M.; Lehman, S.; Morales, S. Bacteriophages and biofilms. Antibiotics 2014, 3, 270–284. [Google Scholar] [CrossRef]
- Vukotic, G.; Obradovic, M.; Novovic, K.; Di Luca, M.; Jovcic, B.; Fira, D.; Neve, H.; Kojic, M.; McAuliffe, O. Characterization, Antibiofilm, and Depolymerizing Activity of Two Phages Active on Carbapenem-Resistant Acinetobacter baumannii. Front. Med. 2020, 7, 426. [Google Scholar] [CrossRef]
- Alves, D.R.; Gaudion, A.; Bean, J.E.; Perez Esteban, P.; Arnot, T.C.; Harper, D.R.; Kot, W.; Hansen, L.H.; Enright, M.C.; Jenkins, A.T.A. Combined use of bacteriophage K and a novel bacteriophage to reduce Staphylococcus aureus biofilm formation. Appl. Environ. Microbiol. 2014, 80, 6694–6703. [Google Scholar] [CrossRef] [Green Version]
- Szymczak, M.; Grygorcewicz, B.; Karczewska-Golec, J.; Decewicz, P.; Pankowski, J.A.; Országh-Szturo, H.; Bącal, P.; Dołęgowska, B.; Golec, P. Characterization of a Unique Bordetella bronchiseptica vB_BbrP_BB8 Bacteriophage and Its Application as an Antibacterial Agent. Int. J. Mol. Sci. 2020, 21, 1403. [Google Scholar] [CrossRef] [Green Version]
- Leshkasheli, L.; Kutateladze, M.; Balarjishvili, N.; Bolkvadze, D.; Save, J.; Oechslin, F.; Que, Y.A.; Resch, G. Efficacy of newly isolated and highly potent bacteriophages in a mouse model of extensively drug-resistant Acinetobacter baumannii bacteraemia. J. Glob. Antimicrob. Resist. 2019, 19, 255–261. [Google Scholar] [CrossRef]
- Desbois, A.P.; Coote, P.J. Wax moth larva (Galleria mellonella): An in vivo model for assessing the efficacy of antistaphylococcal agents. J. Antimicrob. Chemother. 2011, 66, 1785–1790. [Google Scholar] [CrossRef] [Green Version]
- Tsai, C.J.Y.; Loh, J.M.S.; Proft, T. Galleria mellonella infection models for the study of bacterial diseases and for antimicrobial drug testing. Virulence 2016, 7, 214–229. [Google Scholar] [CrossRef] [Green Version]
- Comeau, A.M.; Tétart, F.; Trojet, S.N.; Prère, M.F.; Krisch, H.M. Phage-antibiotic synergy (PAS): β-lactam and quinolone antibiotics stimulate virulent phage growth. PLoS ONE 2007, 2, e799. [Google Scholar] [CrossRef] [Green Version]
- Salmond, G.P.C.; Fineran, P.C. A century of the phage: Past, present and future. Nat. Rev. Microbiol. 2015, 13, 777–786. [Google Scholar] [CrossRef]
- Kamal, F.; Dennis, J.J. Burkholderia cepacia complex phage-antibiotic synergy (PAS): Antibiotics stimulate lytic phage activity. Appl. Environ. Microbiol. 2015, 81, 1132–1138. [Google Scholar] [CrossRef] [Green Version]
- Regeimbal, J.M.; Jacobs, A.C.; Corey, B.W.; Henry, M.S.; Thompson, M.G.; Pavlicek, R.L.; Quinones, J.; Hannah, R.M.; Ghebremedhin, M.; Crane, N.J.; et al. Personalized therapeutic cocktail of wild environmental phages rescues mice from acinetobacter baumannii wound infections. Antimicrob. Agents Chemother. 2016, 60, 5806–5816. [Google Scholar] [CrossRef] [Green Version]
- Li, L.L.; Yu, P.; Wang, X.; Yu, S.S.; Mathieu, J.; Yu, H.Q.; Alvarez, P.J.J. Enhanced biofilm penetration for microbial control by polyvalent phages conjugated with magnetic colloidal nanoparticle clusters (CNCs). Environ. Sci. Nano 2017, 4, 1817–1826. [Google Scholar] [CrossRef]
- Grygorcewicz, B.; Wojciuk, B.; Roszak, M.; Łubowska, N.; Błażejczak, P.; Jursa-Kulesza, J.; Rakoczy, R.; Masiuk, H.; Dołęgowska, B. Environmental Phage-Based Cocktail and Antibiotic Combination Effects on Acinetobacter baumannii Biofilm in a Human Urine Model. Microb. Drug Resist. 2021, 27, 25–35. [Google Scholar] [CrossRef]
- CLSI. M100-S27 Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Seventh Informational Supplement; CLSI: Wayne, PA, USA, 2017. [Google Scholar]
- Cruz, C.D.; Shah, S.; Tammela, P. Defining conditions for biofilm inhibition and eradication assays for Gram-positive clinical reference strains. BMC Microbiol. 2018, 18, 173. [Google Scholar] [CrossRef]
- Junka, A.; Szymczyk, P.; Ziółkowski, G.; Karuga-Kuzniewska, E.; Smutnicka, D.; Bil-Lula, I.; Bartoszewicz, M.; Mahabady, S.; Sedghizadeh, P.P. Bad to the bone: On in vitro and ex vivo microbial biofilm ability to directly destroy colonized bone surfaces without participation of host immunity or osteoclastogenesis. PLoS ONE 2017, 12, e0169565. [Google Scholar] [CrossRef]

| MDRSA Strain | MIC Value [µg × mL−1] | ||||
|---|---|---|---|---|---|
| SXT | GM | TE | FA | VAN | |
| 44 | 2.048 | 0.38 | R | 0.188 | 1.5 |
| 70 | 1.024 | 0.38 | R | 0.188 | 1.5 |
| 110 | 2.048 | 0.25 | 0.094 | 0.125 | 1.5 |
| 113 | 2.048 | 0.19 | 0.094 | 0.188 | 1 |
| 115 | 4.096 | 0.25 | 0.094 | 0.094 | 1.5 |
| 124 | 2.048 | 0.19 | 0.094 | 0.125 | 1 |
| 203 | 2.048 | 0.19 | 0.094 | 0.125 | 1.5 |
| 316 | 2.048 | 0.76 | 0.375 | 0.375 | 1.5 |
| 317 | 2.048 | 0.76 | 0.375 | 0.188 | 1.5 |
| 352 | 2.048 | 0.38 | 0.375 | 0.188 | 1.5 |
| 370 | 2.048 | 0.38 | 0.094 | 0.094 | 1.5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kaźmierczak, N.; Grygorcewicz, B.; Roszak, M.; Bochentyn, B.; Piechowicz, L. Comparative Assessment of Bacteriophage and Antibiotic Activity against Multidrug-Resistant Staphylococcus aureus Biofilms. Int. J. Mol. Sci. 2022, 23, 1274. https://doi.org/10.3390/ijms23031274
Kaźmierczak N, Grygorcewicz B, Roszak M, Bochentyn B, Piechowicz L. Comparative Assessment of Bacteriophage and Antibiotic Activity against Multidrug-Resistant Staphylococcus aureus Biofilms. International Journal of Molecular Sciences. 2022; 23(3):1274. https://doi.org/10.3390/ijms23031274
Chicago/Turabian StyleKaźmierczak, Natalia, Bartłomiej Grygorcewicz, Marta Roszak, Beata Bochentyn, and Lidia Piechowicz. 2022. "Comparative Assessment of Bacteriophage and Antibiotic Activity against Multidrug-Resistant Staphylococcus aureus Biofilms" International Journal of Molecular Sciences 23, no. 3: 1274. https://doi.org/10.3390/ijms23031274






