Identification of a Novel Theranostic Signature of Metabolic and Immune-Inflammatory Dysregulation in Myocardial Infarction, and the Potential Therapeutic Properties of Ovatodiolide, a Diterpenoid Derivative
Abstract
:1. Introduction
2. Results
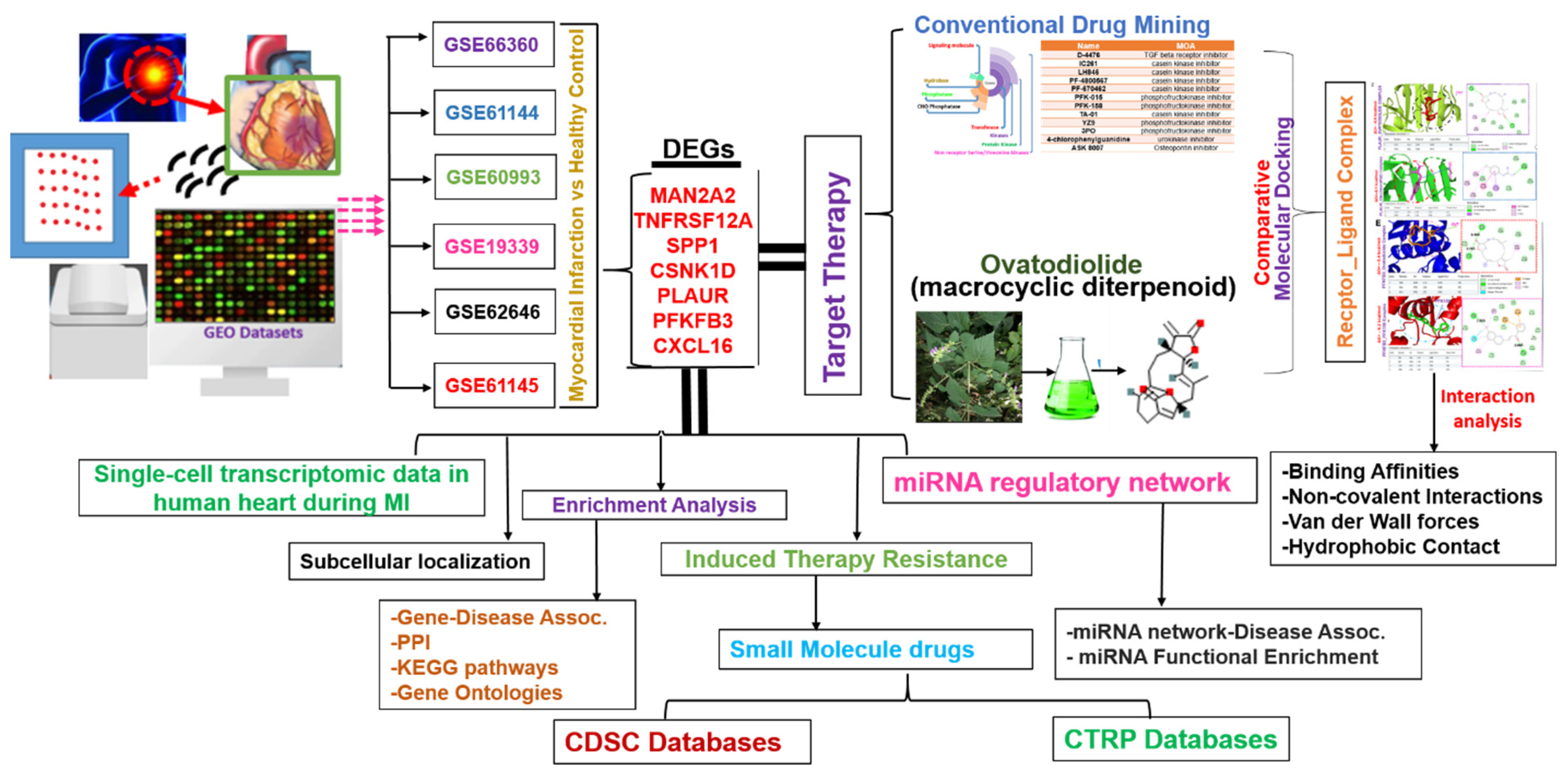
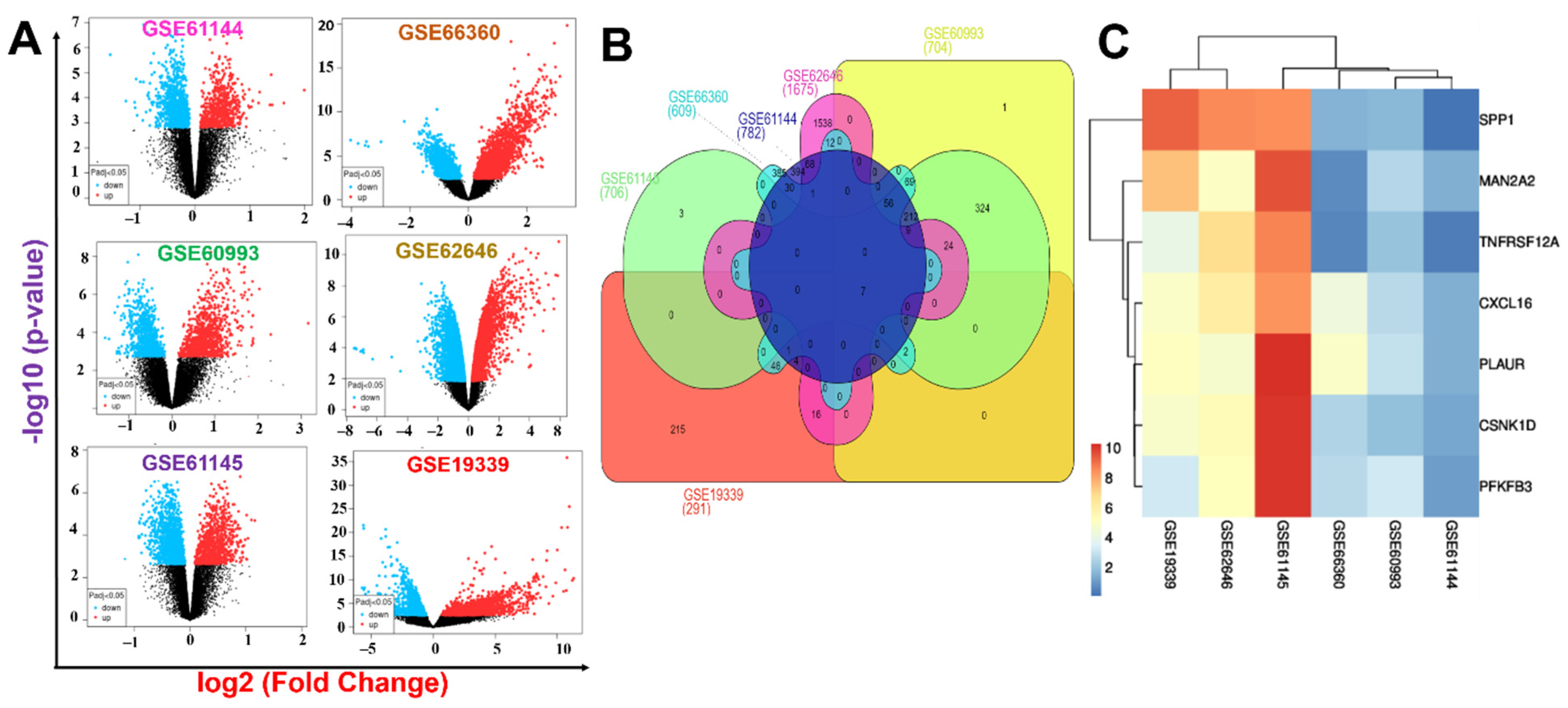
2.1. Identification of MAN2A2/TNFRSF12A/SPP1/CSNK1D/PLAUR/PFKFB3/CXCL16 as a Novel Pathological Signature of Myocardial Infarction
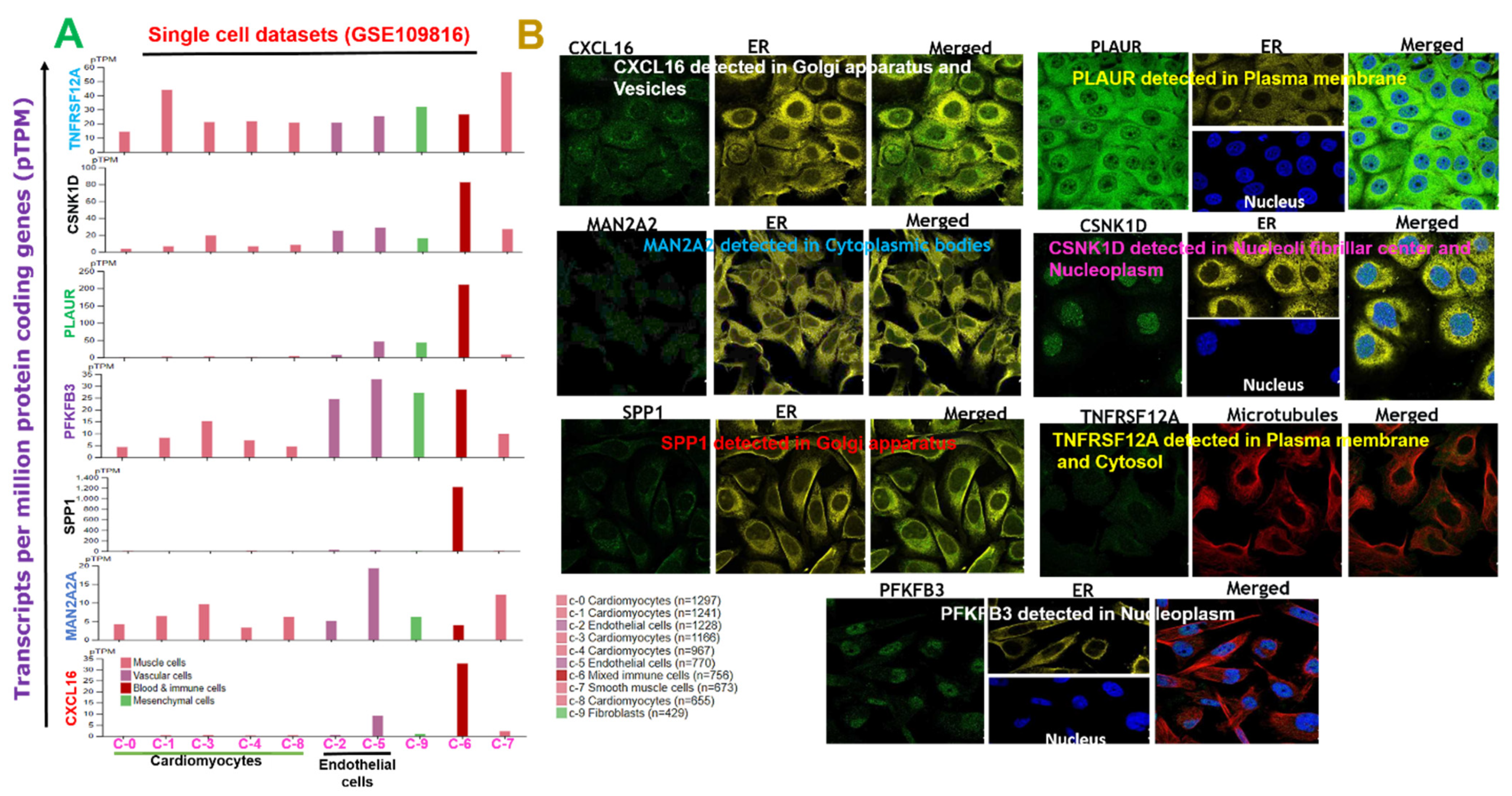
2.2. Subcellular Localization and Single-Cell Transcriptomic Data of DEGs Signature in the Adult Human Heart during MI
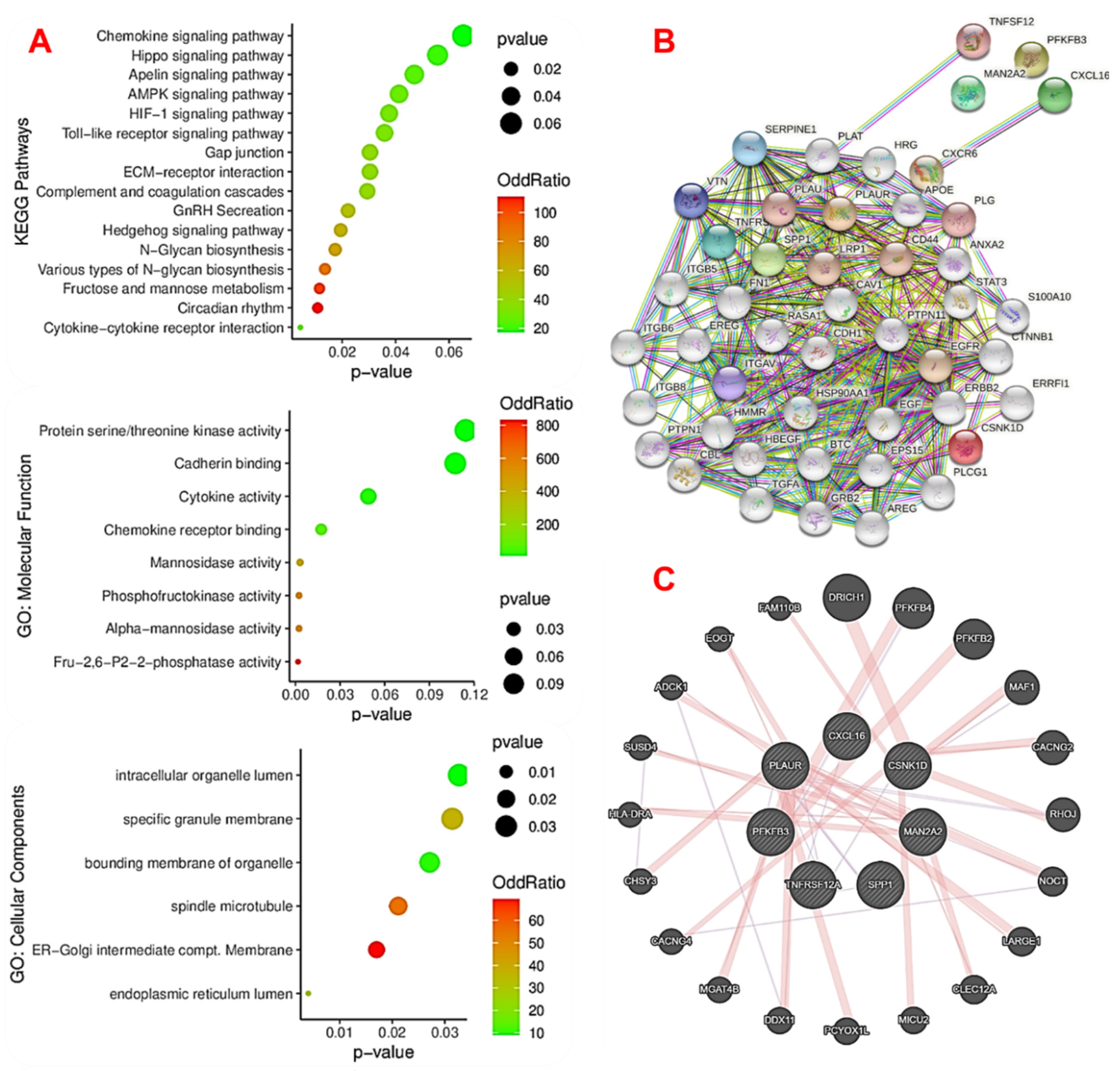
2.3. The Novel MTSCPPC Signature Is Associated with Disruption of Metabolic and Immune-Inflammatory Pathways during the Pathogenesis of MI
2.4. The MTSCPPC Signature Is Implicated in Therapy Resistance and Pathogenesis of Heart Related Diseases
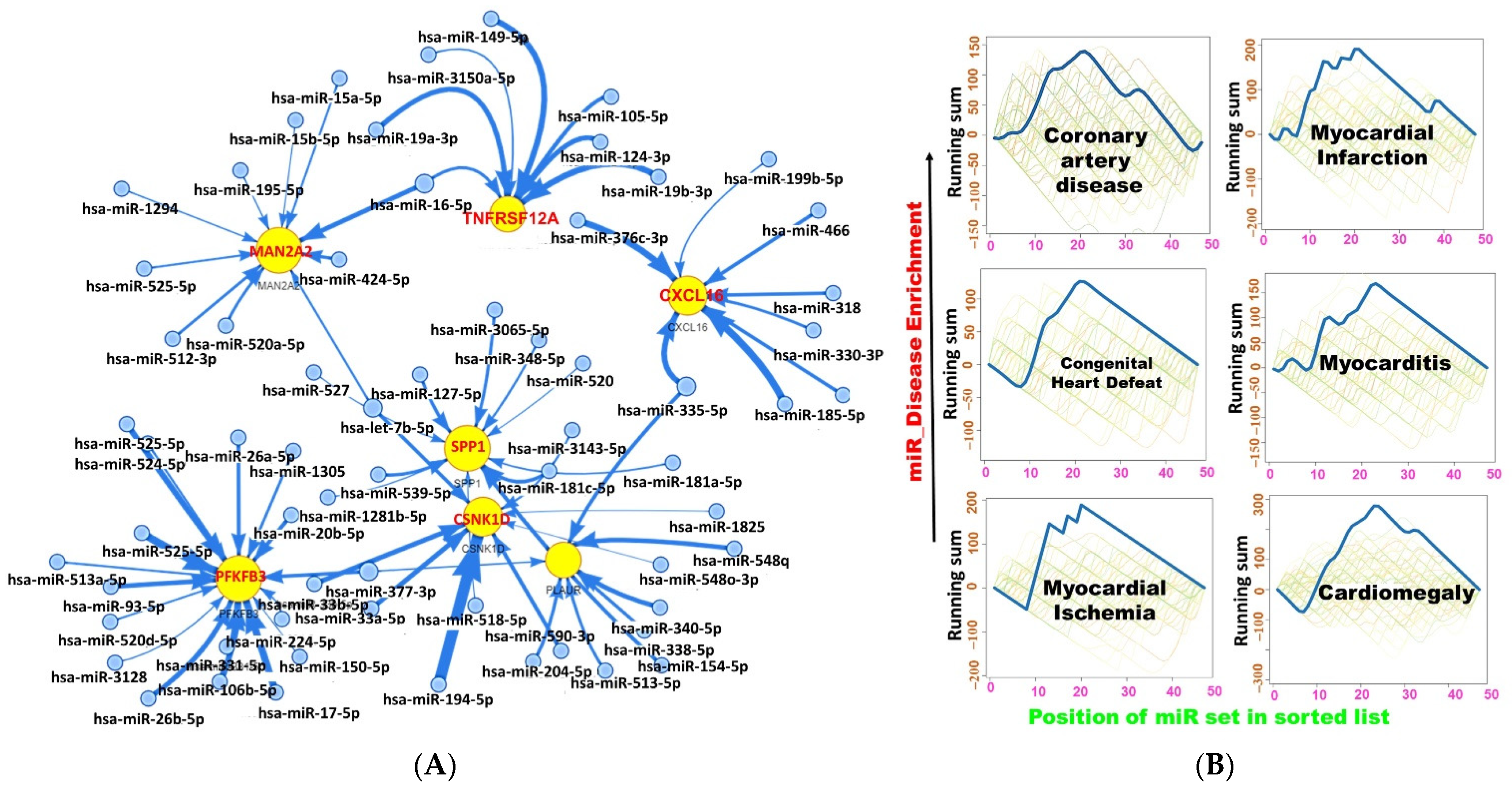
2.5. miRNA Regulatory Network of the DEGs
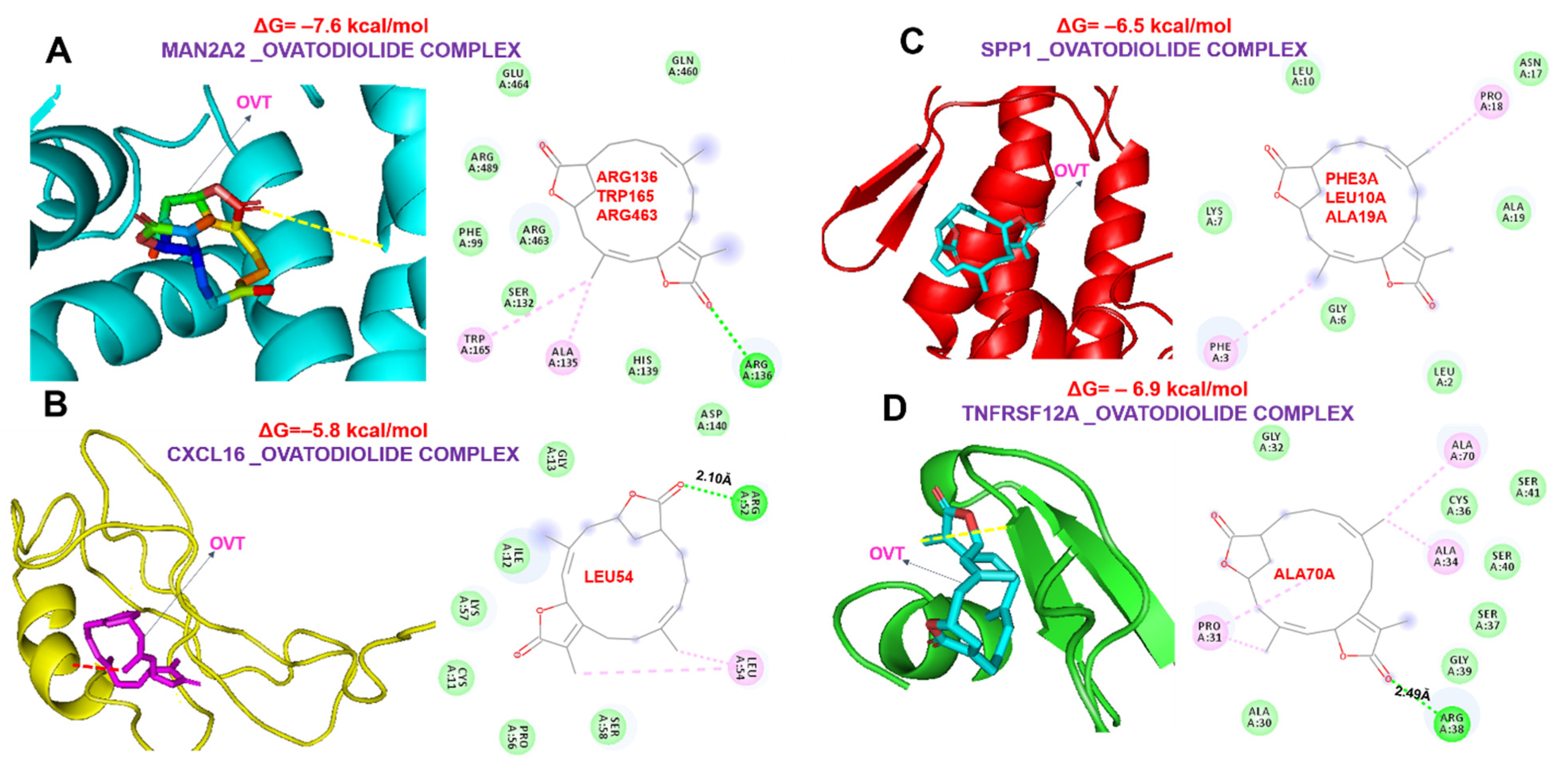
2.6. Ovatodiolide, a Macrocyclic Diterpenoid, a Potential Drug for Targeting the MTSCPPC Signature
3. Discussion
4. Materials and Methods
4.1. Transcriptomic Data Acquisition and Identification of DEGs in MI
4.2. Subcellular Localization and Single-Cell Transcriptomic Data Analysis of the Adult Human Heart during MI
4.3. Interaction and Disease Networks, and Gene Set Enrichment Analysis of the DEGs
4.4. Drug Response and Sensitivity Analysis of the DEGs
4.5. Micro (mi)RNA Regulatory Network and Enrichment Analysis of the DEGs
4.6. Comparative Analysis of Ovatodiolide, a Macrocyclic Diterpenoid and Conventional Drugs for Targeting the DEGs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, R.; Ji, Z.; Qu, Y.; Yang, M.; Su, Y.; Zuo, W.; Zhao, Q.; Ma, G.; Li, Y. Clinical value of ARG1 in acute myocardial infarction patients: Bioinformatics-based approach. Biomed. Pharmacother. 2020, 121, 109590. [Google Scholar] [CrossRef] [PubMed]
- Ojha, N.; Dhamoon, A.S. Myocardial infarction. In StatPearls; StatPearls Publishing LLC.: Treasure Island, FL, USA, 2021. [Google Scholar]
- Murphy, D.A.; Craver, J.M.; Jones, E.L.; King, S.B., 3rd; Curling, P.E.; Douglas, J.S., Jr. Hemodynamic deterioration after coronary angioplasty in the presence of previous left ventricular infarction. Am. J. Cardiol. 1984, 54, 448–450. [Google Scholar] [CrossRef]
- Boateng, S.; Sanborn, T. Acute myocardial infarction. Disease-a-Month 2013, 59, 83–96. [Google Scholar] [CrossRef] [PubMed]
- Bajaj, A.; Sethi, A.; Rathor, P.; Suppogu, N.; Sethi, A. Acute complications of myocardial infarction in the current era: Diagnosis and management. J. Investig. Med. 2015, 63, 844–855. [Google Scholar] [CrossRef] [PubMed]
- De Winter, R.J.; Koster, R.W.; Sturk, A.; Sanders, G.T. Value of myoglobin, troponin T, and CK-MBmass in ruling out an acute myocardial infarction in the emergency room. Circulation 1995, 92, 3401–3407. [Google Scholar] [CrossRef] [PubMed]
- Jaffe, A.S.; Ravkilde, J.; Roberts, R.; Naslund, U.; Apple, F.S.; Galvani, M.; Katus, H. It’s time for a change to a troponin standard. Circulation 2000, 102, 1216–1220. [Google Scholar] [CrossRef] [Green Version]
- Collinson, P.; Hadcocks, L.; Foo, Y.; Rosalki, S.; Stubbs, P.; Morgan, S.; O’Donnell, J. Cardiac troponins in patients with renal dysfunction. Ann. Clin. Biochem. 1998, 35, 380–386. [Google Scholar] [CrossRef]
- Agewall, S.; Giannitsis, E.; Jernberg, T.; Katus, H. Troponin elevation in coronary vs. non-coronary disease. Eur. Heart J. 2011, 32, 404–411. [Google Scholar] [CrossRef]
- Katus, H.; Ziegler, A.; Ekinci, O.; Giannitsis, E.; Stough, W.G.; Achenbach, S.; Blankenberg, S.; Brueckmann, M.; Collinson, P.; Comaniciu, D. Early diagnosis of acute coronary syndrome. Eur. Heart J. 2017, 38, 3049–3055. [Google Scholar] [CrossRef]
- Sarkisian, L.; Saaby, L.; Poulsen, T.S.; Gerke, O.; Hosbond, S.; Jangaard, N.; Diederichsen, A.C.; Thygesen, K.; Mickley, H. Prognostic impact of myocardial injury related to various cardiac and noncardiac conditions. Am. J. Med. 2016, 129, 506–514.e501. [Google Scholar] [CrossRef] [Green Version]
- Thygesen, K.; Alpert, J.S.; Jaffe, A.S.; Chaitman, B.R.; Bax, J.J.; Morrow, D.A.; White, H.D. Fourth Universal Definition of Myocardial Infarction. Circulation 2018, 138, e618–e651. [Google Scholar] [CrossRef] [PubMed]
- Khera, R.; Haimovich, J.; Hurley, N.C.; McNamara, R.; Spertus, J.A.; Desai, N.; Rumsfeld, J.S.; Masoudi, F.A.; Huang, C.; Normand, S.-L.; et al. Use of Machine Learning Models to Predict Death After Acute Myocardial Infarction. JAMA Cardiol. 2021, 6, 633–641. [Google Scholar] [CrossRef] [PubMed]
- Ibanez, B.; James, S.; Agewall, S.; Antunes, M.J.; Bucciarelli-Ducci, C.; Bueno, H.; Caforio, A.L.P.; Crea, F.; Goudevenos, J.A.; Halvorsen, S.; et al. 2017 ESC Guidelines for the management of acute myocardial infarction in patients presenting with ST-segment elevation: The Task Force for the management of acute myocardial infarction in patients presenting with ST-segment elevation of the European Society of Cardiology (ESC). Eur. Heart J. 2018, 39, 119–177. [Google Scholar] [CrossRef] [Green Version]
- Collet, J.-P.; Thiele, H.; Barbato, E.; Barthélémy, O.; Bauersachs, J.; Bhatt, D.L.; Dendale, P.; Dorobantu, M.; Edvardsen, T.; Folliguet, T.; et al. 2020 ESC Guidelines for the management of acute coronary syndromes in patients presenting without persistent ST-segment elevation: The Task Force for the management of acute coronary syndromes in patients presenting without persistent ST-segment elevation of the European Society of Cardiology (ESC). Eur. Heart J. 2021, 42, 1289–1367. [Google Scholar] [CrossRef]
- Zhong, Z.; Wu, H.; Zhong, W.; Zhang, Q.; Yu, Z. Expression profiling and bioinformatics analysis of circulating microRNAs in patients with acute myocardial infarction. J. Clin. Lab. Anal. 2020, 34, e23099. [Google Scholar] [CrossRef]
- Agamah, F.E.; Mazandu, G.K.; Hassan, R.; Bope, C.D.; Thomford, N.E.; Ghansah, A.; Chimusa, E.R. Computational/in silico methods in drug target and lead prediction. Brief. Bioinform. 2020, 21, 1663–1675. [Google Scholar] [CrossRef]
- Khedkar, H.N.; Wang, Y.-C.; Yadav, V.K.; Srivastava, P.; Lawal, B.; Mokgautsi, N.; Sumitra, M.R.; Wu, A.T.H.; Huang, H.-S. In-Silico Evaluation of Genetic Alterations in Ovarian Carcinoma and Therapeutic Efficacy of NSC777201, as a Novel Multi-Target Agent for TTK, NEK2, and CDK1. Int. J. Mol. Sci. 2021, 22, 5895. [Google Scholar] [CrossRef]
- Lawal, B.; Lin, L.-C.; Lee, J.-C.; Chen, J.-H.; Bekaii-Saab, T.S.; Wu, A.T.H.; Ho, C.-L. Multi-Omics Data Analysis of Gene Expressions and Alterations, Cancer-Associated Fibroblast and Immune Infiltrations, Reveals the Onco-Immune Prognostic Relevance of STAT3/CDK2/4/6 in Human Malignancies. Cancers 2021, 13, 954. [Google Scholar] [CrossRef]
- Wu, S.-Y.; Lin, K.-C.; Lawal, B.; Wu, A.T.H.; Wu, C.-Z. MXD3 as an onco-immunological biomarker encompassing the tumor microenvironment, disease staging, prognoses, and therapeutic responses in multiple cancer types. Comput. Struct. Biotechnol. J. 2021, 19, 4970–4983. [Google Scholar] [CrossRef]
- Wu, A.T.H.; Lawal, B.; Wei, L.; Wen, Y.-T.; Tzeng, D.T.W.; Lo, W.-C. Multiomics Identification of Potential Targets for Alzheimer Disease and Antrocin as a Therapeutic Candidate. Pharmaceutics 2021, 13, 1555. [Google Scholar] [CrossRef]
- Lawal, B.; Lo, W.-C.; Mokgautsi, N.; Sumitra, M.R.; Khedkar, H.; Wu, A.T.; Huang, H.-S. A preclinical report of a cobimetinib-inspired novel anticancer small-molecule scaffold of isoflavones, NSC777213, for targeting PI3K/AKT/mTOR/MEK in multiple cancers. Am. J. Cancer Res. 2021, 11, 2590–2617. [Google Scholar] [PubMed]
- Boon, R.A.; Dimmeler, S. MicroRNAs in myocardial infarction. Nat. Rev. Cardiol. 2015, 12, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Lawal, B.; Shittu, O.K.; Oibiokpa, F.I.; Berinyuy, E.B.; Mohammed, H. African natural products with potential antioxidants and hepatoprotectives properties: A review. Clin. Phytoscience 2016, 2, 23. [Google Scholar] [CrossRef] [Green Version]
- Lawal, B.; Shittu, O.K.; Kabiru, A.Y.; Jigam, A.A.; Umar, M.B.; Berinyuy, E.B.; Alozieuwa, B.U. Potential antimalarials from African natural products: A reviw. J. Intercult. Ethnopharmacol. 2015, 4, 318. [Google Scholar] [CrossRef]
- Kim, J.H.; Kismali, G.; Gupta, S.C. Natural Products for the Prevention and Treatment of Chronic Inflammatory Diseases: Integrating Traditional Medicine into Modern Chronic Diseases Care. Evid.-Based Complementary Altern. Med. 2018, 2018, 9837863. [Google Scholar] [CrossRef]
- Onikanni, A.S.; Lawal, B.; Olusola, A.O.; Olugbodi, J.O.; Sani, S.; Ajiboye, B.O.; Ilesanmi, O.B.; Alqarni, M.; Mostafa-Hedeab, G.; Obaidullah, A.J.; et al. Sterculia tragacantha Lindl Leaf Extract Ameliorates STZ-Induced Diabetes, Oxidative Stress, Inflammation and Neuronal Impairment. J. Inflamm. Res. 2021, 14, 6749–6764. [Google Scholar] [CrossRef]
- Chen, Y.-L.; Lan, Y.-H.; Hsieh, P.-W.; Wu, C.-C.; Chen, S.-L.; Yen, C.-T.; Chang, F.-R.; Hung, W.-C.; Wu, Y.-C. Bioactive cembrane diterpenoids of Anisomeles indica. J. Nat. Prod. 2008, 71, 1207–1212. [Google Scholar] [CrossRef]
- Liu, S.-C.; Huang, C.-M.; Chang, Y.-L.; Bamodu, O.A.; Yeh, C.-T.; Wang, H.-W.; Lee, F.-P.; Lin, C.-S. Ovatodiolide suppresses inflammatory response in BEAS-2B cells by regulating the CREB/AQP5 pathway, and sensitizes nasopharyngeal carcinoma cells to radiation therapy. Eur. J. Pharmacol. 2019, 859, 172548. [Google Scholar] [CrossRef]
- Bamodu, O.A.; Huang, W.-C.; Tzeng, D.T.W.; Wu, A.; Wang, L.S.; Yeh, C.-T.; Chao, T.-Y. Ovatodiolide sensitizes aggressive breast cancer cells to doxorubicin, eliminates their cancer stem cell-like phenotype, and reduces doxorubicin-associated toxicity. Cancer Lett. 2015, 364, 125–134. [Google Scholar] [CrossRef]
- Chen, J.-H.; Wu, A.T.H.; Bamodu, O.A.; Yadav, V.K.; Chao, T.-Y.; Tzeng, Y.-M.; Mukhopadhyay, D.; Hsiao, M.; Lee, J.-C. Ovatodiolide Suppresses Oral Cancer Malignancy by Down-Regulating Exosomal Mir-21/STAT3/β-Catenin Cargo and Preventing Oncogenic Transformation of Normal Gingival Fibroblasts. Cancers 2020, 12, 56. [Google Scholar] [CrossRef] [Green Version]
- Lin, C.-S.; Bamodu, O.A.; Kuo, K.-T.; Huang, C.-M.; Liu, S.-C.; Wang, C.-H.; Tzeng, Y.-M.; Chao, T.-Y.; Yeh, C.-T. Investigation of ovatodiolide, a macrocyclic diterpenoid, as a potential inhibitor of oral cancer stem-like cells properties via the inhibition of the JAK2/STAT3/JARID1B signal circuit. Phytomedicine 2018, 46, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Ho, J.-Y.; Hsu, R.-J.; Wu, C.-L.; Chang, W.-L.; Cha, T.-L.; Yu, D.-S.; Yu, C.-P. Ovatodiolide Targets β-Catenin Signaling in Suppressing Tumorigenesis and Overcoming Drug Resistance in Renal Cell Carcinoma. Evid.-Based Complementary Altern. Med. 2013, 2013, 161628. [Google Scholar] [CrossRef] [Green Version]
- Su, Y.-K.; Bamodu, O.A.; Tzeng, Y.-M.; Hsiao, M.; Yeh, C.-T.; Lin, C.-M. Ovatodiolide inhibits the oncogenicity and cancer stem cell-like phenotype of glioblastoma cells, as well as potentiate the anticancer effect of temozolomide. Phytomedicine 2019, 61, 152840. [Google Scholar] [CrossRef] [PubMed]
- Momose, Y.; Nimura, M.; Arisawa, M.; Hayashi, T.; Takeda, R.; Nakanishi, S. Hypotensive activity of ovatodiolides isolated from a Chinese crude drug ‘Fang Feng Cao’. Phytother. Res. 1994, 8, 482–484. [Google Scholar] [CrossRef]
- Chen, J.-H.; Wu, A.T.H.; Lawal, B.; Tzeng, D.T.W.; Lee, J.-C.; Ho, C.-L.; Chao, T.-Y. Identification of Cancer Hub Gene Signatures Associated with Immune-Suppressive Tumor Microenvironment and Ovatodiolide as a Potential Cancer Immunotherapeutic Agent. Cancers 2021, 13, 3847. [Google Scholar] [CrossRef]
- Meng, X.-Y.; Zhang, H.-X.; Mezei, M.; Cui, M. Molecular docking: A powerful approach for structure-based drug discovery. Curr. Comput.-Aided Drug Des. 2011, 7, 146–157. [Google Scholar] [CrossRef]
- Lawal, B.; Liu, Y.-L.; Mokgautsi, N.; Khedkar, H.; Sumitra, M.R.; Wu, A.T.H.; Huang, H.-S. Pharmacoinformatics and Preclinical Studies of NSC765690 and NSC765599, Potential STAT3/CDK2/4/6 Inhibitors with Antitumor Activities against NCI60 Human Tumor Cell Lines. Biomedicines 2021, 9, 92. [Google Scholar] [CrossRef]
- Arslan, F.; de Kleijn, D.P.; Pasterkamp, G. Innate immune signaling in cardiac ischemia. Nat. Rev. Cardiol. 2011, 8, 292–300. [Google Scholar] [CrossRef]
- Frangogiannis, N.G.; Smith, C.W.; Entman, M.L. The inflammatory response in myocardial infarction. Cardiovasc. Res. 2002, 53, 31–47. [Google Scholar] [CrossRef]
- Werns, S.W.; Lucchesi, B.R. Inflammation and myocardial infarction. Br. Med. Bull. 1987, 43, 460–471. [Google Scholar] [CrossRef]
- Timmers, L.; Pasterkamp, G.; de Hoog, V.C.; Arslan, F.; Appelman, Y.; de Kleijn, D.P.V. The innate immune response in reperfused myocardium. Cardiovasc. Res. 2012, 94, 276–283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montecucco, F.; Lenglet, S.; Braunersreuther, V.; Pelli, G.; Pellieux, C.; Montessuit, C.; Lerch, R.; Deruaz, M.; Proudfoot, A.E.; Mach, F. Single administration of the CXC chemokine-binding protein Evasin-3 during ischemia prevents myocardial reperfusion injury in mice. Arterioscler. Thromb. Vasc. Biol. 2010, 30, 1371–1377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mylonas, K.J.; Turner, N.A.; Bageghni, S.A.; Kenyon, C.J.; White, C.I.; McGregor, K.; Kimmitt, R.A.; Sulston, R.; Kelly, V.; Walker, B.R. 11β-HSD1 suppresses cardiac fibroblast CXCL2, CXCL5 and neutrophil recruitment to the heart post MI. J. Endocrinol. 2017, 233, 315. [Google Scholar] [CrossRef] [PubMed]
- Dallacasagrande, V.; Hajjar, K.A. Annexin A2 in Inflammation and Host Defense. Cells 2020, 9, 1499. [Google Scholar] [CrossRef]
- Zong, L.; Wang, W. CircANXA2 Promotes Myocardial Apoptosis in Myocardial Ischemia-Reperfusion Injury via Inhibiting miRNA-133 Expression. BioMed Res. Int. 2020, 2020, 8590861. [Google Scholar] [CrossRef]
- Tanaka, N.; Masamura, K.; Yoshida, M.; Kato, M.; Kawai, Y.; Miyamori, I. A role of heparin-binding epidermal growth factor-like growth factor in cardiac remodeling after myocardial infarction. Biochem. Biophys. Res. Commun. 2002, 297, 375–381. [Google Scholar] [CrossRef]
- Shivshankar, P.; Halade, G.V.; Calhoun, C.; Escobar, G.P.; Mehr, A.J.; Jimenez, F.; Martinez, C.; Bhatnagar, H.; Mjaatvedt, C.H.; Lindsey, M.L.; et al. Caveolin-1 deletion exacerbates cardiac interstitial fibrosis by promoting M2 macrophage activation in mice after myocardial infarction. J. Mol. Cell. Cardiol. 2014, 76, 84–93. [Google Scholar] [CrossRef] [Green Version]
- Mani, R.; Onge, R.P.S.; Hartman, J.L.; Giaever, G.; Roth, F.P. Defining genetic interaction. Proc. Natl. Acad. Sci. USA 2008, 105, 3461–3466. [Google Scholar] [CrossRef] [Green Version]
- Baryshnikova, A.; Costanzo, M.; Myers, C.L.; Andrews, B.; Boone, C. Genetic interaction networks: Toward an understanding of heritability. Annu. Rev. Genom. Hum. Genet. 2013, 14, 111–133. [Google Scholar] [CrossRef]
- Fece de la Cruz, F.; Gapp, B.V.; Nijman, S.M. Synthetic lethal vulnerabilities of cancer. Annu. Rev. Pharmacol. Toxicol. 2015, 55, 513–531. [Google Scholar] [CrossRef]
- Ashworth, A.; Lord, C.J.; Reis-Filho, J.S. Genetic interactions in cancer progression and treatment. Cell 2011, 145, 30–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fong, P.C.; Boss, D.S.; Yap, T.A.; Tutt, A.; Wu, P.; Mergui-Roelvink, M.; Mortimer, P.; Swaisland, H.; Lau, A.; O’Connor, M.J.; et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N. Engl. J. Med. 2009, 361, 123–134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lewis, B.P.; Shih, I.-H.; Jones-Rhoades, M.W.; Bartel, D.P.; Burge, C.B. Prediction of mammalian microRNA targets. Cell 2003, 115, 787–798. [Google Scholar] [CrossRef] [Green Version]
- Pu, M.; Chen, J.; Tao, Z.; Miao, L.; Qi, X.; Wang, Y.; Ren, J. Regulatory network of miRNA on its target: Coordination between transcriptional and post-transcriptional regulation of gene expression. Cell. Mol. Life Sci. 2019, 76, 441–451. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.-K.; Zhu, J.-Q.; Zhang, J.-T.; Li, Q.; Li, Y.; He, J.; Qin, Y.-W.; Jing, Q. Circulating microRNA: A novel potential biomarker for early diagnosis of acute myocardial infarction in humans. Eur. Heart J. 2010, 31, 659–666. [Google Scholar] [CrossRef]
- Meder, B.; Keller, A.; Vogel, B.; Haas, J.; Sedaghat-Hamedani, F.; Kayvanpour, E.; Just, S.; Borries, A.; Rudloff, J.; Leidinger, P. MicroRNA signatures in total peripheral blood as novel biomarkers for acute myocardial infarction. Basic Res. Cardiol. 2011, 106, 13–23. [Google Scholar] [CrossRef]
- Benhamed, M.; Herbig, U.; Ye, T.; Dejean, A.; Bischof, O. Senescence is an endogenous trigger for microRNA-directed transcriptional gene silencing in human cells. Nat. Cell Biol. 2012, 14, 266–275. [Google Scholar] [CrossRef]
- Roberts, T.C. The MicroRNA Biology of the Mammalian Nucleus. Mol. Ther. Nucleic Acids 2014, 3, e188. [Google Scholar] [CrossRef]
- Arthur, D.E.; Uzairu, A. Molecular docking studies on the interaction of NCI anticancer analogues with human Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit. J. King Saud Univ. Sci. 2019, 31, 1151–1166. [Google Scholar] [CrossRef]
- Park, H.J.; Noh, J.H.; Eun, J.W.; Koh, Y.S.; Seo, S.M.; Park, W.S.; Lee, J.Y.; Chang, K.; Seung, K.B.; Kim, P.J.; et al. Assessment and diagnostic relevance of novel serum biomarkers for early decision of ST-elevation myocardial infarction. Oncotarget 2015, 6, 12970–12983. [Google Scholar] [CrossRef] [Green Version]
- Kiliszek, M.; Burzynska, B.; Michalak, M.; Gora, M.; Winkler, A.; Maciejak, A.; Leszczynska, A.; Gajda, E.; Kochanowski, J.; Opolski, G. Altered gene expression pattern in peripheral blood mononuclear cells in patients with acute myocardial infarction. PLoS ONE 2012, 7, e50054. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muse, E.D.; Kramer, E.R.; Wang, H.; Barrett, P.; Parviz, F.; Novotny, M.A.; Lasken, R.S.; Jatkoe, T.A.; Oliveira, G.; Peng, H.; et al. A Whole Blood Molecular Signature for Acute Myocardial Infarction. Sci. Rep. 2017, 7, 12268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Heberle, H.; Meirelles, G.V.; da Silva, F.R.; Telles, G.P.; Minghim, R. InteractiVenn: A web-based tool for the analysis of sets through Venn diagrams. BMC Bioinform. 2015, 16, 169. [Google Scholar] [CrossRef]
- Toro-Domínguez, D.; Martorell-Marugán, J.; López-Domínguez, R.; García-Moreno, A.; González-Rumayor, V.; Alarcón-Riquelme, M.E.; Carmona-Sáez, P. ImaGEO: Integrative gene expression meta-analysis from GEO database. Bioinformatics 2019, 35, 880–882. [Google Scholar] [CrossRef] [PubMed]
- Marot, G.; Foulley, J.-L.; Mayer, C.-D.; Jaffrézic, F. Moderated effect size and P-value combinations for microarray meta-analyses. Bioinformatics 2009, 25, 2692–2699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sullivan, G.M.; Feinn, R. Using Effect Size-or Why the P Value Is Not Enough. J. Grad. Med. Educ. 2012, 4, 279–282. [Google Scholar] [CrossRef] [Green Version]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Wang, L.; Yu, P.; Zhou, B.; Song, J.; Li, Z.; Zhang, M.; Guo, G.; Wang, Y.; Chen, X.; Han, L.; et al. Single-cell reconstruction of the adult human heart during heart failure and recovery reveals the cellular landscape underlying cardiac function. Nat. Cell Biol. 2020, 22, 108–119. [Google Scholar] [CrossRef]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’ayan, A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef] [Green Version]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mostafavi, S.; Ray, D.; Warde-Farley, D.; Grouios, C.; Morris, Q. GeneMANIA: A real-time multiple association network integration algorithm for predicting gene function. Genome Biol. 2008, 9, S4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ochoa, D.; Hercules, A.; Carmona, M.; Suveges, D.; Gonzalez-Uriarte, A.; Malangone, C.; Miranda, A.; Fumis, L.; Carvalho-Silva, D.; Spitzer, M.; et al. Open Targets Platform: Supporting systematic drug–target identification and prioritisation. Nucleic Acids Res. 2020, 49, D1302–D1310. [Google Scholar] [CrossRef]
- Liu, C.-J.; Hu, F.-F.; Xia, M.-X.; Han, L.; Zhang, Q.; Guo, A.-Y. GSCALite: A web server for gene set cancer analysis. Bioinformatics 2018, 34, 3771–3772. [Google Scholar] [CrossRef]
- Kern, F.; Fehlmann, T.; Solomon, J.; Schwed, L.; Grammes, N.; Backes, C.; Van Keuren-Jensen, K.; Craig, D.W.; Meese, E.; Keller, A. miEAA 2.0: Integrating multi-species microRNA enrichment analysis and workflow management systems. Nucleic Acids Res. 2020, 48, W521–W528. [Google Scholar] [CrossRef]
- Cotto, K.C.; Wagner, A.H.; Feng, Y.Y.; Kiwala, S.; Coffman, A.C.; Spies, G.; Wollam, A.; Spies, N.C.; Griffith, O.L.; Griffith, M. DGIdb 3.0: A redesign and expansion of the drug–gene interaction database. Nucleic Acids Res. 2018, 46, D1068–D1073. [Google Scholar] [CrossRef] [Green Version]
- Subramanian, A.; Narayan, R.; Corsello, S.M.; Peck, D.D.; Natoli, T.E.; Lu, X.; Gould, J.; Davis, J.F.; Tubelli, A.A.; Asiedu, J.K. A next generation connectivity map: L1000 platform and the first 1,000,000 profiles. Cell 2017, 171, 1437–1452.e1417. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.-C.; Wu, A.T.H.; Chen, J.-H.; Huang, W.-Y.; Lawal, B.; Mokgautsi, N.; Huang, H.-S.; Ho, C.-L. HNC0014, a Multi-Targeted Small-Molecule, Inhibits Head and Neck Squamous Cell Carcinoma by Suppressing c-Met/STAT3/CD44/PD-L1 Oncoimmune Signature and Eliciting Antitumor Immune Responses. Cancers 2020, 12, 3759. [Google Scholar] [CrossRef]
- Lawal, B.; Lee, C.-Y.; Mokgautsi, N.; Sumitra, M.R.; Khedkar, H.; Wu, A.T.H.; Huang, H.-S. mTOR/EGFR/iNOS/MAP2K1/FGFR/TGFB1 Are Druggable Candidates for N-(2,4-Difluorophenyl)-2′,4′-Difluoro-4-Hydroxybiphenyl-3-Carboxamide (NSC765598), With Consequent Anticancer Implications. Front. Oncol. 2021, 11, 656738. [Google Scholar] [CrossRef] [PubMed]
- Hanwell, M.D.; Curtis, D.E.; Lonie, D.C.; Vandermeersch, T.; Zurek, E.; Hutchison, G.R. Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. J. Cheminformatics 2012, 4, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Visualizer, D.S. BIOVIA, Dassault Systèmes, BIOVIA Workbook, Release 2020; BIOVIA Pipeline Pilot, Release 2020; Dassault Systèmes: San Diego, CA, USA, 2020. [Google Scholar]


| Gene ID | p-Value Combination | Effect Size Combination | Gene_Name | |||
|---|---|---|---|---|---|---|
| fdr_pval | FC_mean | fdr_pval | pval | zval | ||
| MAN2A2 | 0.0005 | 0.09 | 7.5 × 10−7 | 2.9 × 10−8 | 5.5 | mannosidase alpha class 2A member 2 |
| TNFRSF12A | 0.0081 | 0.13 | 1.6 × 10−7 | 5.1 × 10−9 | 5.8 | TNF receptor superfamily member 12A |
| SPP1 | 0.0064 | 0.33 | 3.0 × 10−5 | 2.0 × 10−6 | 4.8 | secreted phosphoprotein 1 |
| CSNK1D | 0.0029 | 0.13 | 1.2 × 10−6 | 5.3 × 10−8 | 5.4 | casein kinase 1 delta |
| PLAUR | 1.5 × 10−9 | 0.22 | 0 | 0 | 10 | plasminogen activator, urokinase receptor |
| PFKFB3 | 0.0004 | 0.13 | 1.4 × 10−9 | 2.1 × 10−11 | 6.7 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase3 |
| CXCL16 | 3.0 × 10−6 | 0.17 | 0 | 0 | 8.9 | C-X-C motif chemokine ligand 16 |
| Index | Name | p-Value | Odds Ratio | Combined Score |
|---|---|---|---|---|
| GO:0071394 | cellular response to testosterone stimulus | 0.001749 | 832.88 | 5287.72 |
| GO:0006003 | fructose 2,6-bisphosphate metabolic process | 0.001749 | 832.88 | 5287.72 |
| GO:0010818 | T-cell chemotaxis | 0.003844 | 333.05 | 1852.16 |
| GO:0032370 | positive regulation of lipid transport | 0.004542 | 277.51 | 1497.03 |
| GO:0006706 | steroid catabolic process | 0.004890 | 256.15 | 1362.86 |
| GO:2000050 | regulation of non-canonical Wnt signaling pathway | 0.005239 | 237.85 | 1249.08 |
| GO:0042730 | fibrinolysis | 0.005239 | 237.85 | 1249.08 |
| GO:0006491 | N-glycan processing | 0.006632 | 184.95 | 927.70 |
| GO:0045821 | positive regulation of glycolytic process | 0.007328 | 166.44 | 818.24 |
| GO:0008209 | androgen metabolic process | 0.007328 | 166.44 | 818.24 |
| GO:1900544 | positive regulation of purine nucleotide metabolic process | 0.007328 | 166.44 | 818.24 |
| GO:0045913 | positive regulation of carbohydrate metabolic process | 0.008371 | 144.71 | 692.15 |
| GO:0022409 | positive regulation of cell-cell adhesion | 0.01634 | 72.27 | 297.35 |
| GO:0009100 | glycoprotein metabolic process | 0.01909 | 61.54 | 243.60 |
| GO:0031401 | positive regulation of protein modification process | 0.002310 | 37.32 | 226.57 |
| GO:0034341 | response to interferon-gamma | 0.02767 | 42.01 | 150.72 |
| GO:0001934 | positive regulation of protein phosphorylation | 0.006776 | 21.27 | 106.24 |
| GEO Accession No. | Platform | Control | MI | |
|---|---|---|---|---|
| GSE60993 | GPL6884 | Illumina HumanWG-6 v3.0 expression beadchip | 7 | 7 |
| GSE62646 | GPL6244 | HuGene-1_0-st] Affymetrix Human Gene 1.0 ST Array [transcript (gene) version]2 | 14 | 14 |
| GSE19339 | GPL570 | [HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array | 4 | 4 |
| GSE61145 | GPL6884 | Illumina HumanWG-6 v3.0 expression beadchip | 7 | 7 |
| GSE61144 | GPL6106 | Sentrix Human-6 v2 Expression beadchip | 10 | 14 |
| GSE66360 | GPL570 | [HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array | 50 | 49 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, A.T.H.; Lawal, B.; Tzeng, Y.-M.; Shih, C.-C.; Shih, C.-M. Identification of a Novel Theranostic Signature of Metabolic and Immune-Inflammatory Dysregulation in Myocardial Infarction, and the Potential Therapeutic Properties of Ovatodiolide, a Diterpenoid Derivative. Int. J. Mol. Sci. 2022, 23, 1281. https://doi.org/10.3390/ijms23031281
Wu ATH, Lawal B, Tzeng Y-M, Shih C-C, Shih C-M. Identification of a Novel Theranostic Signature of Metabolic and Immune-Inflammatory Dysregulation in Myocardial Infarction, and the Potential Therapeutic Properties of Ovatodiolide, a Diterpenoid Derivative. International Journal of Molecular Sciences. 2022; 23(3):1281. https://doi.org/10.3390/ijms23031281
Chicago/Turabian StyleWu, Alexander T. H., Bashir Lawal, Yew-Min Tzeng, Chun-Che Shih, and Chun-Ming Shih. 2022. "Identification of a Novel Theranostic Signature of Metabolic and Immune-Inflammatory Dysregulation in Myocardial Infarction, and the Potential Therapeutic Properties of Ovatodiolide, a Diterpenoid Derivative" International Journal of Molecular Sciences 23, no. 3: 1281. https://doi.org/10.3390/ijms23031281
APA StyleWu, A. T. H., Lawal, B., Tzeng, Y.-M., Shih, C.-C., & Shih, C.-M. (2022). Identification of a Novel Theranostic Signature of Metabolic and Immune-Inflammatory Dysregulation in Myocardial Infarction, and the Potential Therapeutic Properties of Ovatodiolide, a Diterpenoid Derivative. International Journal of Molecular Sciences, 23(3), 1281. https://doi.org/10.3390/ijms23031281







