Cortical Dysplasia and the mTOR Pathway: How the Study of Human Brain Tissue Has Led to Insights into Epileptogenesis
Abstract
1. Introduction and Historical Context
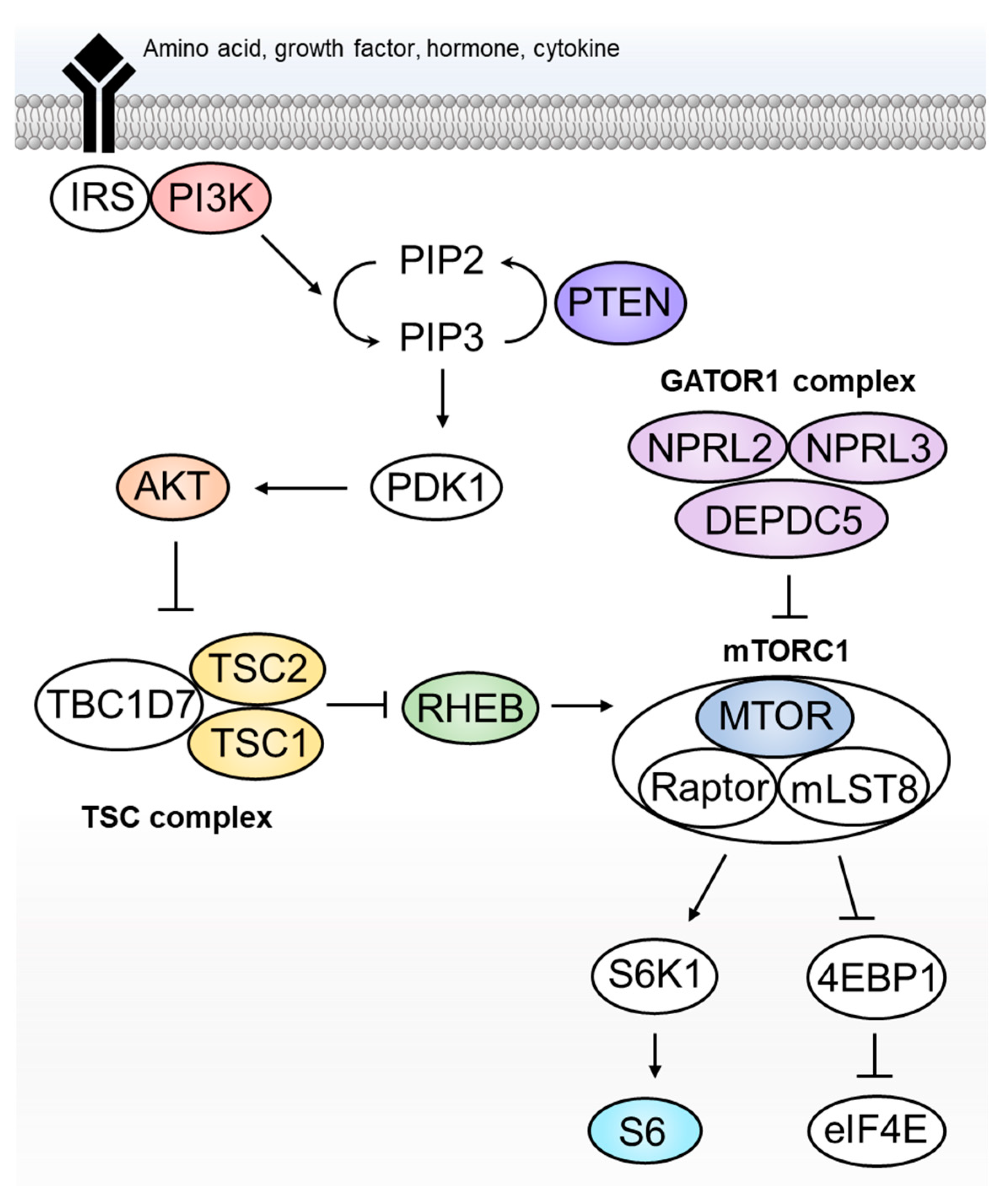
2. The “mTORopathy Hypothesis”—The 2000s
3. Lessons Learnt from Hemispheric Malformations—2012
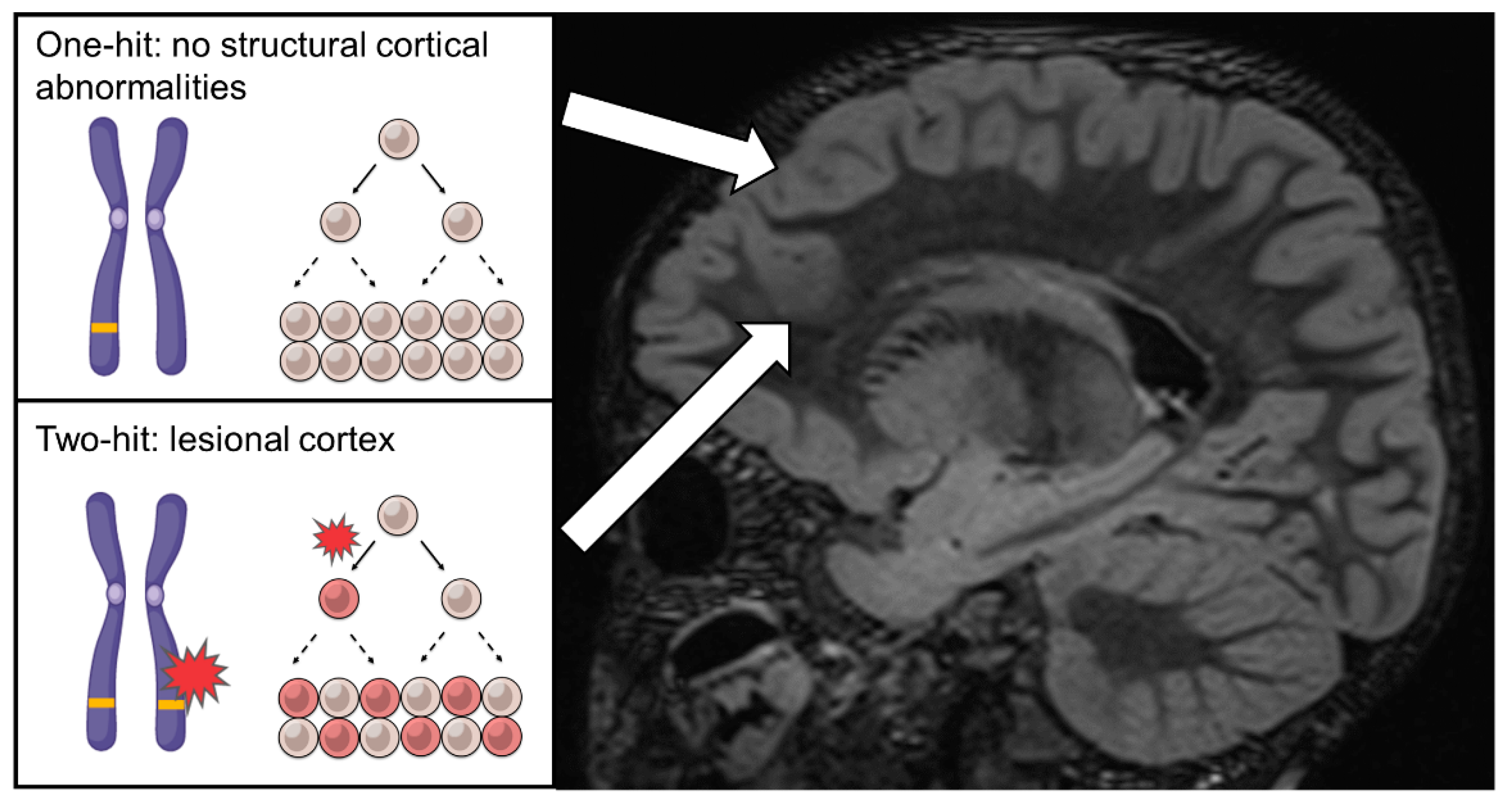
4. The Curious Cases of Familial FCD, Non-Lesional Epilepsy and the Two-Hit Hypothesis—2014
5. First Evidence of Two-Hit Mechanism and Somatic MTOR Variants in FCD—2015~2016
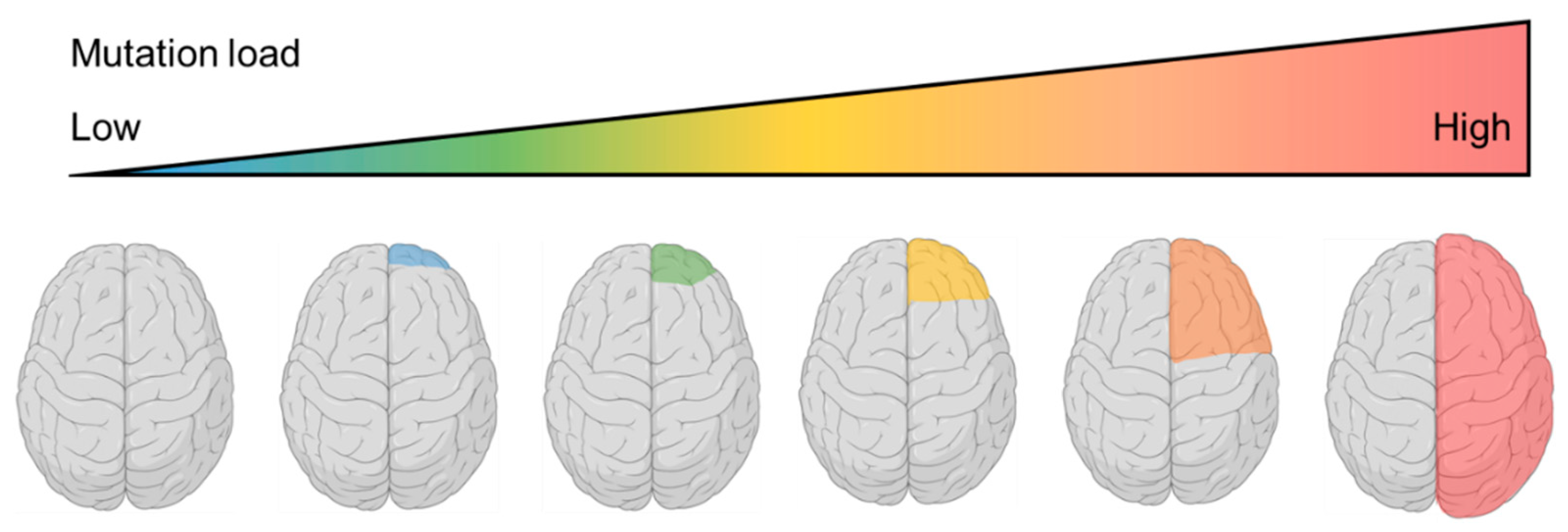
6. Somatic Mosaicism in TSC1/2 and the Continuum of Cortical Dysplasias—2017
7. Confirmation of the Two-Hit Mechanism in FCD and the Identification of Somatic RHEB Variants—2018~2019
8. Pathomechanism of FCDII: What Have We Learnt to Date?
9. Detection of Somatic Mutation: Challenges and Future Directions
10. The Hope of Precision Treatments
11. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Blumcke, I.; Spreafico, R.; Haaker, G.; Coras, R.; Kobow, K.; Bien, C.G.; Pfafflin, M.; Elger, C.; Widman, G.; Schramm, J.; et al. Histopathological Findings in Brain Tissue Obtained during Epilepsy Surgery. N. Engl. J. Med. 2017, 377, 1648–1656. [Google Scholar] [CrossRef] [PubMed]
- Jackson, J.H. A study of convulsions. St Med. Grad. Assoc. Trans. 1869, 22, 162–204. [Google Scholar] [CrossRef]
- Horsley, V. Brain Surgery. Br. Med. J. 1886, 2, 670–677. [Google Scholar]
- Roizin, L. Essay on the origin and evolution of neuropathology; some fundamental neuropathologic contributions to psychiatry. Psychiatr. Q. 1957, 31, 531–555. [Google Scholar] [CrossRef] [PubMed]
- Taylor, D.C.; Falconer, M.A.; Bruton, C.J.; Corsellis, J.A. Focal dysplasia of the cerebral cortex in epilepsy. J. Neurol. Neurosurg. Psychiatry 1971, 34, 369–387. [Google Scholar] [CrossRef] [PubMed]
- Blumcke, I.; Thom, M.; Aronica, E.; Armstrong, D.D.; Vinters, H.V.; Palmini, A.; Jacques, T.S.; Avanzini, G.; Barkovich, A.J.; Battaglia, G.; et al. The clinicopathologic spectrum of focal cortical dysplasias: A consensus classification proposed by an ad hoc Task Force of the ILAE Diagnostic Methods Commission. Epilepsia 2011, 52, 158–174. [Google Scholar] [CrossRef]
- Cheadle, J.P.; Reeve, M.P.; Sampson, J.R.; Kwiatkowski, D.J. Molecular genetic advances in tuberous sclerosis. Hum. Genet. 2000, 107, 97–114. [Google Scholar] [CrossRef]
- Fryer, A.E.; Chalmers, A.; Connor, J.M.; Fraser, I.; Povey, S.; Yates, A.D.; Yates, J.R.; Osborne, J.P. Evidence that the gene for tuberous sclerosis is on chromosome 9. Lancet 1987, 1, 659–661. [Google Scholar] [CrossRef]
- Kandt, R.S.; Haines, J.L.; Smith, M.; Northrup, H.; Gardner, R.J.; Short, M.P.; Dumars, K.; Roach, E.S.; Steingold, S.; Wall, S.; et al. Linkage of an important gene locus for tuberous sclerosis to a chromosome 16 marker for polycystic kidney disease. Nat. Genet. 1992, 2, 37–41. [Google Scholar] [CrossRef]
- Van Slegtenhorst, M.; de Hoogt, R.; Hermans, C.; Nellist, M.; Janssen, B.; Verhoef, S.; Lindhout, D.; van den Ouweland, A.; Halley, D.; Young, J.; et al. Identification of the tuberous sclerosis gene TSC1 on chromosome 9q34. Science 1997, 277, 805–808. [Google Scholar] [CrossRef]
- European Chromosome 16 Tuberous Sclerosis, C. Identification and characterization of the tuberous sclerosis gene on chromosome 16. Cell 1993, 75, 1305–1315. [Google Scholar]
- Potter, C.J.; Huang, H.; Xu, T. Drosophila Tsc1 functions with Tsc2 to antagonize insulin signaling in regulating cell growth, cell proliferation, and organ size. Cell 2001, 105, 357–368. [Google Scholar] [CrossRef]
- Gao, X.; Pan, D. TSC1 and TSC2 tumor suppressors antagonize insulin signaling in cell growth. Genes. Dev. 2001, 15, 1383–1392. [Google Scholar] [CrossRef] [PubMed]
- Tapon, N.; Ito, N.; Dickson, B.J.; Treisman, J.E.; Hariharan, I.K. The Drosophila tuberous sclerosis complex gene homologs restrict cell growth and cell proliferation. Cell 2001, 105, 345–355. [Google Scholar] [CrossRef]
- Brown, E.J.; Albers, M.W.; Shin, T.B.; Ichikawa, K.; Keith, C.T.; Lane, W.S.; Schreiber, S.L. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature 1994, 369, 756–758. [Google Scholar] [CrossRef] [PubMed]
- Sabatini, D.M.; Erdjument-Bromage, H.; Lui, M.; Tempst, P.; Snyder, S.H. RAFT1: A mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell 1994, 78, 35–43. [Google Scholar] [CrossRef]
- Sabers, C.J.; Martin, M.M.; Brunn, G.J.; Williams, J.M.; Dumont, F.J.; Wiederrecht, G.; Abraham, R.T. Isolation of a protein target of the FKBP12-rapamycin complex in mammalian cells. J. Biol. Chem. 1995, 270, 815–822. [Google Scholar] [CrossRef]
- Vezina, C.; Kudelski, A.; Sehgal, S.N. Rapamycin (AY-22,989), a new antifungal antibiotic. I. Taxonomy of the producing streptomycete and isolation of the active principle. J. Antibiot. 1975, 28, 721–726. [Google Scholar] [CrossRef]
- Sabatini, D.M. Twenty-five years of mTOR: Uncovering the link from nutrients to growth. Proc. Natl. Acad. Sci. USA 2017, 114, 11818–11825. [Google Scholar] [CrossRef]
- Liu, G.Y.; Sabatini, D.M. mTOR at the nexus of nutrition, growth, ageing and disease. Nat. Rev. Mol. Cell Biol. 2020, 21, 183–203. [Google Scholar] [CrossRef]
- Baybis, M.; Yu, J.; Lee, A.; Golden, J.A.; Weiner, H.; McKhann, G., 2nd; Aronica, E.; Crino, P.B. mTOR cascade activation distinguishes tubers from focal cortical dysplasia. Ann. Neurol. 2004, 56, 478–487. [Google Scholar] [CrossRef] [PubMed]
- Miyata, H.; Chiang, A.C.; Vinters, H.V. Insulin signaling pathways in cortical dysplasia and TSC-tubers: Tissue microarray analysis. Ann. Neurol. 2004, 56, 510–519. [Google Scholar] [CrossRef] [PubMed]
- Laplante, M.; Sabatini, D.M. mTOR signaling in growth control and disease. Cell 2012, 149, 274–293. [Google Scholar] [CrossRef] [PubMed]
- Sims, J. On Hypertrophy and Atrophy of the Brain. Med.-Chir. Trans. 1835, 19, 315–380. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Barkovich, A.J.; Dobyns, W.B.; Guerrini, R. Malformations of cortical development and epilepsy. Cold Spring Harb. Perspect. Med. 2015, 5, a022392. [Google Scholar] [CrossRef]
- Barkovich, A.J.; Guerrini, R.; Kuzniecky, R.I.; Jackson, G.D.; Dobyns, W.B. A developmental and genetic classification for malformations of cortical development: Update 2012. Brain A J. Neurol. 2012, 135, 1348–1369. [Google Scholar] [CrossRef]
- Crino, P.B. Molecular pathogenesis of focal cortical dysplasia and hemimegalencephaly. J. Child Neurol. 2005, 20, 330–336. [Google Scholar] [CrossRef]
- Ljungberg, M.C.; Bhattacharjee, M.B.; Lu, Y.; Armstrong, D.L.; Yoshor, D.; Swann, J.W.; Sheldon, M.; D’Arcangelo, G. Activation of mammalian target of rapamycin in cytomegalic neurons of human cortical dysplasia. Ann. Neurol. 2006, 60, 420–429. [Google Scholar] [CrossRef]
- Boer, K.; Troost, D.; Spliet, W.G.; Redeker, S.; Crino, P.B.; Aronica, E. A neuropathological study of two autopsy cases of syndromic hemimegalencephaly. Neuropathol. Appl. Neurobiol. 2007, 33, 455–470. [Google Scholar] [CrossRef]
- Aronica, E.; Boer, K.; Baybis, M.; Yu, J.; Crino, P. Co-expression of cyclin D1 and phosphorylated ribosomal S6 proteins in hemimegalencephaly. Acta Neuropathol. 2007, 114, 287–293. [Google Scholar] [CrossRef]
- Crino, P.B. Focal brain malformations: Seizures, signaling, sequencing. Epilepsia 2009, 50 (Suppl. 9), 3–8. [Google Scholar] [CrossRef] [PubMed]
- Salamon, N.; Andres, M.; Chute, D.J.; Nguyen, S.T.; Chang, J.W.; Huynh, M.N.; Chandra, P.S.; Andre, V.M.; Cepeda, C.; Levine, M.S.; et al. Contralateral hemimicrencephaly and clinical-pathological correlations in children with hemimegalencephaly. Brain A J. Neurol. 2006, 129, 352–365. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Huynh, M.; Silhavy, J.L.; Kim, S.; Dixon-Salazar, T.; Heiberg, A.; Scott, E.; Bafna, V.; Hill, K.J.; Collazo, A.; et al. De novo somatic mutations in components of the PI3K-AKT3-mTOR pathway cause hemimegalencephaly. Nat. Genet. 2012, 44, 941–945. [Google Scholar] [CrossRef] [PubMed]
- Poduri, A.; Evrony, G.D.; Cai, X.; Elhosary, P.C.; Beroukhim, R.; Lehtinen, M.K.; Hills, L.B.; Heinzen, E.L.; Hill, A.; Hill, R.S.; et al. Somatic activation of AKT3 causes hemispheric developmental brain malformations. Neuron 2012, 74, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Leventer, R.J.; Jansen, F.E.; Mandelstam, S.A.; Ho, A.; Mohamed, I.; Sarnat, H.B.; Kato, M.; Fukasawa, T.; Saitsu, H.; Matsumoto, N.; et al. Is focal cortical dysplasia sporadic? Family evidence for genetic susceptibility. Epilepsia 2014, 55, e22–e26. [Google Scholar] [CrossRef] [PubMed]
- Bar-Peled, L.; Chantranupong, L.; Cherniack, A.D.; Chen, W.W.; Ottina, K.A.; Grabiner, B.C.; Spear, E.D.; Carter, S.L.; Meyerson, M.; Sabatini, D.M. A Tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science 2013, 340, 1100–1106. [Google Scholar] [CrossRef]
- Dibbens, L.M.; de Vries, B.; Donatello, S.; Heron, S.E.; Hodgson, B.L.; Chintawar, S.; Crompton, D.E.; Hughes, J.N.; Bellows, S.T.; Klein, K.M.; et al. Mutations in DEPDC5 cause familial focal epilepsy with variable foci. Nat. Genet. 2013, 45, 546–551. [Google Scholar] [CrossRef]
- Ishida, S.; Picard, F.; Rudolf, G.; Noe, E.; Achaz, G.; Thomas, P.; Genton, P.; Mundwiller, E.; Wolff, M.; Marescaux, C.; et al. Mutations of DEPDC5 cause autosomal dominant focal epilepsies. Nat. Genet. 2013, 45, 552–555. [Google Scholar] [CrossRef]
- Scheffer, I.E.; Heron, S.E.; Regan, B.M.; Mandelstam, S.; Crompton, D.E.; Hodgson, B.L.; Licchetta, L.; Provini, F.; Bisulli, F.; Vadlamudi, L.; et al. Mutations in mammalian target of rapamycin regulator DEPDC5 cause focal epilepsy with brain malformations. Ann. Neurol. 2014, 75, 782–787. [Google Scholar] [CrossRef]
- Knudson, A.G. Two genetic hits (more or less) to cancer. Nat. Rev. Cancer 2001, 1, 157–162. [Google Scholar] [CrossRef]
- Green, A.J.; Smith, M.; Yates, J.R. Loss of heterozygosity on chromosome 16p13.3 in hamartomas from tuberous sclerosis patients. Nat. Genet. 1994, 6, 193–196. [Google Scholar] [CrossRef] [PubMed]
- Niida, Y.; Stemmer-Rachamimov, A.O.; Logrip, M.; Tapon, D.; Perez, R.; Kwiatkowski, D.J.; Sims, K.; MacCollin, M.; Louis, D.N.; Ramesh, V. Survey of somatic mutations in tuberous sclerosis complex (TSC) hamartomas suggests different genetic mechanisms for pathogenesis of TSC lesions. Am. J. Hum. Genet. 2001, 69, 493–503. [Google Scholar] [CrossRef] [PubMed]
- Martin, K.R.; Zhou, W.; Bowman, M.J.; Shih, J.; Au, K.S.; Dittenhafer-Reed, K.E.; Sisson, K.A.; Koeman, J.; Weisenberger, D.J.; Cottingham, S.L.; et al. The genomic landscape of tuberous sclerosis complex. Nat. Commun. 2017, 8, 15816. [Google Scholar] [CrossRef] [PubMed]
- Carvill, G.L.; Crompton, D.E.; Regan, B.M.; McMahon, J.M.; Saykally, J.; Zemel, M.; Schneider, A.L.; Dibbens, L.; Howell, K.B.; Mandelstam, S.; et al. Epileptic spasms are a feature of DEPDC5 mTORopathy. Neurol. Genet. 2015, 1, e17. [Google Scholar] [CrossRef] [PubMed]
- Baulac, S.; Ishida, S.; Marsan, E.; Miquel, C.; Biraben, A.; Nguyen, D.K.; Nordli, D.; Cossette, P.; Nguyen, S.; Lambrecq, V.; et al. Familial focal epilepsy with focal cortical dysplasia due to DEPDC5 mutations. Ann. Neurol. 2015, 77, 675–683. [Google Scholar] [CrossRef]
- D’Gama, A.M.; Geng, Y.; Couto, J.A.; Martin, B.; Boyle, E.A.; LaCoursiere, C.M.; Hossain, A.; Hatem, N.E.; Barry, B.J.; Kwiatkowski, D.J.; et al. Mammalian target of rapamycin pathway mutations cause hemimegalencephaly and focal cortical dysplasia. Ann. Neurol. 2015, 77, 720–725. [Google Scholar] [CrossRef]
- Scerri, T.; Riseley, J.R.; Gillies, G.; Pope, K.; Burgess, R.; Mandelstam, S.A.; Dibbens, L.; Chow, C.W.; Maixner, W.; Harvey, A.S.; et al. Familial cortical dysplasia type IIA caused by a germline mutation in DEPDC5. Ann. Clin. Transl. Neurol. 2015, 2, 575–580. [Google Scholar] [CrossRef]
- Weckhuysen, S.; Marsan, E.; Lambrecq, V.; Marchal, C.; Morin-Brureau, M.; An-Gourfinkel, I.; Baulac, M.; Fohlen, M.; Kallay Zetchi, C.; Seeck, M.; et al. Involvement of GATOR complex genes in familial focal epilepsies and focal cortical dysplasia. Epilepsia 2016, 57, 994–1003. [Google Scholar] [CrossRef]
- Ricos, M.G.; Hodgson, B.L.; Pippucci, T.; Saidin, A.; Ong, Y.S.; Heron, S.E.; Licchetta, L.; Bisulli, F.; Bayly, M.A.; Hughes, J.; et al. Mutations in the mammalian target of rapamycin pathway regulators NPRL2 and NPRL3 cause focal epilepsy. Ann. Neurol. 2016, 79, 120–131. [Google Scholar] [CrossRef]
- Sim, J.C.; Scerri, T.; Fanjul-Fernandez, M.; Riseley, J.R.; Gillies, G.; Pope, K.; van Roozendaal, H.; Heng, J.I.; Mandelstam, S.A.; McGillivray, G.; et al. Familial cortical dysplasia caused by mutation in the mammalian target of rapamycin regulator NPRL3. Ann. Neurol. 2016, 79, 132–137. [Google Scholar] [CrossRef]
- Mirzaa, G.M.; Campbell, C.D.; Solovieff, N.; Goold, C.; Jansen, L.A.; Menon, S.; Timms, A.E.; Conti, V.; Biag, J.D.; Adams, C.; et al. Association of MTOR Mutations With Developmental Brain Disorders, Including Megalencephaly, Focal Cortical Dysplasia, and Pigmentary Mosaicism. JAMA Neurol. 2016, 73, 836–845. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.S.; Kim, W.I.; Kang, H.C.; Kim, S.H.; Park, A.H.; Park, E.K.; Cho, Y.W.; Kim, S.; Kim, H.M.; Kim, J.A.; et al. Brain somatic mutations in MTOR cause focal cortical dysplasia type II leading to intractable epilepsy. Nat. Med. 2015, 21, 395–400. [Google Scholar] [CrossRef] [PubMed]
- Leventer, R.J.; Scerri, T.; Marsh, A.P.; Pope, K.; Gillies, G.; Maixner, W.; MacGregor, D.; Harvey, A.S.; Delatycki, M.B.; Amor, D.J.; et al. Hemispheric cortical dysplasia secondary to a mosaic somatic mutation in MTOR. Neurology 2015, 84, 2029–2032. [Google Scholar] [CrossRef]
- Nakashima, M.; Saitsu, H.; Takei, N.; Tohyama, J.; Kato, M.; Kitaura, H.; Shiina, M.; Shirozu, H.; Masuda, H.; Watanabe, K.; et al. Somatic Mutations in the MTOR gene cause focal cortical dysplasia type IIb. Ann. Neurol. 2015, 78, 375–386. [Google Scholar] [CrossRef] [PubMed]
- Moller, R.S.; Weckhuysen, S.; Chipaux, M.; Marsan, E.; Taly, V.; Bebin, E.M.; Hiatt, S.M.; Prokop, J.W.; Bowling, K.M.; Mei, D.; et al. Germline and somatic mutations in the MTOR gene in focal cortical dysplasia and epilepsy. Neurol Genet. 2016, 2, e118. [Google Scholar] [CrossRef]
- Grabiner, B.C.; Nardi, V.; Birsoy, K.; Possemato, R.; Shen, K.; Sinha, S.; Jordan, A.; Beck, A.H.; Sabatini, D.M. A diverse array of cancer-associated MTOR mutations are hyperactivating and can predict rapamycin sensitivity. Cancer Discov. 2014, 4, 554–563. [Google Scholar] [CrossRef]
- Jansen, L.A.; Mirzaa, G.M.; Ishak, G.E.; O’Roak, B.J.; Hiatt, J.B.; Roden, W.H.; Gunter, S.A.; Christian, S.L.; Collins, S.; Adams, C.; et al. PI3K/AKT pathway mutations cause a spectrum of brain malformations from megalencephaly to focal cortical dysplasia. Brain A J. Neurol. 2015, 138, 1613–1628. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.S.; Gopalappa, R.; Kim, S.H.; Ramakrishna, S.; Lee, M.; Kim, W.I.; Kim, J.; Park, S.M.; Lee, J.; Oh, J.H.; et al. Somatic Mutations in TSC1 and TSC2 Cause Focal Cortical Dysplasia. Am. J. Hum. Genet. 2017, 100, 454–472. [Google Scholar] [CrossRef]
- D’Gama, A.M.; Woodworth, M.B.; Hossain, A.A.; Bizzotto, S.; Hatem, N.E.; LaCoursiere, C.M.; Najm, I.; Ying, Z.; Yang, E.; Barkovich, A.J.; et al. Somatic Mutations Activating the mTOR Pathway in Dorsal Telencephalic Progenitors Cause a Continuum of Cortical Dysplasias. Cell Rep. 2017, 21, 3754–3766. [Google Scholar] [CrossRef]
- Hoelz, H.; Coppenrath, E.; Hoertnagel, K.; Roser, T.; Tacke, M.; Gerstl, L.; Borggraefe, I. Childhood-Onset Epileptic Encephalopathy Associated With Isolated Focal Cortical Dysplasia and a Novel TSC1 Germline Mutation. Clin. EEG Neurosci. 2018, 49, 187–191. [Google Scholar] [CrossRef]
- Poduri, A.; Evrony, G.D.; Cai, X.; Walsh, C.A. Somatic mutation, genomic variation, and neurological disease. Science 2013, 341, 1237758. [Google Scholar] [CrossRef] [PubMed]
- Ribierre, T.; Deleuze, C.; Bacq, A.; Baldassari, S.; Marsan, E.; Chipaux, M.; Muraca, G.; Roussel, D.; Navarro, V.; Leguern, E.; et al. Second-hit mosaic mutation in mTORC1 repressor DEPDC5 causes focal cortical dysplasia-associated epilepsy. J. Clin. Investig. 2018, 128, 2452–2458. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, F.A.; Borlot, F.; Cossette, P.; Minassian, B.A.; Andrade, D.M. Two definite cases of sudden unexpected death in epilepsy in a family with a DEPDC5 mutation. Neurol. Genet. 2015, 1, e28. [Google Scholar] [CrossRef] [PubMed]
- Bagnall, R.D.; Crompton, D.E.; Petrovski, S.; Lam, L.; Cutmore, C.; Garry, S.I.; Sadleir, L.G.; Dibbens, L.M.; Cairns, A.; Kivity, S.; et al. Exome-based analysis of cardiac arrhythmia, respiratory control, and epilepsy genes in sudden unexpected death in epilepsy. Ann. Neurol. 2016, 79, 522–534. [Google Scholar] [CrossRef] [PubMed]
- Baldassari, S.; Picard, F.; Verbeek, N.E.; van Kempen, M.; Brilstra, E.H.; Lesca, G.; Conti, V.; Guerrini, R.; Bisulli, F.; Licchetta, L.; et al. The landscape of epilepsy-related GATOR1 variants. Genet. Med. Off. J. Am. Coll. Med. Genet. 2019, 21, 398–408. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.S.; Stephenson, S.E.M.; Howell, K.B.; Pope, K.; Gillies, G.; Wray, A.; Maixner, W.; Mandelstam, S.A.; Berkovic, S.F.; Scheffer, I.E.; et al. Second-hit DEPDC5 mutation is limited to dysmorphic neurons in cortical dysplasia type IIA. Ann. Clin. Transl. Neurol. 2019, 6, 1338–1344. [Google Scholar] [CrossRef]
- Baldassari, S.; Ribierre, T.; Marsan, E.; Adle-Biassette, H.; Ferrand-Sorbets, S.; Bulteau, C.; Dorison, N.; Fohlen, M.; Polivka, M.; Weckhuysen, S.; et al. Dissecting the genetic basis of focal cortical dysplasia: A large cohort study. Acta Neuropathol. 2019, 138, 885–900. [Google Scholar] [CrossRef]
- Sim, N.S.; Ko, A.; Kim, W.K.; Kim, S.H.; Kim, J.S.; Shim, K.W.; Aronica, E.; Mijnsbergen, C.; Spliet, W.G.M.; Koh, H.Y.; et al. Precise detection of low-level somatic mutation in resected epilepsy brain tissue. Acta Neuropathol. 2019, 138, 901–912. [Google Scholar] [CrossRef]
- Pelorosso, C.; Watrin, F.; Conti, V.; Buhler, E.; Gelot, A.; Yang, X.; Mei, D.; McEvoy-Venneri, J.; Manent, J.B.; Cetica, V.; et al. Somatic double-hit in MTOR and RPS6 in hemimegalencephaly with intractable epilepsy. Hum. Mol. Genet. 2019, 28, 3755–3765. [Google Scholar] [CrossRef]
- Salinas, V.; Vega, P.; Piccirilli, M.V.; Chicco, C.; Ciraolo, C.; Christiansen, S.; Consalvo, D.; Perez-Maturo, J.; Medina, N.; Gonzalez-Moron, D.; et al. Identification of a somatic mutation in the RHEB gene through high depth and ultra-high depth next generation sequencing in a patient with Hemimegalencephaly and drug resistant Epilepsy. Eur. J. Med. Genet. 2019, 62, 103571. [Google Scholar] [CrossRef]
- Zhao, S.; Li, Z.; Zhang, M.; Zhang, L.; Zheng, H.; Ning, J.; Wang, Y.; Wang, F.; Zhang, X.; Gan, H.; et al. A brain somatic RHEB doublet mutation causes focal cortical dysplasia type II. Exp. Mol. Med. 2019, 51, 84. [Google Scholar] [CrossRef] [PubMed]
- Winawer, M.R.; Griffin, N.G.; Samanamud, J.; Baugh, E.H.; Rathakrishnan, D.; Ramalingam, S.; Zagzag, D.; Schevon, C.A.; Dugan, P.; Hegde, M.; et al. Somatic SLC35A2 variants in the brain are associated with intractable neocortical epilepsy. Ann. Neurol. 2018, 83, 1133–1146. [Google Scholar] [CrossRef] [PubMed]
- Sim, N.S.; Seo, Y.; Lim, J.S.; Kim, W.K.; Son, H.; Kim, H.D.; Kim, S.; An, H.J.; Kang, H.C.; Kim, S.H.; et al. Brain somatic mutations in SLC35A2 cause intractable epilepsy with aberrant N-glycosylation. Neurology. Genet. 2018, 4, e294. [Google Scholar] [CrossRef]
- Miller, K.E.; Koboldt, D.C.; Schieffer, K.M.; Bedrosian, T.A.; Crist, E.; Sheline, A.; Leraas, K.; Magrini, V.; Zhong, H.; Brennan, P.; et al. Somatic SLC35A2 mosaicism correlates with clinical findings in epilepsy brain tissue. Neurology. Genet. 2020, 6, e460. [Google Scholar] [CrossRef]
- Bonduelle, T.; Hartlieb, T.; Baldassari, S.; Sim, N.S.; Kim, S.H.; Kang, H.C.; Kobow, K.; Coras, R.; Chipaux, M.; Dorfmuller, G.; et al. Frequent SLC35A2 brain mosaicism in mild malformation of cortical development with oligodendroglial hyperplasia in epilepsy (MOGHE). Acta Neuropathol. Commun. 2021, 9, 3. [Google Scholar] [CrossRef] [PubMed]
- Cepeda, C.; Andre, V.M.; Vinters, H.V.; Levine, M.S.; Mathern, G.W. Are cytomegalic neurons and balloon cells generators of epileptic activity in pediatric cortical dysplasia? Epilepsia 2005, 46 (Suppl. 5), 82–88. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.S.; Baldassari, S.; Chipaux, M.; Adle-Biassette, H.; Stephenson, S.E.M.; Maixner, W.; Harvey, A.S.; Lockhart, P.J.; Baulac, S.; Leventer, R.J. Gradient of brain mosaic RHEB variants causes a continuum of cortical dysplasia. Ann. Clin. Transl. Neurol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.H.; Mahadeo, T.; Bordey, A. mTOR Hyperactivity Levels Influence the Severity of Epilepsy and Associated Neuropathology in an Experimental Model of Tuberous Sclerosis Complex and Focal Cortical Dysplasia. J. Neurosci. Off. J. Soc. Neurosci. 2019, 39, 2762–2773. [Google Scholar] [CrossRef] [PubMed]
- Park, S.M.; Lim, J.S.; Ramakrishina, S.; Kim, S.H.; Kim, W.K.; Lee, J.; Kang, H.C.; Reiter, J.F.; Kim, D.S.; Kim, H.H.; et al. Brain Somatic Mutations in MTOR Disrupt Neuronal Ciliogenesis, Leading to Focal Cortical Dyslamination. Neuron 2018, 99, 83–97. [Google Scholar] [CrossRef]
- May-Simera, H.L.; Kelley, M.W. Cilia, Wnt signaling, and the cytoskeleton. Cilia 2012, 1, 7. [Google Scholar] [CrossRef]
- Guo, J.; Higginbotham, H.; Li, J.; Nichols, J.; Hirt, J.; Ghukasyan, V.; Anton, E.S. Developmental disruptions underlying brain abnormalities in ciliopathies. Nat. Commun. 2015, 6, 7857. [Google Scholar] [CrossRef] [PubMed]
- Hilgendorf, K.I.; Johnson, C.T.; Jackson, P.K. The primary cilium as a cellular receiver: Organizing ciliary GPCR signaling. Curr. Opin. Cell Biol. 2016, 39, 84–92. [Google Scholar] [CrossRef] [PubMed]
- Bangs, F.; Anderson, K.V. Primary Cilia and Mammalian Hedgehog Signaling. Cold Spring Harb. Perspect. Biol. 2017, 9, a028175. [Google Scholar] [CrossRef] [PubMed]
- Boitard, M.; Bocchi, R.; Egervari, K.; Petrenko, V.; Viale, B.; Gremaud, S.; Zgraggen, E.; Salmon, P.; Kiss, J.Z. Wnt signaling regulates multipolar-to-bipolar transition of migrating neurons in the cerebral cortex. Cell Rep. 2015, 10, 1349–1361. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, L.S.; Wen, J.H.; Claycomb, K.; Huang, Y.; Harrsch, F.A.; Naegele, J.R.; Hyder, F.; Buchanan, G.F.; Bordey, A. Convulsive seizures from experimental focal cortical dysplasia occur independently of cell misplacement. Nat. Commun. 2016, 7, 11753. [Google Scholar] [CrossRef]
- Koh, H.Y.; Jang, J.; Ju, S.H.; Kim, R.; Cho, G.B.; Kim, D.S.; Sohn, J.W.; Paik, S.B.; Lee, J.H. Non-Cell Autonomous Epileptogenesis in Focal Cortical Dysplasia. Ann. Neurol. 2021, 90, 285–299. [Google Scholar] [CrossRef]
- Avansini, S.H.; Torres, F.R.; Vieira, A.S.; Dogini, D.B.; Rogerio, F.; Coan, A.C.; Morita, M.E.; Guerreiro, M.M.; Yasuda, C.L.; Secolin, R.; et al. Dysregulation of NEUROG2 plays a key role in focal cortical dysplasia. Ann. Neurol. 2018, 83, 623–635. [Google Scholar] [CrossRef]
- Lee, J.Y.; Park, A.K.; Lee, E.S.; Park, W.Y.; Park, S.H.; Choi, J.W.; Phi, J.H.; Wang, K.C.; Kim, S.K. miRNA expression analysis in cortical dysplasia: Regulation of mTOR and LIS1 pathway. Epilepsy Res. 2014, 108, 433–441. [Google Scholar] [CrossRef]
- Li, L.; Liu, C.Q.; Li, T.F.; Guan, Y.G.; Zhou, J.; Qi, X.L.; Yang, Y.T.; Deng, J.H.; Xu, Z.Q.; Luan, G.M. Analysis of Altered Micro RNA Expression Profiles in Focal Cortical Dysplasia IIB. J. Child Neurol. 2016, 31, 613–620. [Google Scholar] [CrossRef]
- Enright, N.; Simonato, M.; Henshall, D.C. Discovery and validation of blood microRNAs as molecular biomarkers of epilepsy: Ways to close current knowledge gaps. Epilepsia Open 2018, 3, 427–436. [Google Scholar] [CrossRef]
- Wang, X.; Sun, Y.; Tan, Z.; Che, N.; Ji, A.; Luo, X.; Sun, X.; Li, X.; Yang, K.; Wang, G.; et al. Serum MicroRNA-4521 is a Potential Biomarker for Focal Cortical Dysplasia with Refractory Epilepsy. Neurochem. Res. 2016, 41, 905–912. [Google Scholar] [CrossRef] [PubMed]
- Che, N.; Zu, G.; Zhou, T.; Wang, X.; Sun, Y.; Tan, Z.; Liu, Y.; Wang, D.; Luo, X.; Zhao, Z.; et al. Aberrant Expression of miR-323a-5p in Patients with Refractory Epilepsy Caused by Focal Cortical Dysplasia. Genet. Test. Mol. Biomark. 2017, 21, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Tsai, V.; Parker, W.E.; Aronica, E.; Baybis, M.; Crino, P.B. Detection of human papillomavirus in human focal cortical dysplasia type IIB. Ann. Neurol. 2012, 72, 881–892. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Lu, L.; Cheng, X.; Xu, G.; Yang, H. Viral infection and focal cortical dysplasia. Ann. Neurol. 2014, 75, 614–616. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, V.; Llinares-Benadero, C.; Borrell, V. Cerebral cortex expansion and folding: What have we learned? EMBO J. 2016, 35, 1021–1044. [Google Scholar] [CrossRef] [PubMed]
- Rakic, P. Mode of cell migration to the superficial layers of fetal monkey neocortex. J. Comp. Neurol. 1972, 145, 61–83. [Google Scholar] [CrossRef] [PubMed]
- Anderson, S.A.; Eisenstat, D.D.; Shi, L.; Rubenstein, J.L. Interneuron migration from basal forebrain to neocortex: Dependence on Dlx genes. Science 1997, 278, 474–476. [Google Scholar] [CrossRef]
- Soriano, E.; Dumesnil, N.; Auladell, C.; Cohen-Tannoudji, M.; Sotelo, C. Molecular heterogeneity of progenitors and radial migration in the developing cerebral cortex revealed by transgene expression. Proc. Natl. Acad. Sci. USA 1995, 92, 11676–11680. [Google Scholar] [CrossRef]
- Rossini, L.; Medici, V.; Tassi, L.; Cardinale, F.; Tringali, G.; Bramerio, M.; Villani, F.; Spreafico, R.; Garbelli, R. Layer-specific gene expression in epileptogenic type II focal cortical dysplasia: Normal-looking neurons reveal the presence of a hidden laminar organization. Acta Neuropathol. Commun. 2014, 2, 45. [Google Scholar] [CrossRef]
- Juric-Sekhar, G.; Hevner, R.F. Malformations of Cerebral Cortex Development: Molecules and Mechanisms. Annu. Rev. Pathol. 2019, 14, 293–318. [Google Scholar] [CrossRef] [PubMed]
- Jehi, L. The Epileptogenic Zone: Concept and Definition. Epilepsy Curr. 2018, 18, 12–16. [Google Scholar] [CrossRef] [PubMed]
- Stephenson, S.E.M.; Owens, H.G.; Richards, K.L.; Lee, W.S.; D’Arcy, C.; Barton, S.; Mandelstam, S.A.; Maixner, W.J.; MacGregor, D.; Petrou, S.; et al. Dysmorphic neuron density is highest in the centre of epileptogenic cortical tubers. bioRxiv 2019, 621607. [Google Scholar] [CrossRef]
- Huang, L.; Ma, F.; Chapman, A.; Lu, S.; Xie, X.S. Single-Cell Whole-Genome Amplification and Sequencing: Methodology and Applications. Annu. Rev. Genom. Hum. Genet. 2015, 16, 79–102. [Google Scholar] [CrossRef] [PubMed]
- Lodato, M.A.; Woodworth, M.B.; Lee, S.; Evrony, G.D.; Mehta, B.K.; Karger, A.; Lee, S.; Chittenden, T.W.; D’Gama, A.M.; Cai, X.; et al. Somatic mutation in single human neurons tracks developmental and transcriptional history. Science 2015, 350, 94–98. [Google Scholar] [CrossRef] [PubMed]
- Lodato, M.A.; Walsh, C.A. Genome aging: Somatic mutation in the brain links age-related decline with disease and nominates pathogenic mechanisms. Hum. Mol. Genet. 2019, 28, R197–R206. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, L.S.; Wen, J.H.; Nguyen, L.H.; Zhang, L.; Getz, S.A.; Torres-Reveron, J.; Wang, Y.; Spencer, D.D.; Bordey, A. Ectopic HCN4 expression drives mTOR-dependent epilepsy in mice. Sci. Transl. Med. 2020, 12. [Google Scholar] [CrossRef]
- Zhang, L.; Huang, T.; Teaw, S.; Nguyen, L.H.; Hsieh, L.S.; Gong, X.; Burns, L.H.; Bordey, A. Filamin A inhibition reduces seizure activity in a mouse model of focal cortical malformations. Sci. Transl. Med. 2020, 12. [Google Scholar] [CrossRef]
- Dimartino, P.; Mariani, V.; Marconi, C.; Minardi, R.; Bramerio, M.; Licchetta, L.; Menghi, V.; Morandi, L.; Magini, P.; Mongelli, P.; et al. Accurate Detection of Hot-Spot MTOR Somatic Mutations in Archival Surgical Specimens of Focal Cortical Dysplasia by Molecular Inversion Probes. Mol. Diagn. Ther. 2020, 24, 571–577. [Google Scholar] [CrossRef] [PubMed]
- Evrony, G.D.; Cai, X.; Lee, E.; Hills, L.B.; Elhosary, P.C.; Lehmann, H.S.; Parker, J.J.; Atabay, K.D.; Gilmore, E.C.; Poduri, A.; et al. Single-neuron sequencing analysis of L1 retrotransposition and somatic mutation in the human brain. Cell 2012, 151, 483–496. [Google Scholar] [CrossRef]
- French, J.A.; Lawson, J.A.; Yapici, Z.; Ikeda, H.; Polster, T.; Nabbout, R.; Curatolo, P.; de Vries, P.J.; Dlugos, D.J.; Berkowitz, N.; et al. Adjunctive everolimus therapy for treatment-resistant focal-onset seizures associated with tuberous sclerosis (EXIST-3): A phase 3, randomised, double-blind, placebo-controlled study. Lancet 2016, 388, 2153–2163. [Google Scholar] [CrossRef]
- Xu, Q.; Uliel-Sibony, S.; Dunham, C.; Sarnat, H.; Flores-Sarnat, L.; Brunga, L.; Davidson, S.; Lo, W.; Shlien, A.; Connolly, M.; et al. mTOR Inhibitors as a New Therapeutic Strategy in Treatment Resistant Epilepsy in Hemimegalencephaly: A Case Report. J. Child Neurol. 2019, 34, 132–138. [Google Scholar] [CrossRef] [PubMed]
- Hadouiri, N.; Darmency, V.; Guibaud, L.; Arzimanoglou, A.; Sorlin, A.; Carmignac, V.; Riviere, J.B.; Huet, F.; Luu, M.; Bardou, M.; et al. Compassionate use of everolimus for refractory epilepsy in a patient with MTOR mosaic mutation. Eur. J. Med. Genet. 2020, 63, 104036. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.K.; Cho, J.; Kim, S.H.; Kang, H.C.; Kim, D.S.; Kim, V.N.; Lee, J.H. Brain somatic mutations in MTOR reveal translational dysregulations underlying intractable focal epilepsy. J. Clin. Investig. 2019, 129, 4207–4223. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Baldassari, S.; Sim, N.S.; Chipaux, M.; Dorfmuller, G.; Kim, D.S.; Chang, W.S.; Taly, V.; Lee, J.H.; Baulac, S. Detection of Brain Somatic Mutations in Cerebrospinal Fluid from Refractory Epilepsy Patients. Ann. Neurol. 2021, 89, 1248–1252. [Google Scholar] [CrossRef]
- Ye, Z.; Chatterton, Z.; Pflueger, J.; Damiano, J.A.; McQuillan, L.; Harvey, A.S.; Malone, S.; Do, H.; Maixner, W.; Schneider, A.; et al. Cerebrospinal fluid liquid biopsy for detecting somatic mosaicism in brain. Brain Commun. 2021, 3, fcaa235. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, W.S.; Baldassari, S.; Stephenson, S.E.M.; Lockhart, P.J.; Baulac, S.; Leventer, R.J. Cortical Dysplasia and the mTOR Pathway: How the Study of Human Brain Tissue Has Led to Insights into Epileptogenesis. Int. J. Mol. Sci. 2022, 23, 1344. https://doi.org/10.3390/ijms23031344
Lee WS, Baldassari S, Stephenson SEM, Lockhart PJ, Baulac S, Leventer RJ. Cortical Dysplasia and the mTOR Pathway: How the Study of Human Brain Tissue Has Led to Insights into Epileptogenesis. International Journal of Molecular Sciences. 2022; 23(3):1344. https://doi.org/10.3390/ijms23031344
Chicago/Turabian StyleLee, Wei Shern, Sara Baldassari, Sarah E. M. Stephenson, Paul J. Lockhart, Stéphanie Baulac, and Richard J. Leventer. 2022. "Cortical Dysplasia and the mTOR Pathway: How the Study of Human Brain Tissue Has Led to Insights into Epileptogenesis" International Journal of Molecular Sciences 23, no. 3: 1344. https://doi.org/10.3390/ijms23031344
APA StyleLee, W. S., Baldassari, S., Stephenson, S. E. M., Lockhart, P. J., Baulac, S., & Leventer, R. J. (2022). Cortical Dysplasia and the mTOR Pathway: How the Study of Human Brain Tissue Has Led to Insights into Epileptogenesis. International Journal of Molecular Sciences, 23(3), 1344. https://doi.org/10.3390/ijms23031344





