A Putative Effector CcSp84 of Cytospora chrysosperma Localizes to the Plant Nucleus to Trigger Plant Immunity
Abstract
:1. Introduction
2. Results
2.1. Two NLS Motif Containing Candidate Effectors Were Induced during Infection Processes
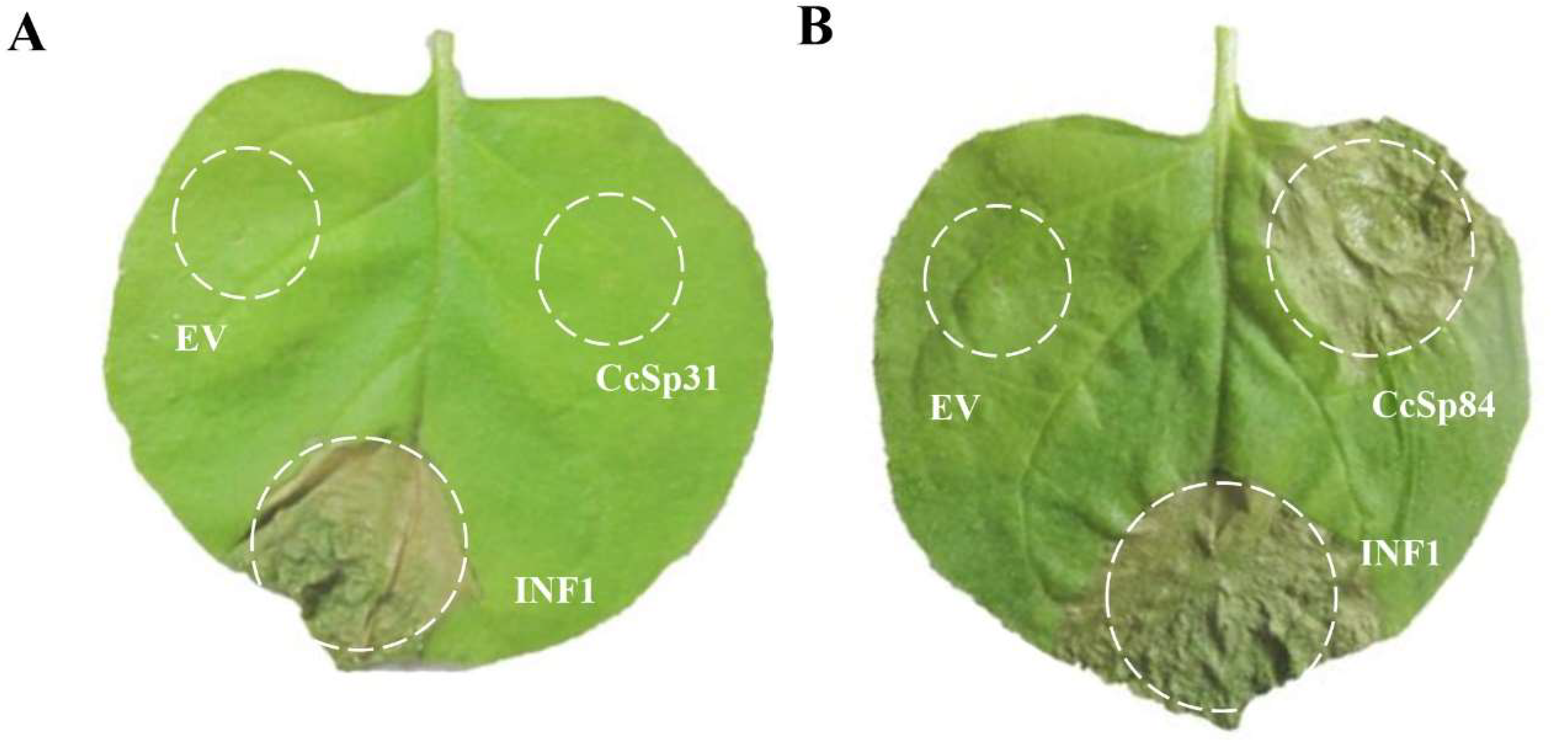
2.2. CcSp84 Induced Cell Death in N. benthamiana Leaves
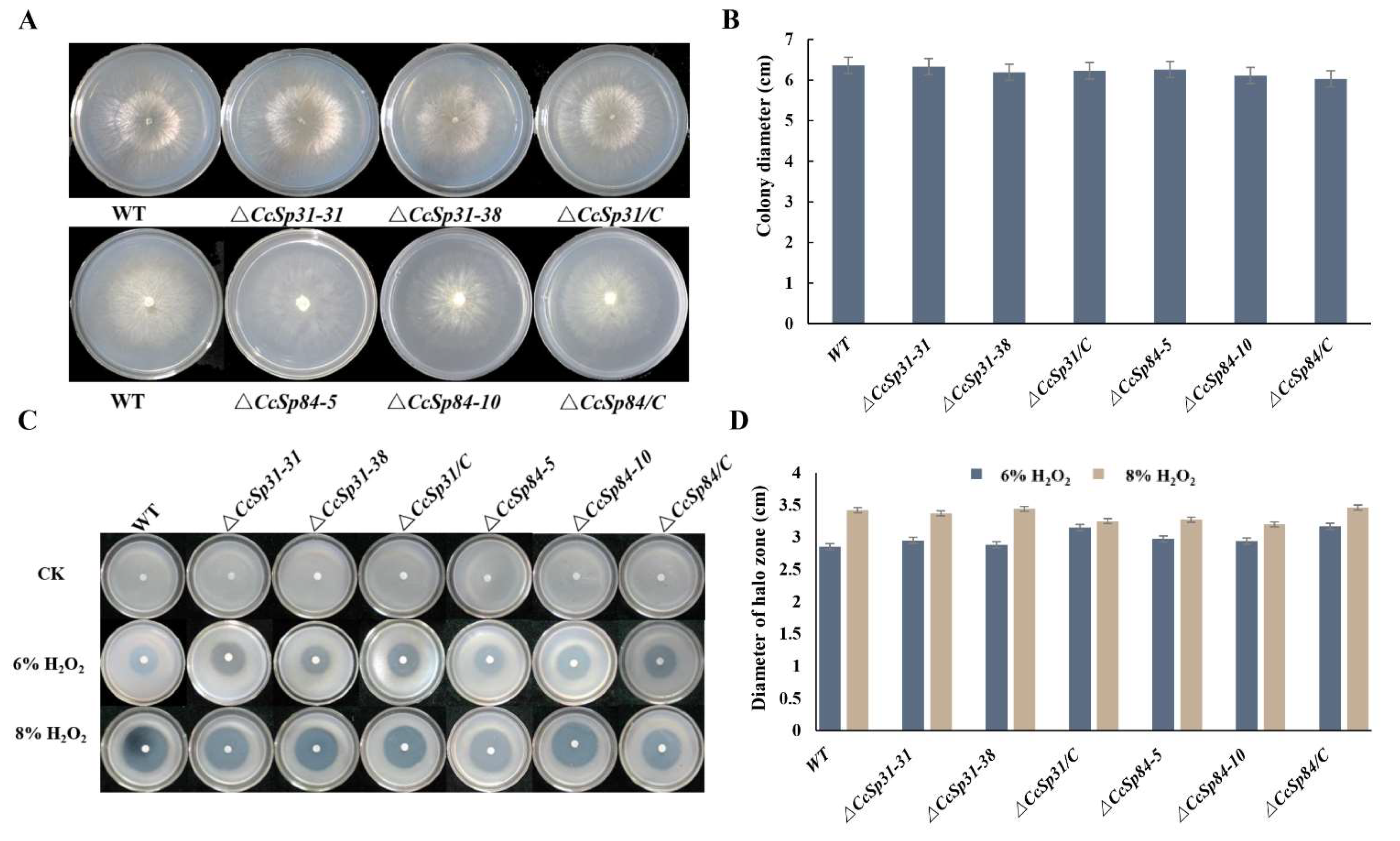
2.3. CcSp31 and CcSp84 Were Not Required for Fungal Growth and the Resistance to H2O2
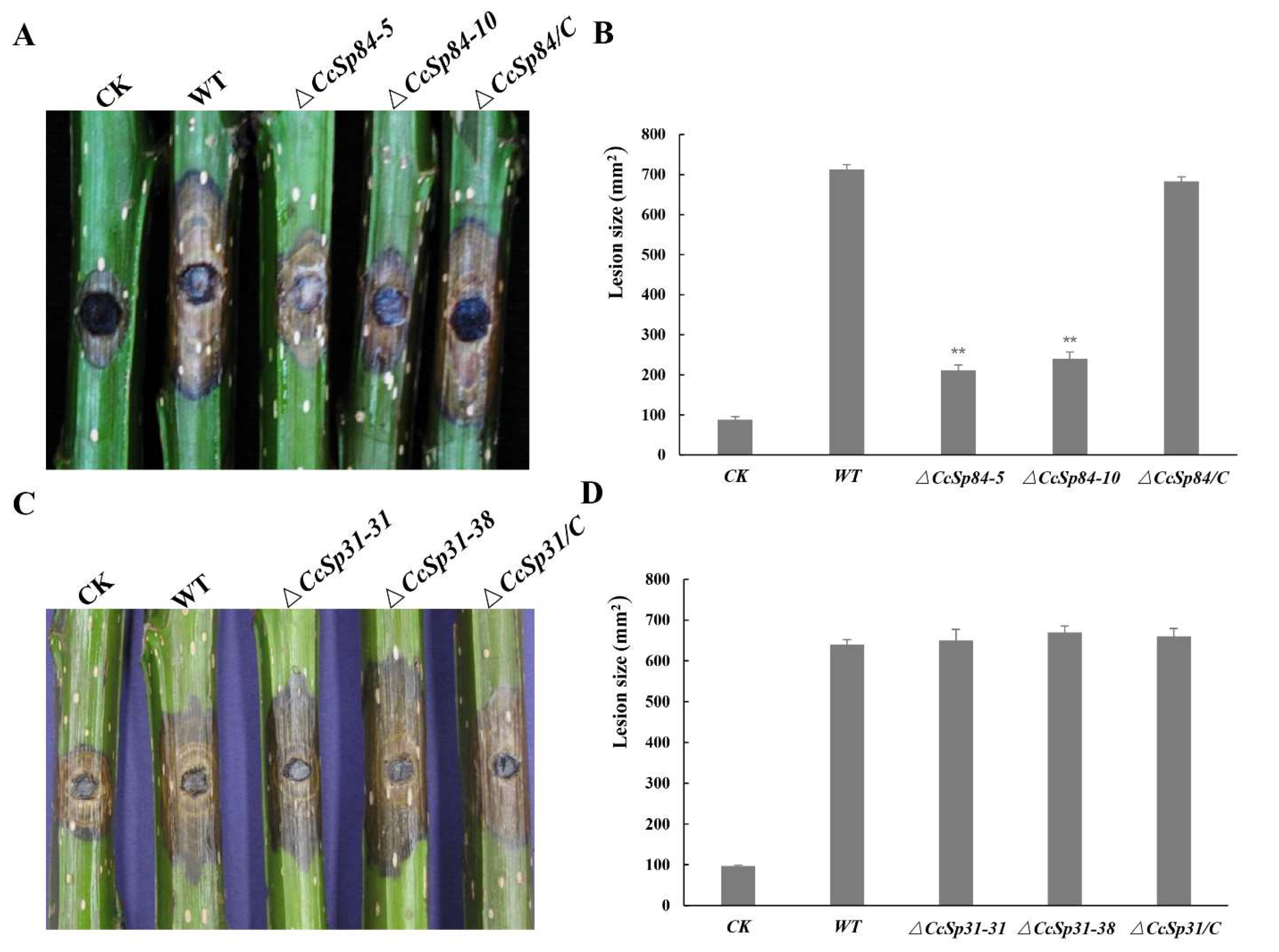
2.4. CcSp84 Was Essential for Fungal Pathogenicity
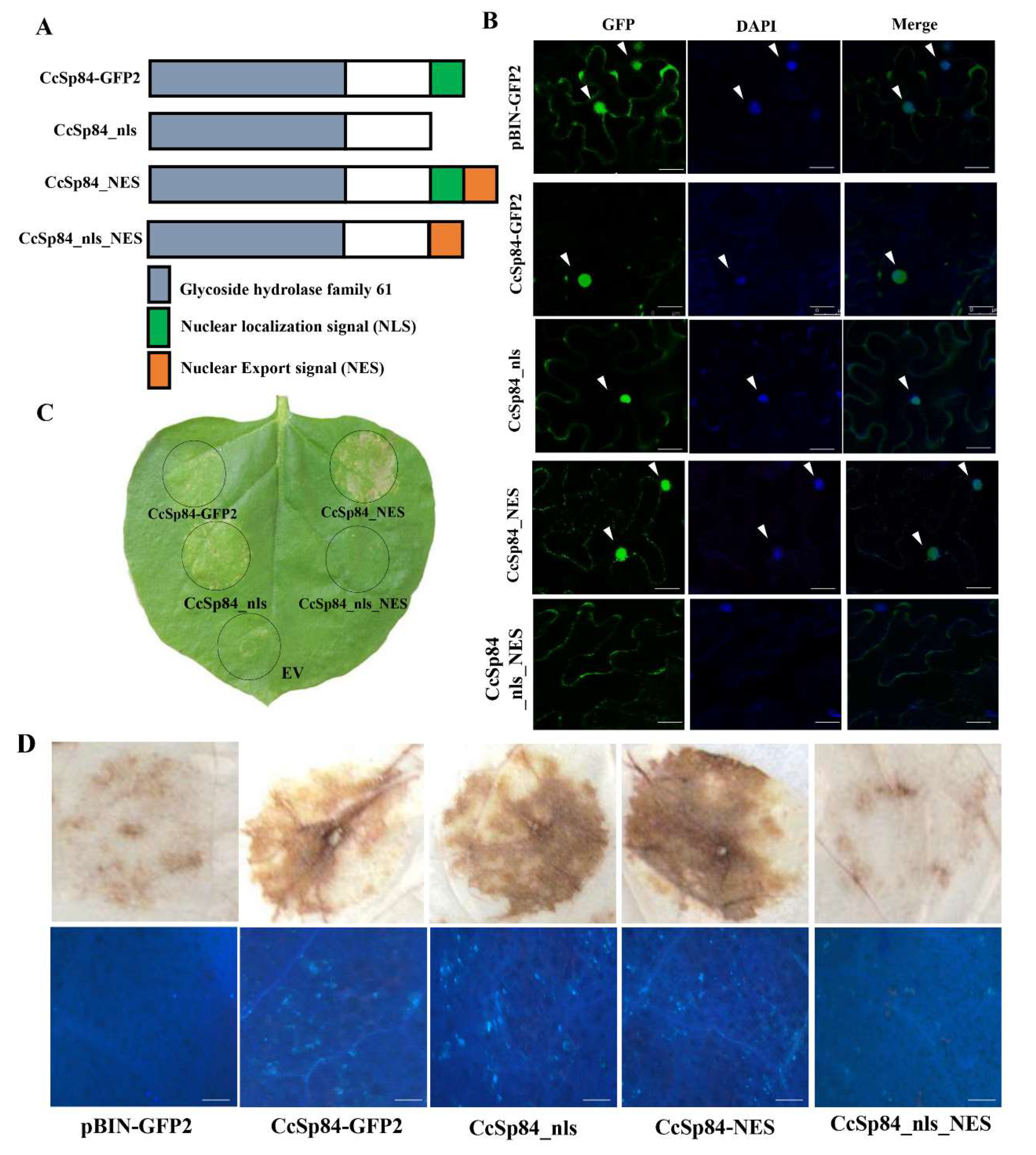
2.5. The Nuclear Localization of CcSp84 Was Required to Induce Plant Defense Responses
2.6. Expression of CcSp84 Could Induce the Expression of Defense-Related Genes in N. benthamiana
3. Discussion
4. Material and Methods
4.1. Fungal Strains and Plants Growth Conditions
4.2. Bioinformatics Analysis
4.3. Gene Knockout and Complementation
4.4. Plasmid Construction and Transient Expression
4.5. Confocal Fluorescent Analysis
4.6. Detection of Reactive Oxygen and Callosum
4.7. Pathogenicity Assay
4.8. RNA Extraction and qRT-PCR Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kepley, J.B.; Reeves, F.B.; Jacobi, W.R.; Adams, G.C. Species associated with cytospora canker on Populus tremuloides. Mycotaxon 2015, 130, 783–805. [Google Scholar] [CrossRef]
- Han, Z.; Xiong, D.; Xu, Z.; Liu, T.; Tian, C. The Cytospora chrysosperma Virulence Effector CcCAP1 Mainly Localizes to the Plant Nucleus to Suppress Plant Immune Responses. mSphere 2021, 6, 1. [Google Scholar] [CrossRef] [PubMed]
- Tao, D.; Li, P.H.; Carter, J.V.; Ostry, M.E. Relationship of Environmental Stress and Cytospora chrysosperma Infection to Spring Dieback of Poplar Shoots. For. Sci. 1984, 30, 645–651. [Google Scholar]
- Biggs, A.R.; Davis, D.D.; Merrill, W. Histopathology of cankers on Populus caused by Cytospora chrysosperma. Can. J. Bot. 1983, 61, 563–574. [Google Scholar] [CrossRef]
- Yu, L.; Xiong, D.; Han, Z.; Liang, Y.; Tian, C. The mitogen-activated protein kinase gene CcPmk1 is required for fungal growth, cell wall integrity and pathogenicity in Cytospora chrysosperma. Fungal Genet. Biol. 2019, 128, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Guyon, J.C.; Jacobi, W.R.; Mcintyre, G.A. Effects of environmental stress on the development of cytospora canker of aspen. Plant Dis. 1996, 80, 1320–1326. [Google Scholar] [CrossRef]
- Fan, X.-L.; Liang, Y.-M.; Ma, R.; Tian, C.-M. Morphological and phylogenetic studies of Cytospora (Valsaceae, Diaporthales) isolates from Chinese scholar tree, with description of a new species. Mycoscience 2014, 55, 252–259. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Y. Oxalic acid metabolism contributes to full virulence and pycnidial development in the poplar canker fungus Cytospora chrysosperma. Phytopathology 2020, 110, 1319–1325. [Google Scholar] [CrossRef]
- Xiong, D.; Yu, L.; Shan, H.; Tian, C. CcPmk1 is a regulator of pathogenicity in Cytospora chrysosperma and can be used as a potential target for disease control. Mol. Plant Pathol. 2021, 22, 710–726. [Google Scholar] [CrossRef]
- Han, Z.; Yu, R.; Xiong, D.; Tian, C. A Sge1 homolog in Cytospora chrysosperma governs conidiation, virulence and the expression of putative effectors. Gene 2021, 778, 145474. [Google Scholar] [CrossRef]
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dodds, P.N.; Rathjen, J.P. Plant immunity: Towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 2010, 11, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Chisholm, S.T.; Coaker, G.; Day, B.; Staskawicz, B.J. Host-Microbe Interactions: Shaping the Evolution of the Plant Immune Response. Cell 2006, 124, 803–814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kunze, G.; Zipfel, C.; Robatzek, S.; Niehaus, K.; Boller, T.; Felix, G. The N Terminus of Bacterial Elongation Factor Tu Elicits Innate Immunity in Arabidopsis Plants. Plant Cell 2004, 16, 3496–3507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fliegmann, J.; Mithofer, A.; Wanner, G.; Ebel, J. An ancient enzyme domain hidden in the putative beta-glucan elicitor receptor of soybean may play an active part in the perception of pathogen-associated molecular patterns during broad host resistance. J. Biol. Chem. 2004, 279, 1132–1140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.; Han, C.; Ferreira, A.O.; Yu, X.; Ye, W.; Tripathy, S.; Kale, S.D.; Gu, B.; Sheng, Y.; Sui, Y.; et al. Transcriptional Programming and Functional Interactions within the Phytophthora sojae RXLR Effector Repertoire. Plant Cell 2011, 23, 2064–2086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Białas, A.; Zess, E.K.; De la Concepcion, J.C.; Franceschetti, M.; Pennington, H.G.; Yoshida, K.; Upson, J.L.; Chanclud, E.; Wu, C.-H.; Langner, T.; et al. Lessons in Effector and NLR Biology of Plant-Microbe Systems. Mol. Plant-Microbe Interact. 2018, 31, 34–45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Hu, L.; Sun, L.; Lin, B.; Huang, K.; Zhuo, K.; Liao, J. A novel Meloidogyne graminicola effector, MgMO237, interacts with multiple host defence-related proteins to manipulate plant basal immunity and promote parasitism. Mol. Plant Pathol. 2018, 19, 1942–1955. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dou, D.; Zhou, J.-M. Phytopathogen Effectors Subverting Host Immunity: Different Foes, Similar Battleground. Cell Host Microbe 2012, 12, 484–495. [Google Scholar] [CrossRef] [Green Version]
- Yuan, M.; Jiang, Z.; Bi, G.; Nomura, K.; Liu, M.; Wang, Y.; Cai, B.; Zhou, J.-M.; He, S.Y.; Xin, X.-F. Pattern-recognition receptors are required for NLR-mediated plant immunity. Nature 2021, 592, 105–109. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.; Nguyen, Q.-M.; Iswanto, A.B.B.; Hong, J.C.; Bhattacharjee, S.; Gassmann, W.; Kim, S.H. Nuclear Localization of HopA1Pss61 Is Required for Effector-Triggered Immunity. Plants 2021, 10, 888. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Ni, H.; Du, X.; Wang, S.; Ma, X.; Nürnberger, T.; Guo, H.; Hua, C. The Verticillium-specific protein VdSCP7 localizes to the plant nucleus and modulates immunity to fungal infections. New Phytol. 2017, 215, 368–381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boevink, P.C.; Wang, X.; McLellan, H.; He, Q.; Naqvi, S.; Armstrong, M.R.; Zhang, W.; Hein, I.; Gilroy, E.M.; Tian, Z.; et al. A Phytophthora infestans RXLR effector targets plant PP1c isoforms that promote late blight disease. Nat. Commun. 2016, 7, 10311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, T.; Peng, J.; Yin, X.; Li, M.; Xiang, G.; Wang, Y.; Lei, Y.; Xu, Y. Importin-αs are required for the nuclear localization and function of the Plasmopara viticola effector PvAVH53. Hortic. Res. 2021, 8, 1–12. [Google Scholar] [CrossRef]
- Franceschetti, M.; Maqbool, A.; Jiménez-Dalmaroni, M.J.; Pennington, H.G.; Kamoun, S.; Banfield, M.J. Effectors of Filamentous Plant Pathogens: Commonalities amid Diversity. Microbiol. Mol. Biol. Rev. 2017, 81, e00066-16. [Google Scholar] [CrossRef] [Green Version]
- Sperschneider, J.; Dodds, P.N.; Gardiner, D.M.; Singh, K.B.; Taylor, J.M. Improved prediction of fungal effector proteins from secretomes with EffectorP 2.0. Mol. Plant Pathol. 2018, 19, 2094–2110. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Tian, L.; Zhang, D.; Song, J.; Song, S.; Yin, C.; Zhou, L.; Liu, Y.; Wang, B.; Kong, Z.; et al. Functional analyses of small secreted cysteine-rich proteins identified candidate effectors in Verticillium dahliae. Mol. Plant Pathol. 2020, 21, 667–685. [Google Scholar] [CrossRef] [Green Version]
- Chapman, S.; Kavanagh, T.; Baulcombe, D. Potato virus X as a vector for gene expression in plants. Plant J. 1992, 2, 549–557. [Google Scholar]
- Broekaert, W.F.; Delauré, S.L.; De Bolle, M.F.; Cammue, B.P. The Role of Ethylene in Host-Pathogen Interactions. Annu. Rev. Phytopathol. 2006, 44, 393–416. [Google Scholar] [CrossRef]
- Gupta, A.; Bhardwaj, M.; Tran, L.-S. Jasmonic Acid at the Crossroads of Plant Immunity and Pseudomonas syringae Virulence. Int. J. Mol. Sci. 2020, 21, 7482. [Google Scholar] [CrossRef]
- Situ, J.; Jiang, L.; Fan, X.; Yang, W.; Li, W.; Xi, P.; Deng, Y.; Kong, G.; Jiang, Z. An RXLR effector PlAvh142 from Peronophythora litchii triggers plant cell death and contributes to virulence. Mol. Plant Pathol. 2020, 21, 415–428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pieterse, C.M.J.; Van der Does, D.; Zamioudis, C.; Leon-Reyes, A.; Van Wees, S.C.M. Hormonal Modulation of Plant Immunity. Annu. Rev. Cell Dev. Biol. 2012, 28, 489–521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, C.J.; Ronald, P.C. Cleavage and nuclear localization of the rice XA21 immune receptor. Nat. Commun. 2012, 3, 920. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deslandes, L.; Rivas, S. The plant cell nucleus: A true arena for the fight between plants and pathogens. Plant Signal. Behav. 2011, 6, 42–48. [Google Scholar] [CrossRef] [Green Version]
- Jeon, H.; Kim, W.; Kim, B.; Lee, S.; Jayaraman, J.; Jung, G.; Choi, S.; Sohn, K.H.; Segonzac, C. Ralstonia solanacearum Type III Effectors with Predicted Nuclear Localization Signal Localize to Various Cell Compartments and Modulate Immune Responses in Nicotiana spp. Plant Pathol. J. 2020, 36, 43–53. [Google Scholar] [CrossRef]
- Cui, H.; Tsuda, K.; Parker, J.E. Effector-Triggered Immunity: From Pathogen Perception to Robust Defense. Annu. Rev. Plant Biol. 2015, 66, 487–511. [Google Scholar] [CrossRef]
- Gui, Y.-J.; Chen, J.-Y.; Zhang, D.-D.; Li, N.-Y.; Li, T.-G.; Zhang, W.-Q.; Wang, X.-Y.; Short, D.P.G.; Li, L.; Guo, W.; et al. Verticillium dahliae manipulates plant immunity by glycoside hydrolase 12 proteins in conjunction with carbohydrate-binding module 1. Environ. Microbiol. 2017, 19, 1914–1932. [Google Scholar] [CrossRef] [Green Version]
- Qi, T.; Guo, J.; Liu, P.; He, F.; Wan, C.; Islam, A.; Tyler, B.M.; Kang, Z.; Guo, J. Stripe Rust Effector PstGSRE1 Disrupts Nuclear Localization of ROS-Promoting Transcription Factor TaLOL2 to Defeat ROS-Induced Defense in Wheat. Mol. Plant 2019, 12, 1624–1638. [Google Scholar] [CrossRef]
- Ma, Z.; Song, T.; Zhu, L.; Ye, W.; Wang, Y.; Shao, Y.; Dong, S.; Zhang, Z.; Dou, D.; Zheng, X.; et al. A Phytophthora sojae Glycoside Hydrolase 12 Protein Is a Major Virulence Factor during Soybean Infection and Is Recognized as a PAMP. Plant Cell 2015, 27, 2057–2072. [Google Scholar] [CrossRef] [Green Version]
- Zhu, W.; Ronen, M.; Gur, Y.; Minz-Dub, A.; Masrati, G.; Ben-Tal, N.; Savidor, A.; Sharon, I.; Eizner, E.; Valerius, O.; et al. BcXYG1, a Secreted Xyloglucanase from Botrytis cinerea, Triggers Both Cell Death and Plant Immune Responses. Plant Physiol. 2017, 175, 438–456. [Google Scholar] [CrossRef] [Green Version]
- Wu, C.-H.; Derevnina, L.; Kamoun, S. Receptor networks underpin plant immunity. Science 2018, 360, 1300–1301. [Google Scholar] [CrossRef] [PubMed]
- Doehlemann, G.; Ökmen, B.; Zhu, W.; Sharon, A. Plant pathogenic fungi. Microbiol. Spectr. 2017, 5, 14. [Google Scholar] [CrossRef] [PubMed]
- Kanja, C.; Hammond-Kosack, K.E. Proteinaceous effector discovery and characterization in filamentous plant pathogens. Mol. Plant Pathol. 2020, 21, 1353–1376. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Tang, C.; Wang, X.; Sun, S.; Zhao, J.; Kang, Z.; Wang, X. An effector protein of the wheat stripe rust fungus targets chloroplasts and suppresses chloroplast function. Nat. Commun. 2019, 10, 5571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, H.; MacKenzie, C.I.; Rodriguez-Moreno, L.; Berg, G.C.M.V.D.; Chen, H.; Rudd, J.J.; Mesters, J.R.; Thomma, B.P.H.J. Three LysM effectors of Zymoseptoria tritici collectively disarm chitin-triggered plant immunity. Mol. Plant Pathol. 2021, 22, 683–693. [Google Scholar] [CrossRef] [PubMed]
- Yi, S.Y.; Shirasu, K.; Moon, J.S.; Lee, S.-G.; Kwon, S.-Y. The Activated SA and JA Signaling Pathways Have an Influence on flg22-Triggered Oxidative Burst and Callose Deposition. PLoS ONE 2014, 9, e88951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- An, C.; Mou, Z. Salicylic Acid and its Function in Plant Immunity. J. Integr. Plant Biol. 2011, 53, 412–428. [Google Scholar] [CrossRef]
- Betsuyaku, S.; Katou, S.; Takebayashi, Y.; Sakakibara, H.; Nomura, N.; Fukuda, H. Salicylic acid and jasmonic acid pathways are activated in spatially different domains around the infection site during effector-triggered immunity in Arabidopsis thaliana. Plant Cell Physiol. 2018, 59, 439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gimenez-Ibanez, S.; Boter, M.; Fernández-Barbero, G.; Chini, A.; Rathjen, J.; Solano, R. The Bacterial Effector HopX1 Targets JAZ Transcriptional Repressors to Activate Jasmonate Signaling and Promote Infection in Arabidopsis. PLoS Biol. 2014, 12, e1001792. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Z.; Wu, Y.; Yang, Y.; Du, M.; Zhang, X.; Guo, Y.; Li, C.; Zhou, J.-M. An Arabidopsis Plasma Membrane Proton ATPase Modulates JA Signaling and Is Exploited by the Pseudomonas syringae Effector Protein AvrB for Stomatal Invasion. Plant Cell 2015, 27, 2032–2041. [Google Scholar] [CrossRef] [Green Version]
- Thordal-Christensen, H.; Birch, P.R.J.; Spanu, P.D.; Panstruga, R. Why did filamentous plant pathogens evolve the potential to secrete hundreds of effectors to enable disease? Mol. Plant Pathol. 2018, 19, 781–785. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Catlett, N.L.; Lee, B.N.; Yoder, O.C.; Turgeon, B.G. Split-marker recombination for efficient targeted deletion of fungal genes. Fungal Genet. Rep. 2003, 50, 9–11. [Google Scholar] [CrossRef] [Green Version]
- Goswami, R.S. Targeted Gene Replacement in Fungi Using a Split-Marker Approach. Programmed Necrosis 2012, 835, 255–269. [Google Scholar]
- Li, W.; Cao, J.; Xu, Y.; Cai, X. Artificial Agrobacterium tumefaciens strains exhibit diverse mechanisms to repress Xanthomonas oryzae pv. oryzae-induced hypersensitive response and nonhost resistance in Nicotiana benthamiana. Mol. Plant Pathol. 2016, 18, 489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schenk, S.T.; Hernández-Reyes, C.; Samans, B.; Stein, E.; Neumann, C.; Schikora, M.; Reichelt, M.; Mithöfer, A.; Becker, A.; Kogel, K.-H.; et al. N-Acyl-Homoserine Lactone Primes Plants for Cell Wall Reinforcement and Induces Resistance to Bacterial Pathogens via the Salicylic Acid/Oxylipin Pathway. Plant Cell 2014, 26, 2708–2723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
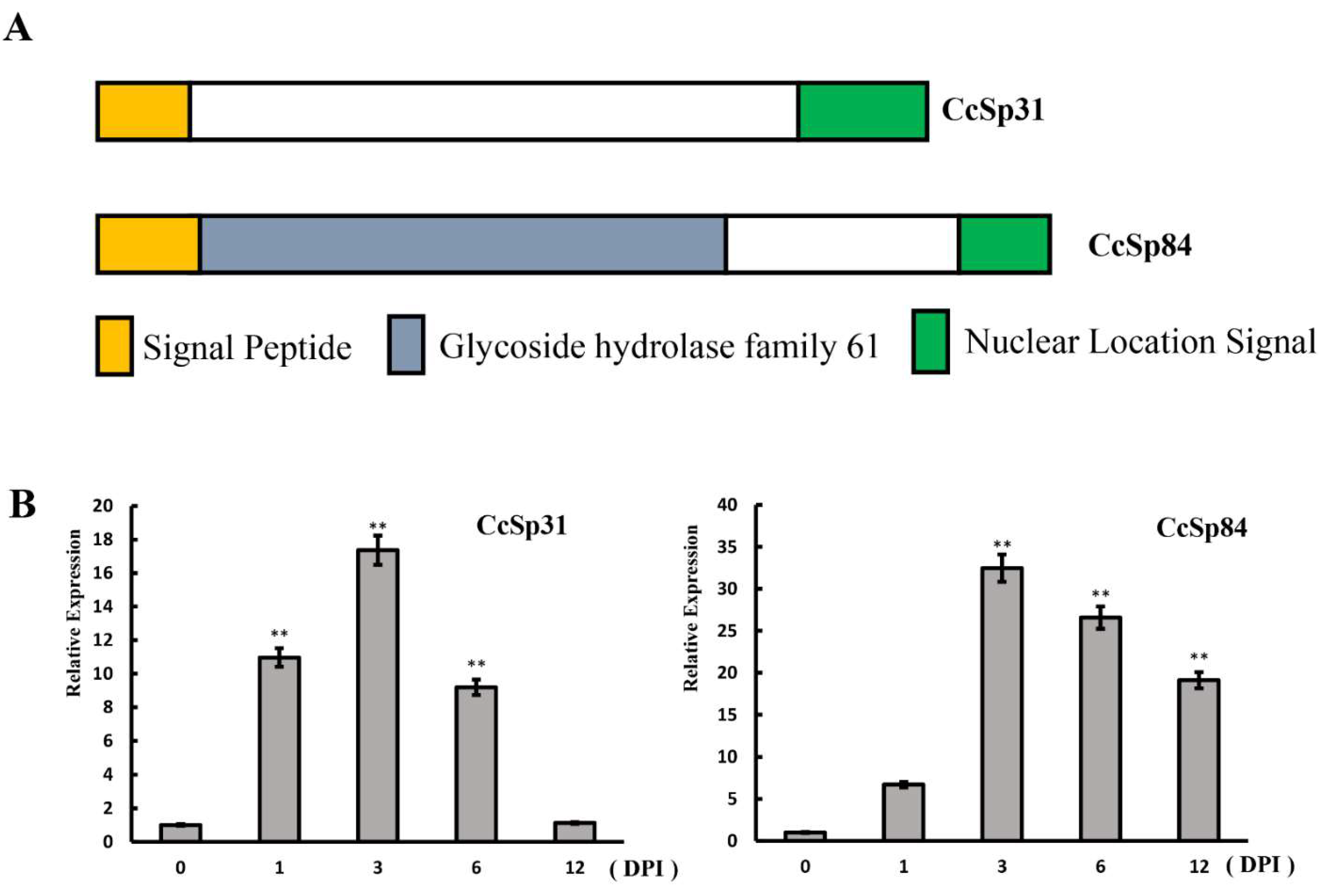


| Gene Name | CcSp31 | CcSp84 |
|---|---|---|
| Gene ID | GME4592_g | GME8128_g |
| Signal Peptide | Y | Y |
| Protein length (SP truncated) | 277 (260) | 315 (293) |
| Cysteines number | 5 | 3 |
| homologous gene | Conserved in fungi | Conserved in fungi |
| NLS sequence | KKMRKRHSDNGVRMPWKKVKR | PSKCKKRRHARD |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Z.; Xiong, D.; Han, Z.; Tian, C. A Putative Effector CcSp84 of Cytospora chrysosperma Localizes to the Plant Nucleus to Trigger Plant Immunity. Int. J. Mol. Sci. 2022, 23, 1614. https://doi.org/10.3390/ijms23031614
Xu Z, Xiong D, Han Z, Tian C. A Putative Effector CcSp84 of Cytospora chrysosperma Localizes to the Plant Nucleus to Trigger Plant Immunity. International Journal of Molecular Sciences. 2022; 23(3):1614. https://doi.org/10.3390/ijms23031614
Chicago/Turabian StyleXu, Zhiye, Dianguang Xiong, Zhu Han, and Chengming Tian. 2022. "A Putative Effector CcSp84 of Cytospora chrysosperma Localizes to the Plant Nucleus to Trigger Plant Immunity" International Journal of Molecular Sciences 23, no. 3: 1614. https://doi.org/10.3390/ijms23031614
APA StyleXu, Z., Xiong, D., Han, Z., & Tian, C. (2022). A Putative Effector CcSp84 of Cytospora chrysosperma Localizes to the Plant Nucleus to Trigger Plant Immunity. International Journal of Molecular Sciences, 23(3), 1614. https://doi.org/10.3390/ijms23031614






