Clinical Utility of a Unique Genome-Wide DNA Methylation Signature for KMT2A-Related Syndrome
Abstract
:1. Introduction
2. Results
2.1. Demographic and Molecular Characteristics of Patients
2.2. Detection and Verification of an Episignature for WDSTS
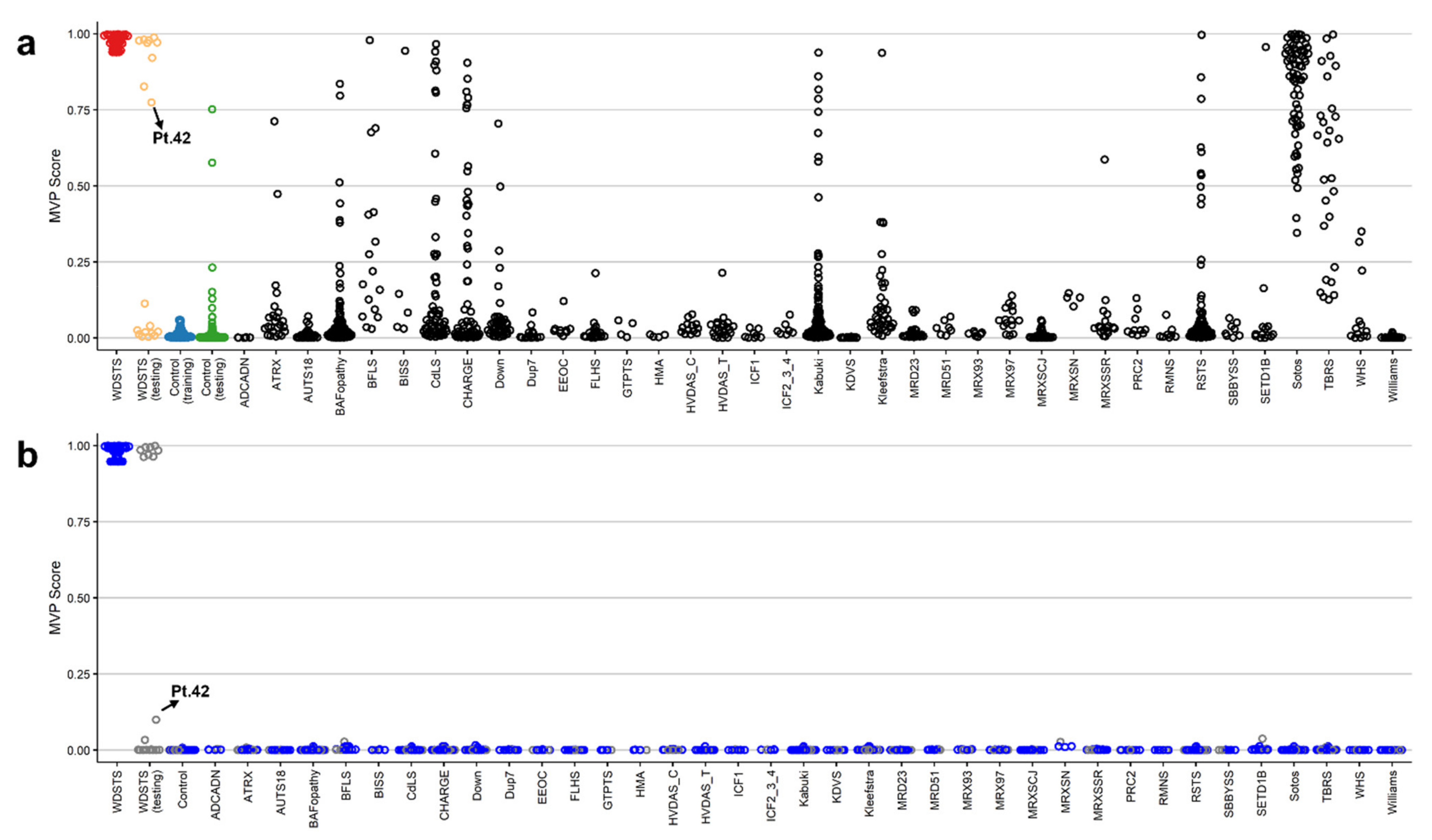
2.3. Construction of the Binary Prediction Model
2.4. Validation of WDSTS Signature Using Testing Cohort and Comparison to Kabuki1 Samples
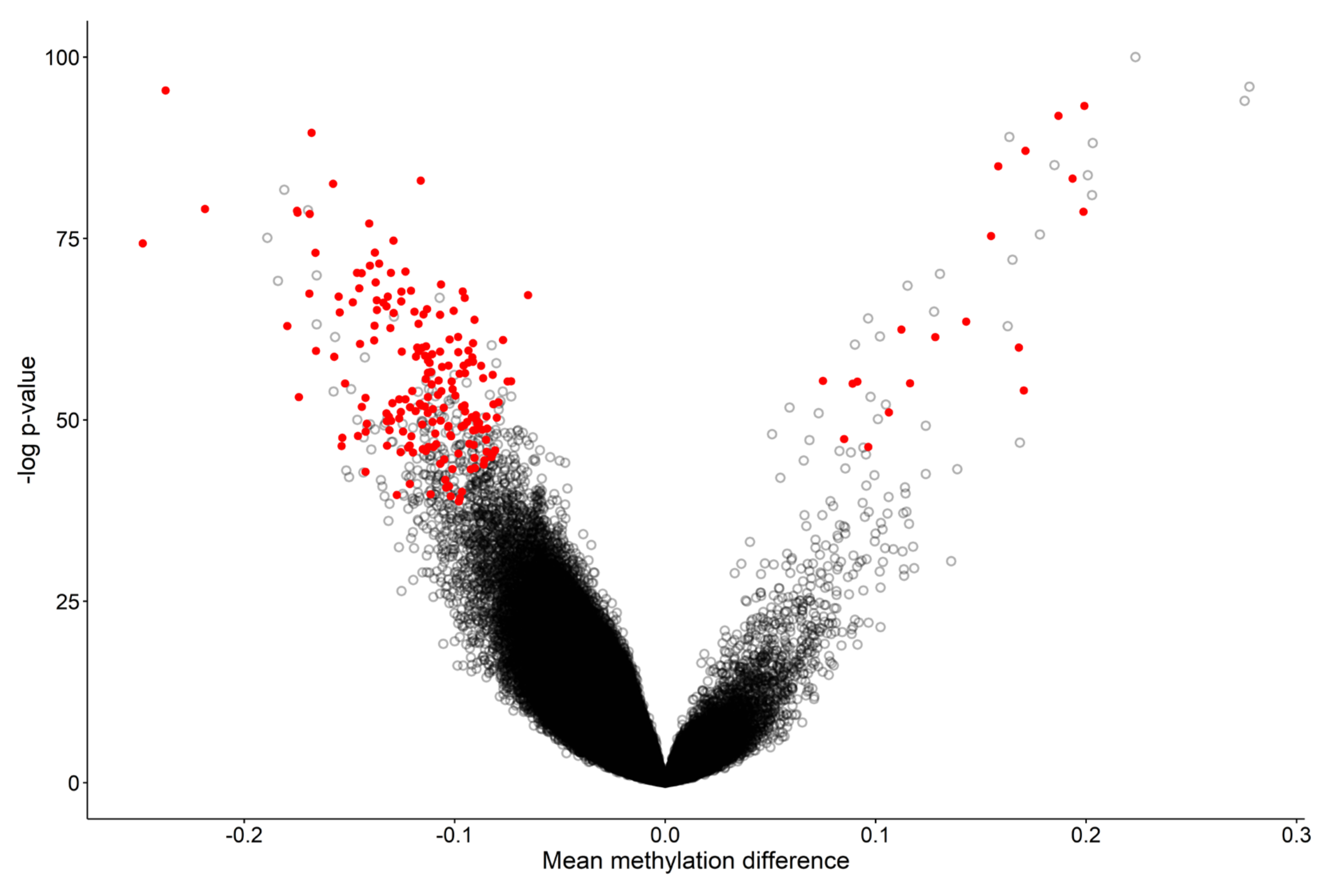
2.5. Identification of Differentially Methylated Regions
2.6. Definitive Classification of KMT2A Variants
3. Discussion
4. Materials and Methods
4.1. Study Cohort
4.2. Methylation Experiment and Selection of Matched Control Subjects
4.3. DNA Methylation Profiling of WDSTS Syndrome
4.4. Construction of a Classification Model for WDSTS Syndrome
4.5. Classification of Kabuki1 Samples with Pathogenic KMT2D Variants as well as WDSTS (Testing) Samples
4.6. Identification of the Differentially Methylated Regions of WDSTS Syndrome
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Aggarwal, A.; Rodriguez-Buritica, D.F.; Northrup, H. Wiedemann-Steiner syndrome: Novel pathogenic variant and review of literature. Eur. J. Med Genet. 2017, 60, 285–288. [Google Scholar] [CrossRef]
- Baer, S.; Afenjar, A.; Smol, T.; Piton, A.; Gérard, B.; Alembik, Y.; Bienvenu, T.; Boursier, G.; Boute, O.; Colson, C.; et al. Wiedemann-Steiner syndrome as a major cause of syndromic intellectual disability: A study of 33 French cases. Clin. Genet. 2018, 94, 141–152. [Google Scholar] [CrossRef] [PubMed]
- Lebrun, N.; Giurgea, I.; Goldenberg, A.; Dieux, A.; Afenjar, A.; Ghoumid, J.; Diebold, B.; Mietton, L.; Briand-Suleau, A.; Billuart, P.; et al. Molecular and cellular issues of KMT2A variants involved in Wiedemann-Steiner syndrome. Eur. J. Hum. Genet. 2017, 26, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Wang, Y.; Yang, Y.; Wang, P.; Huang, H.; Xiong, S.; Sun, L.; Cheng, M.; Song, C.; Cheng, X.; et al. Description of the molecular and phenotypic spectrum of Wiedemann-Steiner syndrome in Chinese patients. Orphanet J. Rare Dis. 2018, 13, 178. [Google Scholar] [CrossRef] [PubMed]
- Chan, A.J.S.; Cytrynbaum, C.; Hoang, N.; Ambrozewicz, P.M.; Weksberg, R.; Drmic, I.; Ritzema, A.; Schachar, R.; Walker, S.; Uddin, M.; et al. Expanding the neurodevelopmental phenotypes of individuals with de novo KMT2A variants. npj Genom. Med. 2019, 4, 9. [Google Scholar] [CrossRef] [PubMed]
- Giangiobbe, S.; Caraffi, S.G.; Ivanovski, I.; Maini, I.; Pollazzon, M.; Rosato, S.; Trimarchi, G.; Lauriello, A.; Marinelli, M.; Nicoli, D.; et al. Expanding the phenotype of Wiedemann-Steiner syndrome: Craniovertebral junction anomalies. Am. J. Med. Genet. Part A 2020, 182, 2877–2886. [Google Scholar] [CrossRef] [PubMed]
- Fontana, P.; Passaretti, F.F.; Maioli, M.; Cantalupo, G.; Scarano, F.; Lonardo, F. Clinical and molecular spectrum of Wiedemann-Steiner syndrome, an emerging member of the chromatinopathy family. World J. Med. Genet. 2020, 9, 1–11. [Google Scholar] [CrossRef]
- Sheppard, S.E.; Campbell, I.M.; Harr, M.H.; Gold, N.; Li, D.; Bjornsson, H.T.; Cohen, J.S.; Fahrner, J.A.; Fatemi, A.; Harris, J.R.; et al. Expanding the genotypic and phenotypic spectrum in a diverse cohort of 104 individuals with Wiedemann-Steiner syndrome. Am. J. Med Genet. Part A 2021, 185, 1649–1665. [Google Scholar] [CrossRef]
- Squeo, G.M.; Augello, B.; Massa, V.; Milani, D.; Colombo, E.A.; Mazza, T.; Castellana, S.; Piccione, M.; Maitz, S.; Petracca, A.; et al. Customised next-generation sequencing multigene panel to screen a large cohort of individuals with chromatin-related disorder. J. Med. Genet. 2020, 57, 760–768. [Google Scholar] [CrossRef]
- Köhler, S.; Gargano, M.; Matentzoglu, N.; Carmody, L.C.; Lewis-Smith, D.; Vasilevsky, N.A.; Danis, D.; Balagura, G.; Baynam, G.; Brower, A.M.; et al. The Human Phenotype Ontology in 2021. Nucleic Acids Res. 2020, 49, D1207–D1217. [Google Scholar] [CrossRef]
- Douzgou, S.; Clayton-Smith, J.; Gardner, S.; Day, R.; Griffiths, P.; Strong, K.; the DYSCERNE Expert Panel. Dysmorphology at a distance: Results of a web-based diagnostic service. Eur. J. Hum. Genet. 2014, 22, 327–332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, D.T.; Adam, M.P.; Aradhya, S.; Biesecker, L.G.; Brothman, A.R.; Carter, N.P.; Church, D.M.; Crolla, J.A.; Eichler, E.E.; Epstein, C.J.; et al. Consensus Statement: Chromosomal Microarray Is a First-Tier Clinical Diagnostic Test for Individuals with Developmental Disabilities or Congenital Anomalies. Am. J. Hum. Genet. 2010, 86, 749–764. [Google Scholar] [CrossRef] [PubMed]
- El Chaer, F.; Keng, M.; Ballen, K.K. MLL-Rearranged Acute Lymphoblastic Leukemia. Curr. Hematol. Malign. Rep. 2020, 15, 83–89. [Google Scholar] [CrossRef] [PubMed]
- Dou, Y.; Milne, T.A.; Tackett, A.J.; Smith, E.R.; Fukuda, A.; Wysocka, J.; Allis, C.D.; Chait, B.T.; Hess, J.L.; Roeder, R.G. Physical Association and Coordinate Function of the H3 K4 Methyltransferase MLL1 and the H4 K16 Acetyltransferase MOF. Cell 2005, 121, 873–885. [Google Scholar] [CrossRef] [Green Version]
- Milne, T.A.; Briggs, S.D.; Brock, H.W.; Martin, M.E.; Gibbs, D.; Allis, C.D.; Hess, J.L. MLL Targets SET Domain Methyltransferase Activity to Hox Gene Promoters. Mol. Cell 2002, 10, 1107–1117. [Google Scholar] [CrossRef]
- Vallianatos, C.N.; Raines, B.; Porter, R.S.; Bonefas, K.M.; Wu, M.C.; Garay, P.M.; Collette, K.M.; Seo, Y.A.; Dou, Y.; Keegan, C.E.; et al. Mutually suppressive roles of KMT2A and KDM5C in behaviour, neuronal structure, and histone H3K4 methylation. Commun. Biol. 2020, 3, 278. [Google Scholar] [CrossRef]
- Pang, D.; Thompson, D.N.P. Embryology and bony malformations of the craniovertebral junction. Child’s Nerv. Syst. 2011, 27, 523–564. [Google Scholar] [CrossRef] [Green Version]
- Gonçalves, C.S.; Le Boiteux, E.; Arnaud, P.; Costa, B.M. HOX gene cluster (de)regulation in brain: From neurodevelopment to malignant glial tumours. Cell Mol. Life Sci. 2020, 77, 3797–3821. [Google Scholar] [CrossRef]
- Hyun, K.; Jeon, J.; Park, K.; Kim, J. Writing, erasing and reading histone lysine methylations. Exp. Mol. Med. 2017, 49, e324. [Google Scholar] [CrossRef] [Green Version]
- Cosgrove, M.S.; Patel, A. Mixed lineage leukemia: A structure-function perspective of the MLL1 protein. FEBS J. 2010, 277, 1832–1842. [Google Scholar] [CrossRef] [Green Version]
- Zhang, P.; Bergamin, E.; Couture, J.-F. The many facets of MLL1 regulation. Biopolymers 2013, 99, 136–145. [Google Scholar] [CrossRef]
- Aref-Eshghi, E.; Kerkhof, J.; Pedro, V.P.; Barat-Houari, M.; Ruiz-Pallares, N.; Andrau, J.-C.; Lacombe, D.; Van-Gils, J.; Fergelot, P.; Dubourg, C.; et al. Evaluation of DNA Methylation Episignatures for Diagnosis and Phenotype Correlations in 42 Mendelian Neurodevelopmental Disorders. Am. J. Hum. Genet. 2020, 106, 356–370. [Google Scholar] [CrossRef] [PubMed]
- Aref-Eshghi, E.; Bend, E.G.; Colaiacovo, S.; Caudle, M.; Chakrabarti, R.; Napier, M.; Brick, L.; Brady, L.; Carere, D.A.; Levy, M.A.; et al. Diagnostic Utility of Genome-wide DNA Methylation Testing in Genetically Unsolved Individuals with Suspected Hereditary Conditions. Am. J. Hum. Genet. 2019, 104, 685–700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ciolfi, A.; Aref-Eshghi, E.; Pizzi, S.; Pedace, L.; Miele, E.; Kerkhof, J.; Flex, E.; Martinelli, S.; Radio, F.C.; Ruivenkamp, C.A.L.; et al. Frameshift mutations at the C-terminus of HIST1H1E result in a specific DNA hypomethylation signature. Clin. Epigenet. 2020, 12, 7. [Google Scholar] [CrossRef]
- Ciolfi, A.; Foroutan, A.; Capuano, A.; Pedace, L.; Travaglini, L.; Pizzi, S.; Andreani, M.; Miele, E.; Invernizzi, F.; Reale, C.; et al. Childhood-onset dystonia-causing KMT2B variants result in a distinctive genomic hypermethylation profile. Clin. Epigenet. 2021, 13, 157. [Google Scholar] [CrossRef] [PubMed]
- Sadikovic, B.; Levy, M.A.; Kerkhof, J.; Aref-Eshghi, E.; Schenkel, L.; Stuart, A.; McConkey, H.; Henneman, P.; Venema, A.; Schwartz, C.E.; et al. Clinical epigenomics: Genome-wide DNA methylation analysis for the diagnosis of Mendelian disorders. Genet. Med. 2021, 23, 1065–1074. [Google Scholar] [CrossRef] [PubMed]
- Sobreira, N.; Brucato, M.; Zhang, L.; Ladd-Acosta, C.; Ongaco, C.; Romm, J.; Doheny, K.F.; Netto, R.C.M.; Bertola, D.; Kim, A.C.; et al. Patients with a Kabuki syndrome phenotype demonstrate DNA methylation abnormalities. Eur. J. Hum. Genet. 2017, 25, 1335–1344. [Google Scholar] [CrossRef]
- Barbosa, M.; Joshi, R.S.; Garg, P.; Martin-Trujillo, A.; Patel, N.; Jadhav, B.; Watson, C.T.; Gibson, W.; Chetnik, K.; Tessereau, C.; et al. Identification of rare de novo epigenetic variations in congenital disorders. Nat. Commun. 2018, 9, 2064. [Google Scholar] [CrossRef] [Green Version]
- Dennis, G.; Sherman, B.T.; Hosack, D.A.; Yang, J.; Gao, W.; Lane, H.C.; Lempicki, R.A. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003, 4, P3. [Google Scholar] [CrossRef]
- Baux, D.; Van Goethem, C.; Ardouin, O.; Guignard, T.; Bergougnoux, A.; Koenig, M.; Roux, A.-F. MobiDetails: Online DNA variants interpretation. Eur. J. Hum. Genet. 2020, 29, 356–360. [Google Scholar] [CrossRef]
- Baux, D. Mobidetails_KMT2A. Available online: https://mobidetails.iurc.montp.inserm.fr/MD/auth/variant_list/EpiSignature_KMT2A (accessed on 14 December 2021).
- Courraud, J.; Chater-Diehl, E.; Durand, B.; Vincent, M.; Moreno, M.D.M.M.; Boujelbene, I.; Drouot, N.; Genschik, L.; Schaefer, E.; Nizon, M.; et al. Integrative approach to interpret DYRK1A variants, leading to a frequent neurodevelopmental disorder. Genet. Med. 2021, 23, 2150–2159. [Google Scholar] [CrossRef] [PubMed]
- Jones, W.D.; Dafou, D.; McEntagart, M.; Woollard, W.J.; Elmslie, F.V.; Holder-Espinasse, M.; Irving, M.; Saggar, A.K.; Smithson, S.; Trembath, R.; et al. De Novo Mutations in MLL Cause Wiedemann-Steiner Syndrome. Am. J. Hum. Genet. 2012, 91, 358–364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takeda, S.; Chen, D.Y.; Westergard, T.D.; Fisher, J.K.; Rubens, J.A.; Sasagawa, S.; Kan, J.T.; Korsmeyer, S.J.; Cheng, E.H.-Y.; Hsieh, J.J.-D. Proteolysis of MLL family proteins is essential for Taspase1-orchestrated cell cycle progression. Genes Dev. 2006, 20, 2397–2409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Collins, B.; Greer, C.B.; Coleman, B.C.; Sweatt, J.D. Histone H3 lysine K4 methylation and its role in learning and memory. Epigenetics Chromatin 2019, 12, 7. [Google Scholar] [CrossRef]
- Hsieh, J.J.-D.; Ernst, P.; Erdjument-Bromage, H.; Tempst, P.; Korsmeyer, S.J. Proteolytic cleavage of MLL generates a complex of N- and C-terminal fragments that confers protein stability and subnuclear localization. Mol. Cell. Biol. 2003, 23, 186–194. [Google Scholar] [CrossRef] [Green Version]
- Terranova, R.; Agherbi, H.; Boned, A.; Meresse, S.; Djabali, M. Histone and DNA methylation defects at Hox genes in mice expressing a SET domain-truncated form of Mll. Proc. Natl. Acad. Sci. USA 2006, 103, 6629–6634. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Song, Y. Structure, function and inhibition of critical protein–protein interactions involving mixed lineage leukemia 1 and its fusion oncoproteins. J. Hematol. Oncol. 2021, 14, 56. [Google Scholar] [CrossRef]
- Philippidou, P.; Dasen, J.S. Hox genes: Choreographers in neural development, architects of circuit organization. Neuron 2013, 80, 12–34. [Google Scholar] [CrossRef] [Green Version]
- Liu, G.; Liu, Z.; Sun, X.; Xia, X.; Liu, Y.; Liu, L. Pan-cancer genome-wide DNA methylation analyses revealed that hypermethylation influences 3D architecture and gene expression dysregulation in HOXA locus during carcinogenesis of cancers. Front. Cell Dev. Biol. 2021, 9, 649168. [Google Scholar] [CrossRef]
- Aryee, M.J.; Jaffe, A.E.; Corrada-Bravo, H.; Ladd-Acosta, C.; Feinberg, A.P.; Hansen, K.D.; Irizarry, R.A. Minfi: A flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 2014, 30, 1363–1369. [Google Scholar] [CrossRef] [Green Version]
- Pidsley, R.; Wong, C.C.Y.; Volta, M.; Lunnon, K.; Mill, J.; Schalkwyk, L.C. A data-driven approach to preprocessing Illumina 450K methylation array data. BMC Genom. 2013, 14, 293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ho, D.E.; Imai, K.; King, G.; Stuart, E.A. Matching as nonparametric preprocessing for reducing model dependence in parametric causal inference. Political Anal. 2007, 15, 199–236. [Google Scholar] [CrossRef] [Green Version]
- Stuart, E.A.; King, G.; Imai, K.; Ho, D. MatchIt: Nonparametric preprocessing for parametric causal inference. J. Stat. Softw. 2011, 42, 1–28. [Google Scholar]
- Sadikovic, B.; Levy, M.A.; Aref-Eshghi, E. Functional annotation of genomic variation: DNA methylation episignatures in neurodevelopmental Mendelian disorders. Hum. Mol. Genet. 2020, 29, R27–R32. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Aref-Eshghi, E.; Rodenhiser, D.I.; Schenkel, L.C.; Lin, H.; Skinner, C.; Ainsworth, P.; Paré, G.; Hood, R.L.; Bulman, D.E.; Kernohan, K.D.; et al. Genomic DNA Methylation Signatures Enable Concurrent Diagnosis and Clinical Genetic Variant Classification in Neurodevelopmental Syndromes. Am. J. Hum. Genet. 2018, 102, 156–174. [Google Scholar] [CrossRef] [Green Version]
- Peters, T.J.; Buckley, M.J.; Statham, A.L.; Pidsley, R.; Samaras, K.; Lord, R.V.; Clark, S.J.; Molloy, P.L. De novo identification of differentially methylated regions in the human genome. Epigenet. Chromatin 2015, 8, 6. [Google Scholar] [CrossRef] [Green Version]


| ID | Sex | Age | Genetic Change in the KMT2A Gene (NM_001197104.2 §) | Cohort_Array Type |
|---|---|---|---|---|
| Pt.1 | m | 2 | c.5312G > A, p.(Trp1771 *) | WDSTS_EPIC |
| Pt.2 | m | 13 | c.2647G > T, p.(Glu883 *) | WDSTS_EPIC |
| Pt.3 | f | 29 | c.3635-1G > A, p.? | WDSTS_EPIC |
| Pt.4 | m | 5 | c.9068del, p.(Gln3023Argfs *3) | WDSTS_EPIC |
| Pt.5 | m | 9 | c.5572C > T, p.(Arg1858 *) | WDSTS_EPIC |
| Pt.6 | f | 4.5 | c.9001del, p.(His3001Thrfs * 15) | WDSTS_EPIC |
| Pt.7 | m | 3 | c.3464G > A, p.(Cys1155Tyr) | WDSTS_EPIC |
| Pt.8 | m | 6 | c.3740_3741del, p.(Ser1247Cysfs * 12) | WDSTS_EPIC |
| Pt.9 | f | 4 | c.3790C > T, p.(Arg1264 *) | WDSTS_EPIC |
| Pt.10 | m | 10 | c.5251A > T, p.(Lys1751 *) | WDSTS_EPIC |
| Pt.11 | m | 12 | c.3634 + 1G > A, p.? | WDSTS_EPIC |
| Pt.12 | f | 7 | c.10837C > T, p.(Gln3613 *) | WDSTS_EPIC |
| Pt.13 | m | 12 | c.3895_3896del, p.(Ser1299Profs * 26) | WDSTS_EPIC |
| Pt.14 | m | 13 | c.478C > T, p.(Arg160 *) | WDSTS_EPIC |
| Pt.15 | m * | 21.8 # | c.6735dup, p.(Val2246Serfs *2) | WDSTS_EPIC |
| Pt.16 | m * | 4 # | c.2318_2319del, p.(Pro773Leufs * 12) | WDSTS_EPIC |
| Pt.17 | f * | 3.9 # | c.3460C > T, p.(Arg1154Trp) | WDSTS_EPIC |
| Pt.18 | f * | 23.7 # | c.8532_8533del, p.(Cys2844Trpfs * 24) | WDSTS_EPIC |
| Pt.19 | m * | 14.3 # | c.11001dup, p.(Pro3668Thrfs * 8) | WDSTS_EPIC |
| Pt.20 | f * | 26.5 # | c.2605G > T, p.(Glu869 *) | WDSTS_EPIC |
| Pt.21 | m * | 15.2 # | c.10498C > T, p.(Gln3500 *) | WDSTS_EPIC |
| Pt.22 | m * | 17.2 # | c.7630G > T, p.(Glu2544 *) | WDSTS_EPIC |
| Pt.23 | m * | 6.1 # | c.10900 + 1G > A, p.? | WDSTS_EPIC |
| Pt.24 | m * | 1.1 # | c.4256G > A, p.(Gly1419Asp) | WDSTS_EPIC |
| Pt.25 | m * | 9.1 # | c.1539del, p.(Ile515Phefs * 52) | WDSTS_EPIC |
| Pt.26 | m | 17.6 # | c.3460C > T, p.(Arg1154Trp) | WDSTS_EPIC |
| Pt.27 | m | 25.7 # | c.2318dup, p.(Ser774Valfs * 12) | WDSTS_EPIC |
| Pt.28 | m | 67 | c.5431C > T, p.(Arg1811 *) | WDSTS_EPIC |
| Pt.29 | f | 10 | c.1128dup, p.(Gln377Thrfs * 12) | WDSTS_EPIC |
| Pt.30 | f | 1.9 # | c.7975C > T, p.(Arg2659 *) | WDSTS_EPIC |
| Pt.31 | f | 34.9 # | c.9538_9539del, p.(Ile3180Glnfs * 55) | WDSTS_EPIC |
| Pt.32 | m | 15.7 # | c.7438C > T, p.(Arg2480 *) | WDSTS_EPIC |
| Pt.33 | m | 10 # | c.3301C > T, p.(Arg1101 *) | WDSTS_EPIC |
| Pt.34 | m | 14 | c.4727dup, p.(Tyr1576 *) | WDSTS_EPIC |
| Pt.35 | m | 19 | c.3629_3634 + 1del, p.(Lys1211_Ala1212del) | WDSTS_EPIC |
| Pt.36 | m | 27 | c.1821_1825del, p.(Arg608Ilefs * 9) | WDSTS_EPIC |
| Pt.37 | m | 22 | c.3451C > T, p.(Arg1151 *) | WDSTS_EPIC |
| Pt.38 | m | 7 | c.7150C > T, p.(Gln2384 *) | WDSTS_EPIC |
| Pt.39 | f | 2 | c.7324G > T, p.(Glu2442 *) | WDSTS_EPIC |
| Pt.40 | f | 3 | c.10736del, p.(Leu3580 *) | WDSTS_EPIC |
| Pt.41 | m | 17 | c.4018G > T, p.(Glu1340 *) | WDSTS_EPIC |
| Pt.42 ¥,₢ | m | 19.5 # | Not available | WDSTS (testing)_ 450k |
| Pt.43 ¥ | f | 9.1 # | c.5803-1G > A, p.?+ | WDSTS (testing)_ 450k |
| Pt.44 ₢ | m | 13 | Not available | WDSTS (testing)_ 450k |
| Pt.45 ₢ | f | 2 | Not available | WDSTS (testing)_ 450k |
| Pt.46 | m | 21 | c.5806T > C, p.(Cys1936Arg) | WDSTS (testing)_EPIC |
| Pt.47 | f | 2 | c.4426T > G, p.(Cys1476Gly) | WDSTS (testing)_EPIC |
| Pt.48 | m | 4 | c.4432_4434del, p.(Arg1478del) | WDSTS (testing)_EPIC |
| Pt.49 | f | 2 | c.4171C > T, p.(Gln1391 *) | WDSTS (testing)_EPIC |
| Pt.50 €,₢ | m | 6.5 # | Not available | WDSTS (testing)_ 450k |
| Pt.51 ¥ | m | 8.5 # | c.3019G > T, p.(Gly1007Cys)+ | WDSTS (testing)_ 450k |
| Pt.52 | m | 3 | c.9575A > C, p.(Gln3192Pro) | WDSTS (testing)_EPIC |
| Pt.53 | M * | 9.4 # | c.29C > T, p.(Pro10Leu) | WDSTS (testing)_EPIC |
| Pt.54 | f | 7 | c.8545C > G, p.(Pro2849Ala) | WDSTS (testing)_EPIC |
| Pt.55 | m | 2 | c.352G > T, p.(Val118Phe) | WDSTS (testing)_EPIC |
| Pt.56 | f | 3 | c.11347_11376del, p.(Phe3783_Pro3792del) | WDSTS (testing)_EPIC |
| Pt.57 | m | 10 | c.8387G > T, p.(Gly2796Val) | WDSTS (testing)_EPIC |
| Pt.58 | m | 2 | c.100C > G, p.(Arg34Gly) | WDSTS (testing)_EPIC |
| Pt.59 | f | 25 | c.10315_10316delinsAC, p.(Gly3439Thr) | WDSTS (testing)_EPIC |
| Pt.60 | m | 2 | c.3379C > T, p.(Pro1127Ser) | WDSTS (testing)_EPIC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Foroutan, A.; Haghshenas, S.; Bhai, P.; Levy, M.A.; Kerkhof, J.; McConkey, H.; Niceta, M.; Ciolfi, A.; Pedace, L.; Miele, E.; et al. Clinical Utility of a Unique Genome-Wide DNA Methylation Signature for KMT2A-Related Syndrome. Int. J. Mol. Sci. 2022, 23, 1815. https://doi.org/10.3390/ijms23031815
Foroutan A, Haghshenas S, Bhai P, Levy MA, Kerkhof J, McConkey H, Niceta M, Ciolfi A, Pedace L, Miele E, et al. Clinical Utility of a Unique Genome-Wide DNA Methylation Signature for KMT2A-Related Syndrome. International Journal of Molecular Sciences. 2022; 23(3):1815. https://doi.org/10.3390/ijms23031815
Chicago/Turabian StyleForoutan, Aidin, Sadegheh Haghshenas, Pratibha Bhai, Michael A. Levy, Jennifer Kerkhof, Haley McConkey, Marcello Niceta, Andrea Ciolfi, Lucia Pedace, Evelina Miele, and et al. 2022. "Clinical Utility of a Unique Genome-Wide DNA Methylation Signature for KMT2A-Related Syndrome" International Journal of Molecular Sciences 23, no. 3: 1815. https://doi.org/10.3390/ijms23031815
APA StyleForoutan, A., Haghshenas, S., Bhai, P., Levy, M. A., Kerkhof, J., McConkey, H., Niceta, M., Ciolfi, A., Pedace, L., Miele, E., Genevieve, D., Heide, S., Alders, M., Zampino, G., Merla, G., Fradin, M., Bieth, E., Bonneau, D., Dieterich, K., ... Lebre, A.-S. (2022). Clinical Utility of a Unique Genome-Wide DNA Methylation Signature for KMT2A-Related Syndrome. International Journal of Molecular Sciences, 23(3), 1815. https://doi.org/10.3390/ijms23031815









