Complex Analysis of Vanillin and Syringic Acid as Natural Antimicrobial Agents against Staphylococcus epidermidis Biofilms
Abstract
:1. Introduction
2. Results
2.1. Selection of Phenolic Compounds
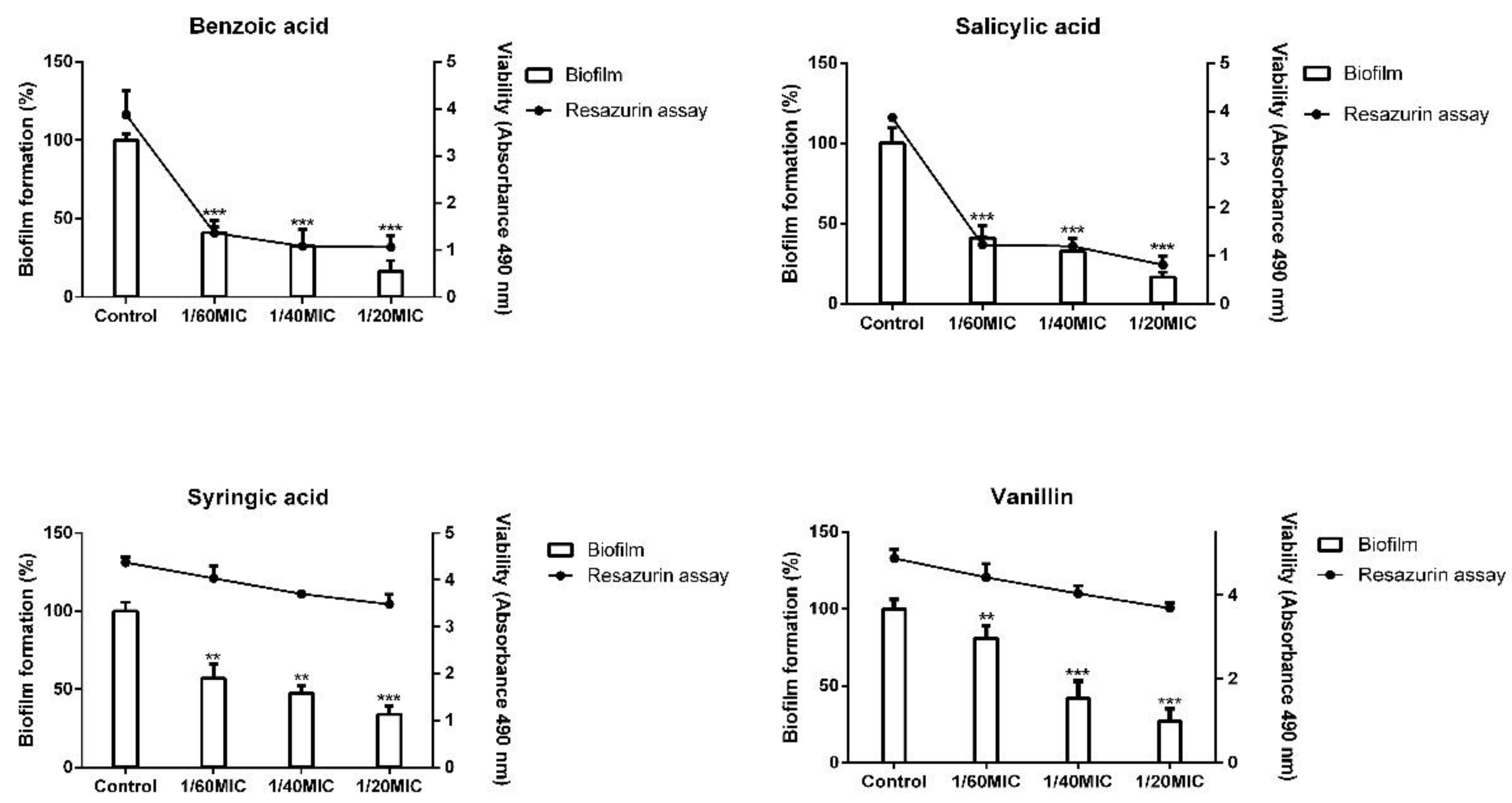
2.2. Biofilm Formation Inhibition Using Phenolic Compounds
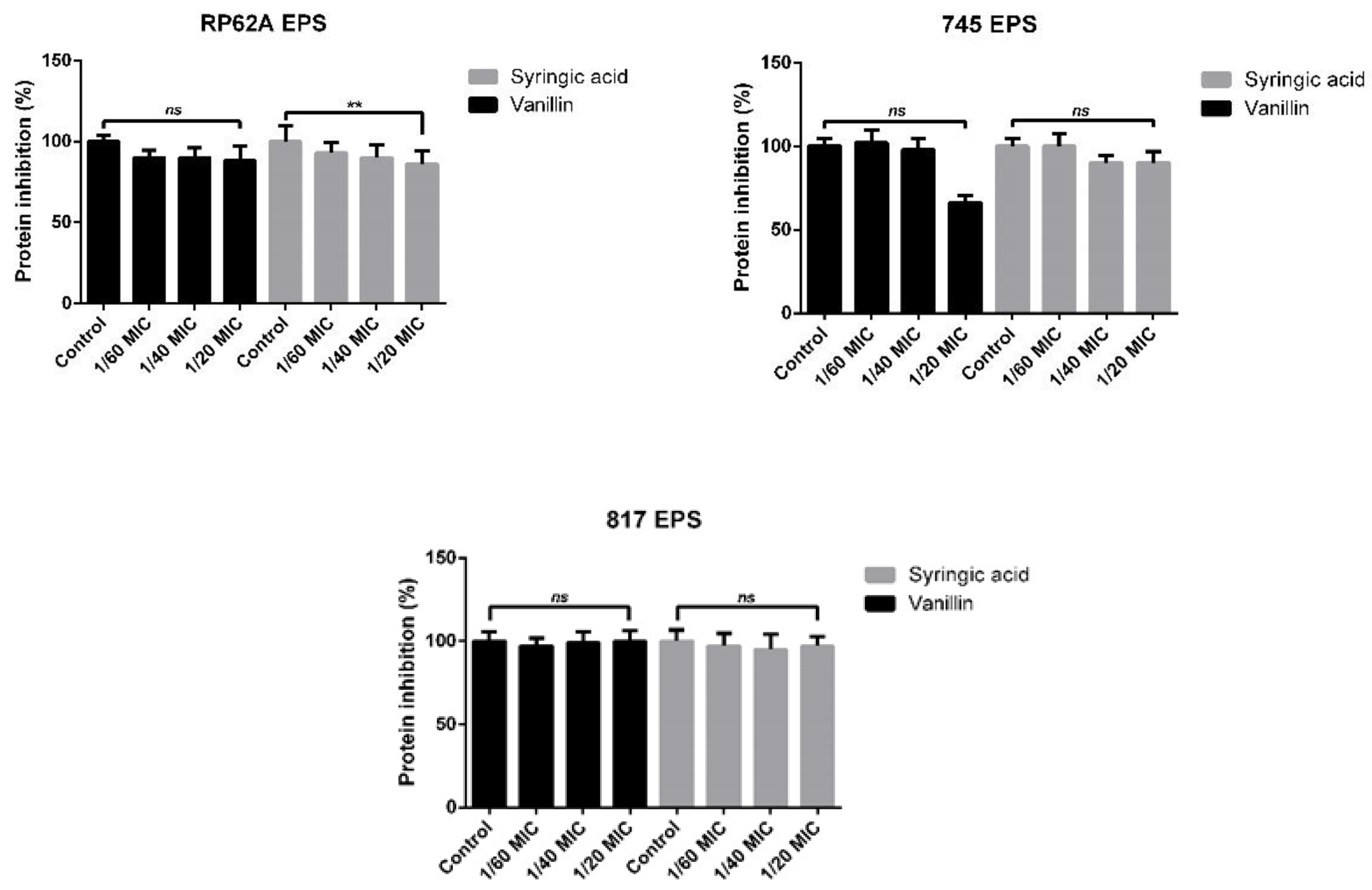
2.3. EPS Formation Inhibition Using Phenolic Compounds
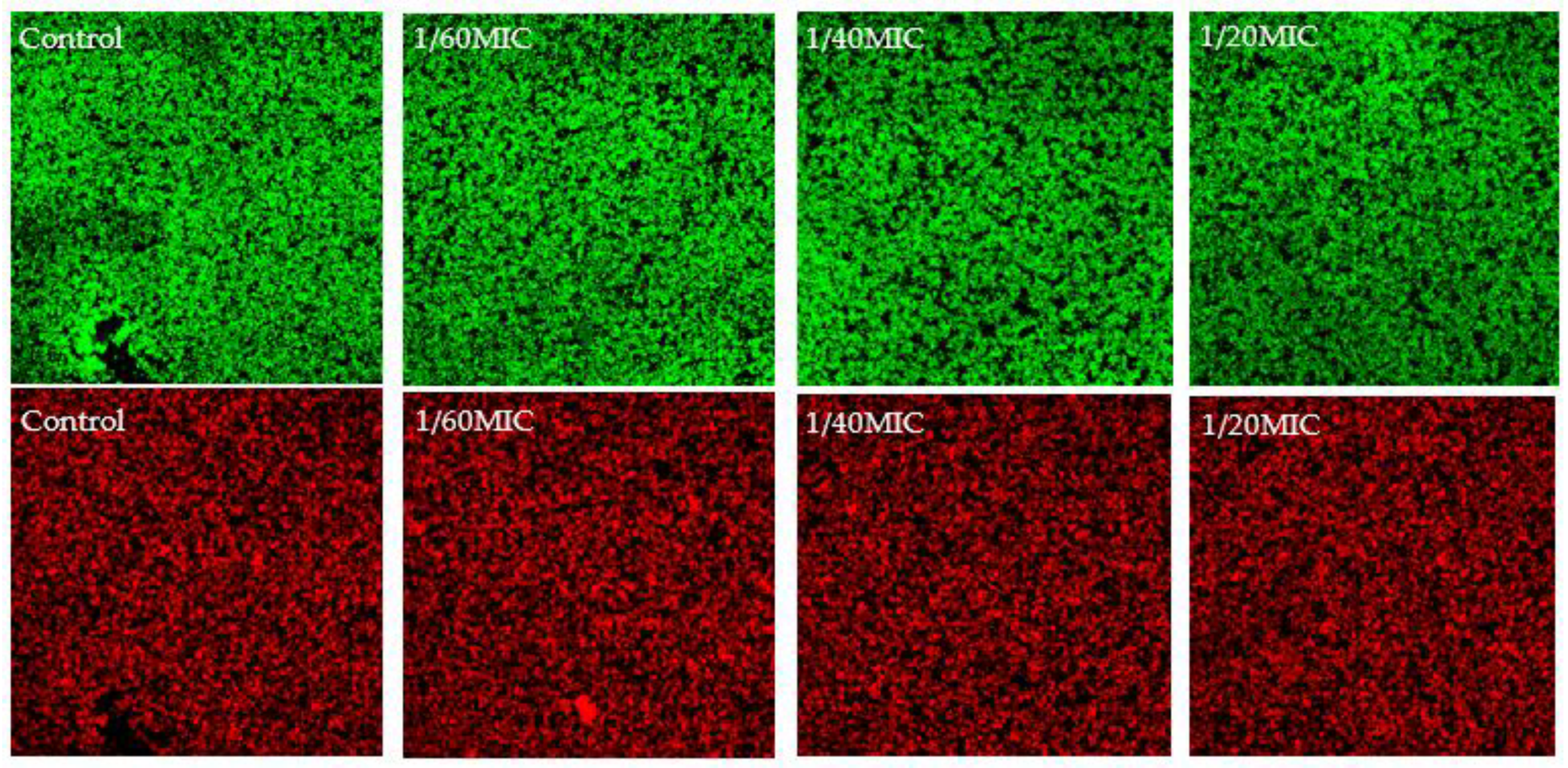
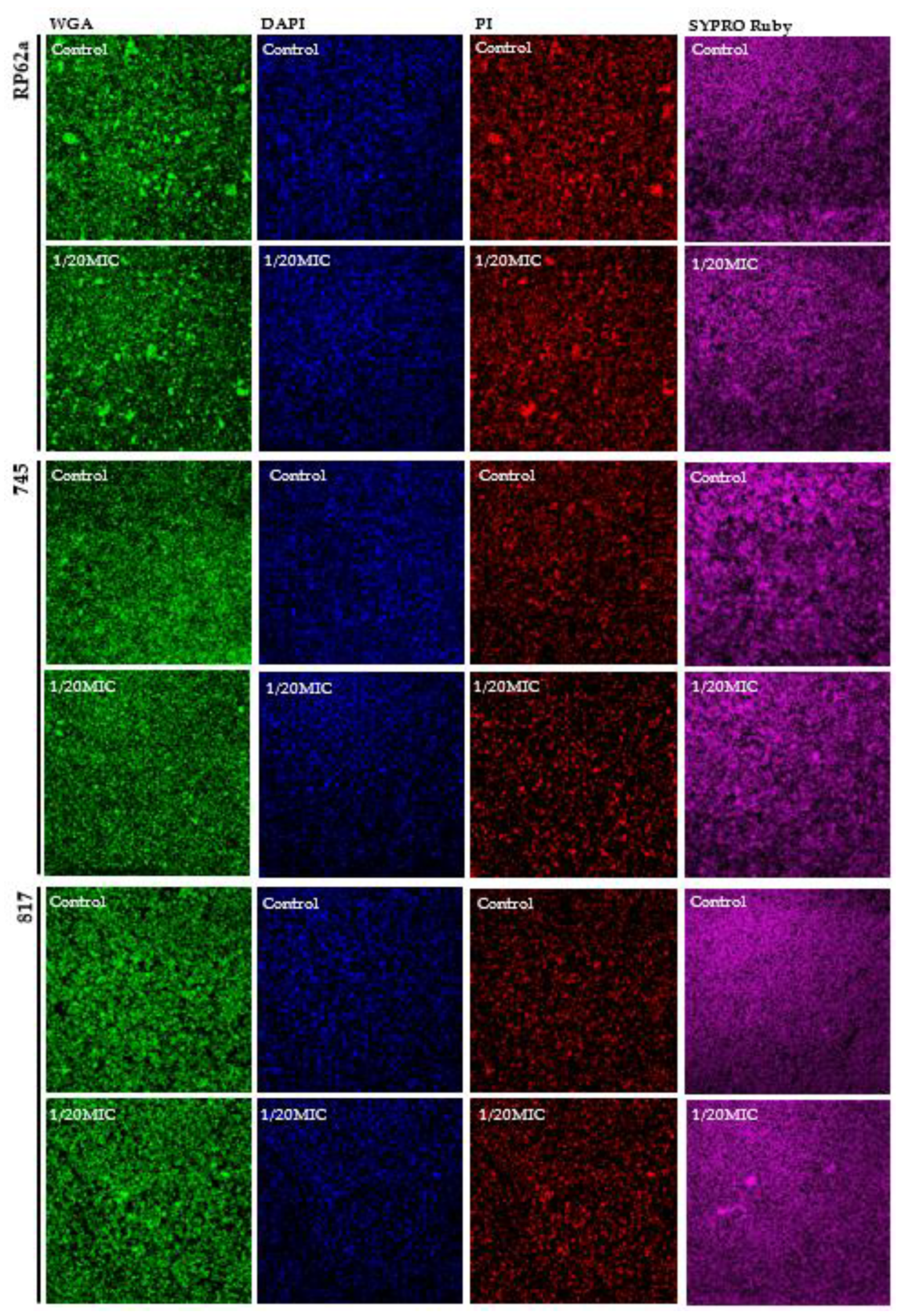
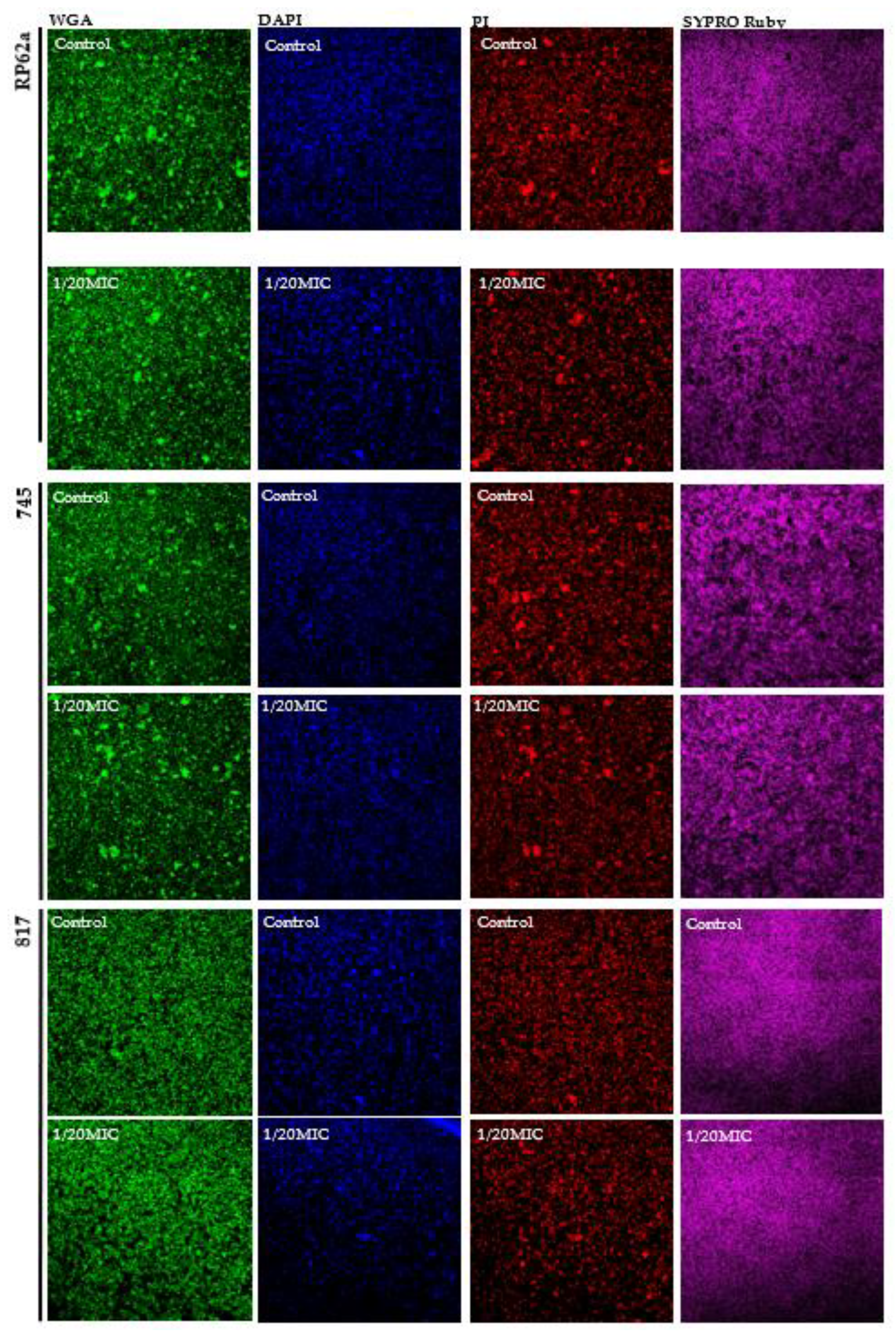
2.4. Complex Analysis of EPS Components
2.5. Real-Time qPCR of Biofilm Determinants
2.5.1. RNAIII-Dependent Genes
2.5.2. RNAIII-Independent Genes
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Culture Conditions
4.2. Phenolic Compounds
4.3. Assessment of Metabolic Activity of Biofilm by Resazurin
4.4. Assessment of Biofilm Formation by Crystal Violet Staining
4.5. EPS Inhibition Analysis by WGA Assay
4.6. EPS Isolation
4.7. Bradford Protein Assay
4.8. Calorimetric DuBois Method
4.9. Confocal Laser Scanning Microscopy (CLSM)
4.10. Phenol-Chloroform RNA Extraction
4.11. Real-Time qPCR
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rogers, K.L.; Fey, P.D.; Rupp, M.E. Coagulase-negative staphylococcal infections. Infect. Dis. Clin. N. Am. 2009, 3, 73–98. [Google Scholar] [CrossRef] [PubMed]
- Aparna, M.S.; Yadav, S. Biofilms: Microbes and disease. Braz. J. Infect. Dis. 2008, 12, 526–530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hall-Stoodley, L.; Costerton, J.W.; Stoodley, P. Bacterial biofilms: From the natural environment to infectious diseases. Nat. Rev. Microbiol. 2004, 2, 95–108. [Google Scholar] [CrossRef]
- Høiby, N.; Ciofu, O.; Johansen, H.K.; Song, Z.J.; Moser, C.; Jensen, P.Ø.; Molin, S.; Givskov, M.; Tolker-Nielsen, T.; Bjarnsholt, T. The clinical impact of bacterial biofilms. Int. J. Oral Sci. 2011, 3, 55–65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Otto, M. Staphylococcus epidermidis-the ‘accidental’ pathogen. Nat. Rev. Microbiol. 2009, 7, 555–567. [Google Scholar] [CrossRef] [Green Version]
- Zapotoczna, M.; O’Neill, E.; O’Gara, J.P. Untangling the Diverse and Redundant Mechanisms of Staphylococcus aureus Biofilm Formation. PLoS Pathog. 2016, 12, e1005671. [Google Scholar] [CrossRef]
- Waters, C.M.; Bassler, B.L. Quorum sensing: Cell-to-cell communication in bacteria. Annu. Rev. Cell. Dev. Biol. 2005, 21, 319–346. [Google Scholar] [CrossRef] [Green Version]
- Antunes, L.C.M.; Ferreira, R.B.R.; Buckner, M.M.C.; Finlay, B.B. Quorum sensing in bacterial virulence. Microbiology 2010, 156 Pt 8, 2271–2282. [Google Scholar] [CrossRef] [Green Version]
- Castillo-Juárez, I.; Maeda, T.; Mandujano-Tinoco, E.A.; Tomás, M.; Pérez-Eretza, B.; García-Contreras, S.J.; Wood, T.K.; García-Contreras, R. Role of quorum sensing in bacterial infections. World J. Clin. Cases 2015, 3, 575–598. [Google Scholar] [CrossRef]
- Diggle, S.P.; Griffin, A.S.; Campbell, G.S.; West, S.A. Cooperation and conflict in quorum-sensing bacterial populations. Nature 2007, 450, 411–414. [Google Scholar] [CrossRef]
- Thoendel, M.; Kavanaugh, J.S.; Flack, C.E.; Horswill, A.R. Peptide signaling in the staphylococci. Chem. Rev. 2011, 111, 117–151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Novick, R.P. Genetic systems in staphylococci. Methods Enzymol. 1991, 204, 587–636. [Google Scholar] [PubMed]
- Janzon, L.; Arvidson, S. The role of the delta-lysin gene (hld) in the regulation of virulence genes by the accessory gene regulator (agr) in Staphylococcus aureus. EMBO J. 1990, 9, 1391–1399. [Google Scholar] [CrossRef]
- Novick, R.P.; Ross, H.F.; Projan, S.J.; Kornblum, J.; Kreiswirth, B.; Moghazeh, S. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 1993, 12, 3967–3975. [Google Scholar] [CrossRef] [PubMed]
- Martínez-García, S.; Rodríguez-Martínez, S.; Cancino-Diaz, M.E.; Cancino-Diaz, J.C. Extracellular proteases of Staphylococcus epidermidis: Roles as virulence factors and their participation in biofilm. APMIS 2018, 126, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Janzon, L.; Löfdahl, S.; Arvidson, S. Identification and nucleotide sequence of the delta-lysin gene, hld, adjacent to the accessory gene regulator (agr) of Staphylococcus aureus. Mol. Gen. Genet. 1989, 219, 480–485. [Google Scholar] [CrossRef]
- Rosenthal, M.E.; Dever, L.L.; Moucha, C.S.; Chavda, K.D.; Otto, M.; Kreiswirth, B.N. Molecular characterization of an early invasive Staphylococcus epidermidis prosthetic joint infection. Microb. Drug Resist. 2011, 17, 345–350. [Google Scholar] [CrossRef] [Green Version]
- Tan, L.; Li, S.R.; Jiang, B.; Hu, X.M.; Li, S. Therapeutic Targeting of the Staphylococcus aureus Accessory Gene Regulator (agr) System. Front. Microbiol. 2018, 9, 55. [Google Scholar] [CrossRef]
- Queck, S.Y.; Jameson-Lee, M.; Villaruz, A.E.; Bach, T.H.; Khan, B.A.; Sturdevant, D.E.; Ricklefs, S.M.; Li, M.; Otto, M. RNAIII-independent target gene control by the agr quorum-sensing system: Insight into the evolution of virulence regulation in Staphylococcus aureus. Mol. Cell 2008, 32, 150–158. [Google Scholar] [CrossRef] [Green Version]
- Koenig, R.L.; Ray, J.L.; Maleki, S.J.; Smeltzer, M.S.; Hurlburt, B.K. Staphylococcus aureus AgrA binding to the RNAIII-agr regulatory region. J. Bacteriol. 2004, 186, 7549–7555. [Google Scholar] [CrossRef] [Green Version]
- Periasamy, S.; Joo, H.S.; Duong, A.C.; Bach, T.H.; Tan, V.Y.; Chatterjee, S.S.; Cheung, G.Y.; Otto, M. How Staphylococcus aureus biofilms develop their characteristic structure. Proc. Natl. Acad. Sci. USA 2012, 109, 1281–1286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roberts, A.L. Identification of Staphylococcus epidermidis in the Clinical Microbiology Laboratory by Molecular Methods. In Staphylococcus epidermidis: Methods and Protocols; Fey, P.D., Ed.; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2014; Volume 1106. [Google Scholar]
- Guarrera, P.M. Traditional phytotherapy in Central Italy (Marche, Abruzzo, and Latium). Fitoterapia 2005, 76, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Silva, L.N.; Zimmer, K.R.; Macedo, A.J.; Trentin, D.S. Plant Natural Products Targeting Bacterial Virulence Factors. Chem. Rev. 2016, 116, 9162–9236. [Google Scholar] [CrossRef] [PubMed]
- Farhadi, K.; Esmaeilzadeh, F.; Hatami, M.; Forough, M.; Molaie, R. Determination of phenolic compounds content and antioxidant activity in skin, pulp, seed, cane and leaf of five native grape cultivars in West Azerbaijan province. Iran. Food Chem. 2016, 199, 847–855. [Google Scholar] [CrossRef]
- Choi, J.S.; Islam, M.N.; Ali, M.Y.; Kim, E.J.; Kim, Y.M.; Jung, H.A. Effects of C-glycosylation on anti-diabetic, anti-Alzheimer’s disease and anti-inflammatory potential of apigenin. Food Chem. Toxicol. 2014, 64, 27–33. [Google Scholar] [CrossRef]
- Spencer, J.P.; Abd El Mohsen, M.M.; Minihane, A.M.; Mathers, J.C. Biomarkers of the intake of dietary polyphenols: Strengths, limitations and application in nutrition research. Br. J. Nutr. 2008, 99, 12–22. [Google Scholar] [CrossRef] [Green Version]
- Xing, M.; Shen, F.; Liu, L.; Chen, Z.; Guo, N.; Wang, X.; Wang, W.; Zhang, K.; Wum, X.; Wang, X.; et al. Antimicrobial efficacy of the alkaloid harmaline alone and in combination with chlorhexidine digluconate against clinical isolates of Staphylococcus aureus grown in planktonic and biofilm cultures. Lett. Appl. Microbiol. 2012, 54, 475–482. [Google Scholar] [CrossRef]
- Waditzer, M.; Bucar, F. Flavonoids as Inhibitors of Bacterial Efflux Pumps. Molecules 2021, 26, 6904. [Google Scholar] [CrossRef]
- Spellberg, B.; Bartlett, J.G.; Gilbert, D.N. The future of antibiotics and resistance. N. Engl. J. Med. 2013, 368, 299–302. [Google Scholar] [CrossRef] [Green Version]
- Laxminarayan, R.; Duse, A.; Wattal, C.; Zaidi, A.K.; Wertheim, H.F.; Sumpradit, N.; Vlieghe, E.; Hara, G.L.; Gould, I.M.; Goossens, H.; et al. Antibiotic resistance-the need for global solutions. Lancet Infect. Dis. 2013, 13, 1057–1098. [Google Scholar] [CrossRef] [Green Version]
- Kannappan, A.; Sivaranjani, M.; Srinivasan, R.; Rathna, J.; Pandian, S.K.; Ravi, A.V. Inhibitory efficacy of geraniol on biofilm formation and development of adaptive resistance in Staphylococcus epidermidis RP62A. J. Med. Microbiol. 2017, 66, 1506–1515. [Google Scholar] [CrossRef] [PubMed]
- Malladi, V.L.; Sobczak, A.J.; Maricic, N.; Murugapiran, S.K.; Schneper, L.; Makemson, J.; Mathee, K.; Wnuk, S.F. Substituted lactam and cyclic azahemiacetals modulate Pseudomonas aeruginosa quorum sensing. Bioorganic Med. Chem. 2011, 19, 5500–5506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riss, T.L.; Moravec, R.A.; Niles, A.L.; Duellman, S.; Benink, H.A.; Worzella, T.J.; Minor, L. Cell Viability Assays. In Assay Guidance Manual; Markossian, S., Grossman, A., Brimacombe, K., Arkin, M., Auld, D., Austin, C.P., Baell, J., Chung, T.D.Y., Coussens, N.P., Dahlin, J.L., et al., Eds.; Eli Lilly & Company and the National Center for Advancing Translational Sciences: Bethesda, MD, USA, 2004. [Google Scholar]
- Guerin, T.F.; Mondido, M.; McClenn, B.; Parsley, B. Application of resazurin for estimating abundance of contaminant-degrading microorganisms. Lett. Appl. Microbiol. 2001, 32, 340–345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skogman, M.E.; Vuorela, P.M.; Fallarero, A. Combining biofilm matrix measurements with biomass and viability assays in susceptibility assessments of antimicrobials against Staphylococcus aureus biofilms. J. Antibiot. 2012, 65, 453–459. [Google Scholar] [CrossRef] [Green Version]
- Rosenberg, M.; Azevedo, N.F.; Ivask, A. Propidium iodide staining underestimates viability of adherent bacterial cells. Sci. Rep. 2019, 9, 6483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, T.; Mei, Y.; He, B.; Sun, X.; Li, J. Reducing Quorum Sensing-Mediated Virulence Factor Expression and Biofilm Formation in Hafnia alvei by Using the Potential Quorum Sensing Inhibitor L-Carvone. Front. Microbiol. 2019, 9, 3324. [Google Scholar] [CrossRef] [PubMed]
- Shastry, R.P.; Ghate, S.D.; Kumar, B.S.; Srinath, B.S.; Kumar, V. Vanillin derivative inhibits quorum sensing and biofilm formation in Pseudomonas aeruginosa: A study in a Caenorhabditis elegans infection model. Nat. Prod. Res. 2021, 22, 1–6. [Google Scholar] [CrossRef]
- Raut, J.S.; Rajput, S.B.; Shinde, R.B.; Surwase, B.S.; Karuppayil, S.M. Vanillin Inhibits Growth, Morphogenesis and Biofilm Formation by Candida albicans. J. Biol Act. Prod. Nat. 2013, 3, 130–138. [Google Scholar]
- Chiba, A.; Sugimoto, S.; Sato, F.; Hori, S.; Mizunoe, Y. A refined technique for extraction of extracellular matrices from bacterial biofilms and its applicability. Microb. Biotechnol. 2014, 8, 392–403. [Google Scholar] [CrossRef]
- DuBois, M.; Gilles, K.; Hamilton, J.K.; Rebers, P.A.; Smith, F. A colorimetric method for the determination of sugars. Nature 1951, 168, 167. [Google Scholar] [CrossRef]
- Mathew, S.; Abraham, T.E.; Zakaria, Z.A. Reactivity of phenolic compounds towards free radicals under in vitro conditions. J. Food Sci. Technol. 2015, 52, 5790–5798. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anku, W.W.; Mamo, M.A.; Govender, P.P. Phenolic Compounds in Water: Sources, Reactivity, Toxicity and Treatment Methods. In Phenolic Compounds—Natural Sources, Importance and Applications; InTech: Rijeka, Croatia, 2017. [Google Scholar] [CrossRef] [Green Version]
- Ozdal, T.; Capanoglu, E.; Altay, F. A review on protein–phenolic interactions and associated changes. Food Res. Int. 2013, 51, 954–970. [Google Scholar] [CrossRef]
- Jakobek, L. Interactions of polyphenols with carbohydrates, lipids and proteins. Food Chem. 2015, 175, 556–567. [Google Scholar] [CrossRef] [PubMed]
- Stiefel, P.; Schmidt-Emrich, S.; Maniura-Weber, K. Critical aspects of using bacterial cell viability assays with the fluorophores SYTO9 and propidium iodide. BMC Microbiol. 2015, 15, 36. [Google Scholar] [CrossRef] [Green Version]
- Sugimoto, S.; Sato, F.; Miyakawa, R. Broad impact of extracellular DNA on biofilm formation by clinically isolated Methicillin-resistant and -sensitive strains of Staphylococcus aureus. Sci. Rep. 2018, 8, 2254. [Google Scholar] [CrossRef]
- Mann, E.E.; Rice, K.C.; Boles, B.R.; Endres, J.L.; Ranjit, D.; Chandramohan, L.; Tsang, L.H.; Smeltzer, M.S.; Horswill, A.R.; Bayles, K.W. Modulation of eDNA release and degradation affects Staphylococcus aureus biofilm maturation. PLoS ONE 2009, 4, e5822. [Google Scholar] [CrossRef] [Green Version]
- Duarte, A.; Alves, A.C.; Ferreira, S.; Silva, F.; Domingues, F.C. Resveratrol inclusion complexes: Antibacterial and anti-biofilm activity against Campylobacter spp. and Arcobacter butzleri. Food Res. Int. 2015, 77, 244–250. [Google Scholar] [CrossRef]
- Quecan, B.X.V.; Santos, J.T.C.; Rivera, M.L.C.; Hassimotto, N.M.A.; Almeida, F.A.; Pinto, U.M. Effect of Quercetin Rich Onion Extracts on Bacterial Quorum Sensing. Front. Microbiol. 2019, 10, 867. [Google Scholar] [CrossRef] [Green Version]
- Santos, C.A.; Lima, E.; Franco, B.; Pinto, U.M. Exploring Phenolic Compounds as Quorum Sensing Inhibitors in Foodborne Bacteria. Front. Microbiol. 2021, 12, 735931. [Google Scholar] [CrossRef]
- Hossain, M.A.; Lee, S.J.; Park, N.H.; Mechesso, A.F.; Birhanu, B.T.; Kang, J.; Reza, M.A.; Suh, J.W.; Park, S.C. Impact of phenolic compounds in the acyl homoserine lactone-mediated quorum sensing regulatory pathways. Sci. Rep. 2017, 7, 10618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stepanović, S.; Vuković, D.; Dakić, I.; Savić, B.; Švabić-Vlahović, M. A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J. Microbiol. 2000, 40, 175–179. [Google Scholar] [CrossRef]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Minich, A.; Levarski, Z.; Mikulášová, M.; Straka, M.; Liptáková, A.; Stuchlík, S. Complex Analysis of Vanillin and Syringic Acid as Natural Antimicrobial Agents against Staphylococcus epidermidis Biofilms. Int. J. Mol. Sci. 2022, 23, 1816. https://doi.org/10.3390/ijms23031816
Minich A, Levarski Z, Mikulášová M, Straka M, Liptáková A, Stuchlík S. Complex Analysis of Vanillin and Syringic Acid as Natural Antimicrobial Agents against Staphylococcus epidermidis Biofilms. International Journal of Molecular Sciences. 2022; 23(3):1816. https://doi.org/10.3390/ijms23031816
Chicago/Turabian StyleMinich, Andrej, Zdenko Levarski, Mária Mikulášová, Marek Straka, Adriána Liptáková, and Stanislav Stuchlík. 2022. "Complex Analysis of Vanillin and Syringic Acid as Natural Antimicrobial Agents against Staphylococcus epidermidis Biofilms" International Journal of Molecular Sciences 23, no. 3: 1816. https://doi.org/10.3390/ijms23031816





