SPDL1 Is an Independent Predictor of Patient Outcome in Colorectal Cancer
Abstract
:1. Introduction
2. Results
2.1. Immunoexpression of SPDL1 in Tumor and Normal Adjacent Tissue and Its Clinicopathological Associations
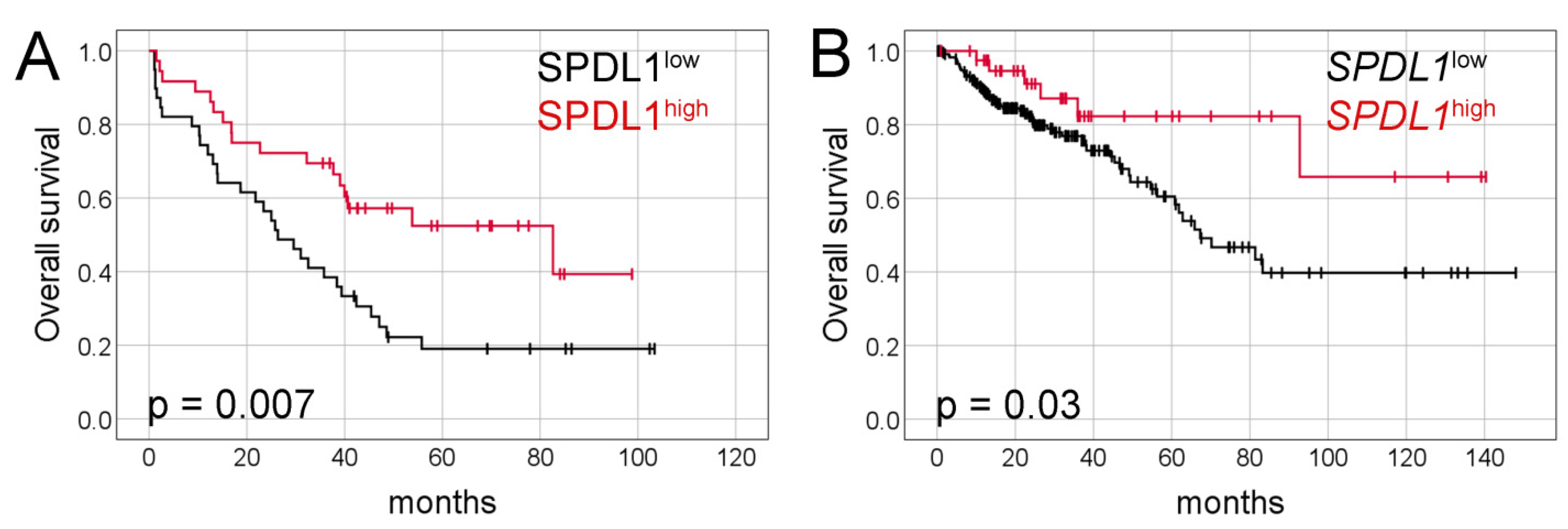
2.2. Prognostic Value of SPDL1 Immunoexpression in Predicting the Overall Survival of CRC Patients
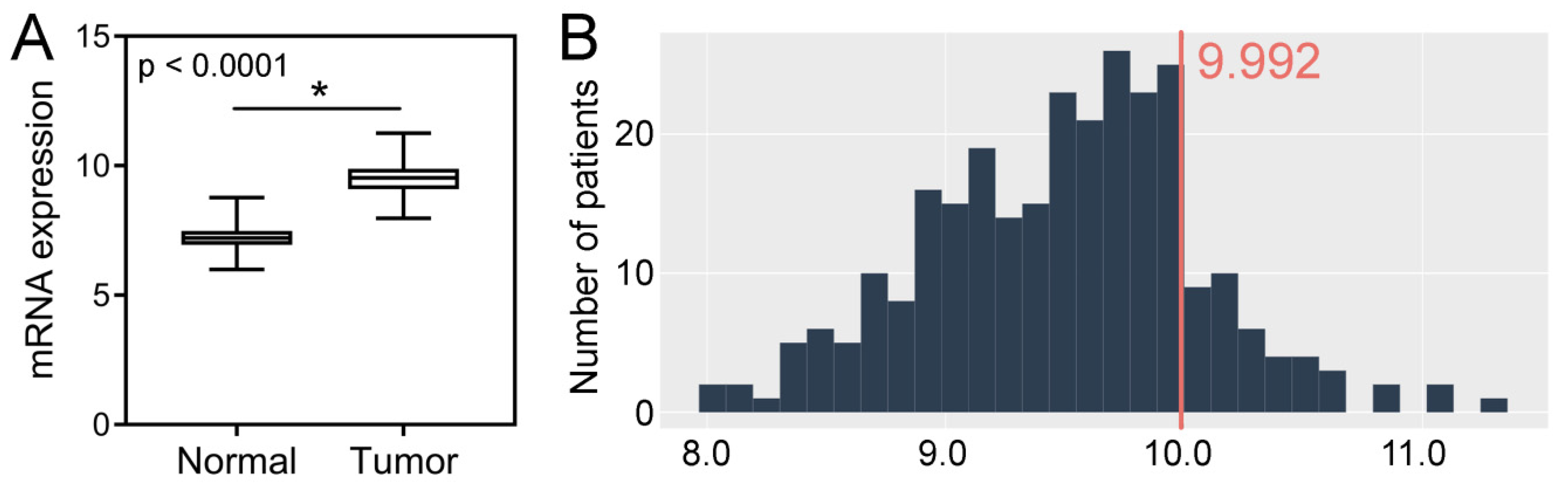
2.3. TCGA CRC Cohort Analysis–Tissue Expression, Clinicopathological and Survival Associations, Correlations with GIN
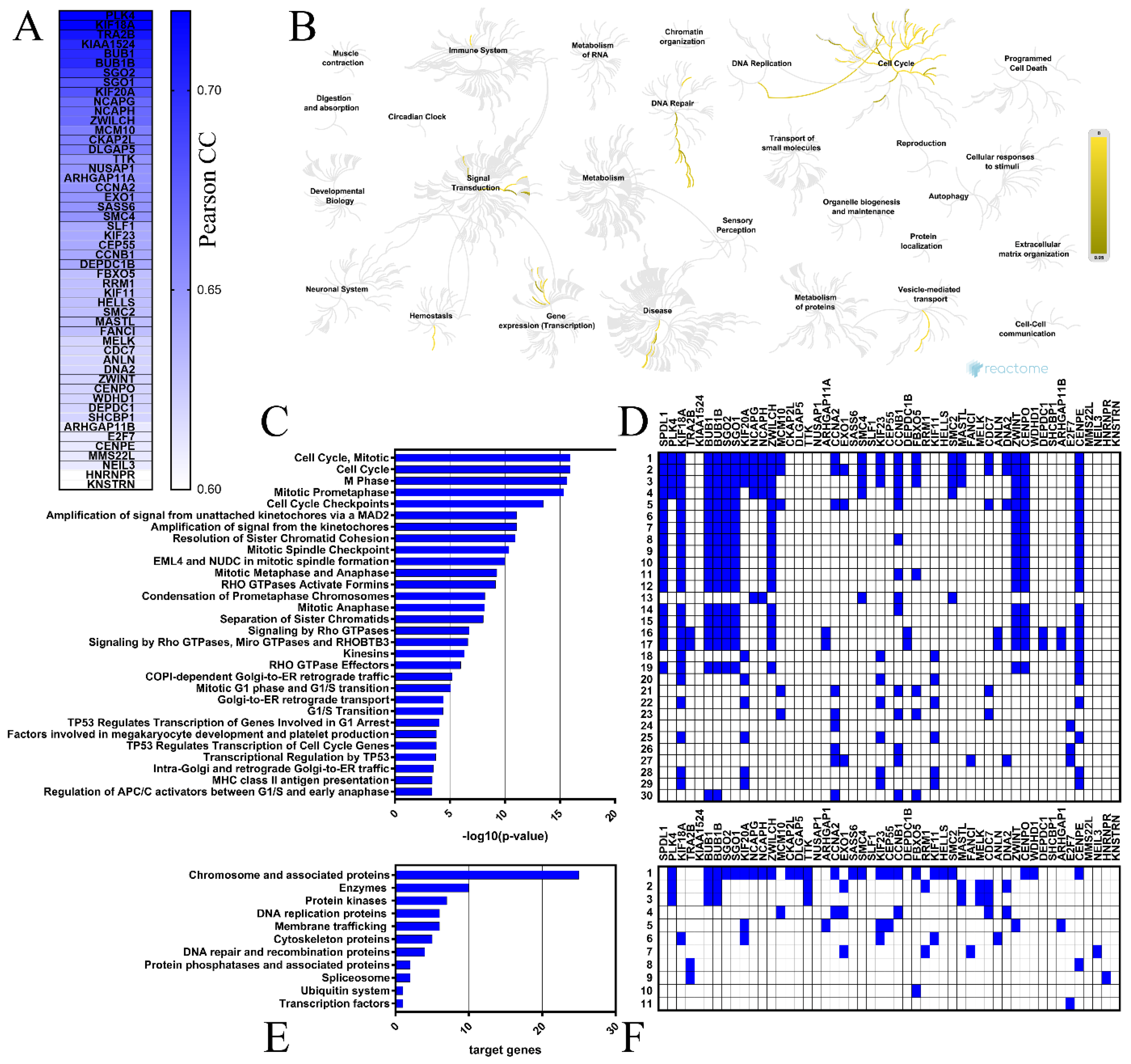
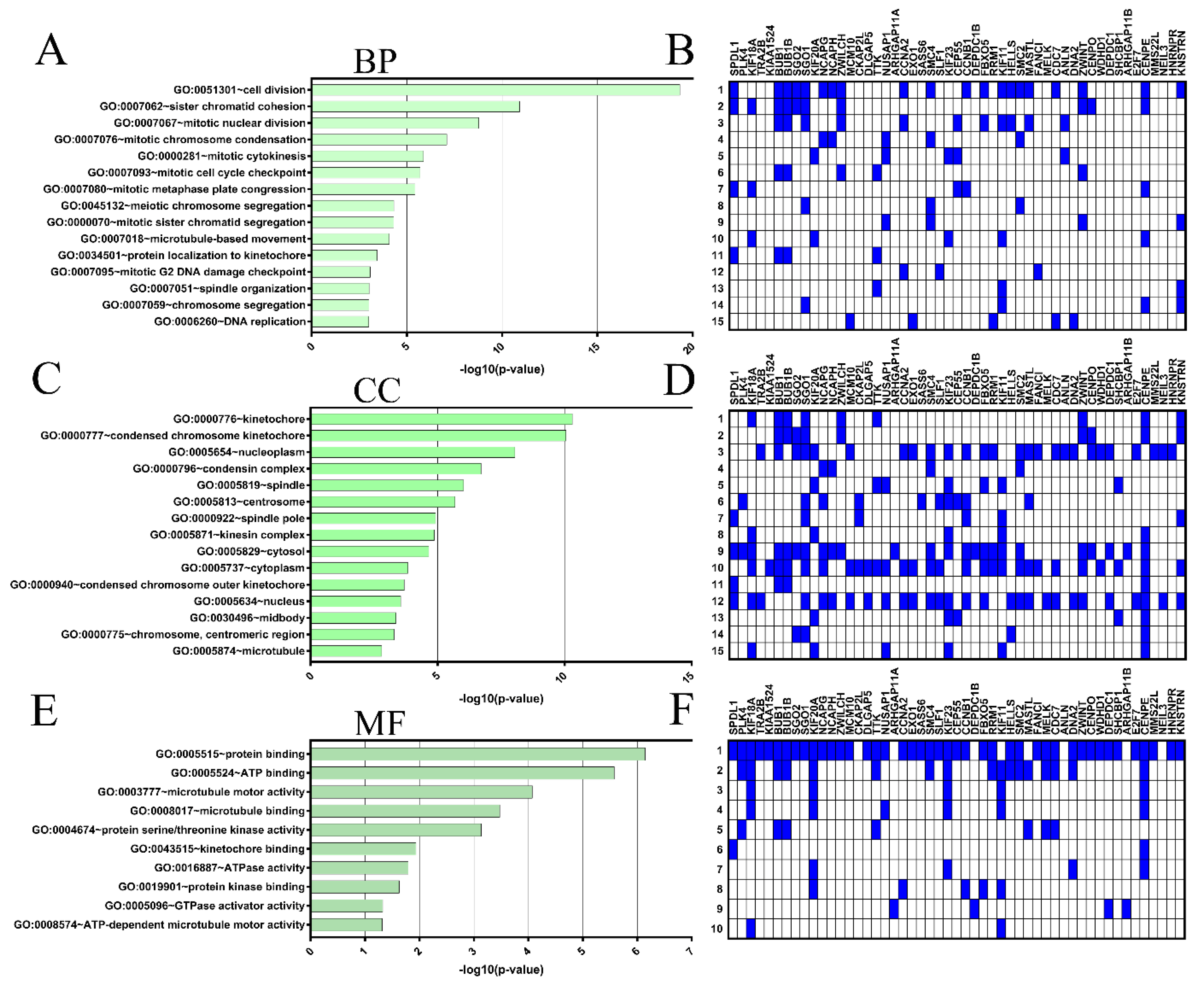
2.4. Coexpressed Genes and Functional Enrichment Analyses
3. Discussion
4. Materials and Methods
4.1. Patients and Tissue Material
4.2. Tissue Microarrays and Immunohistochemical Staining
4.3. Evaluation of Immunohistochemical Staining
4.4. Publicly Available Transcriptomic Data
4.5. Functional Enrichment Analysis
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021. [Google Scholar] [CrossRef] [PubMed]
- Ogunwobi, O.O.; Mahmood, F.; Akingboye, A. Biomarkers in Colorectal Cancer: Current Research and Future Prospects. Int. J. Mol. Sci. 2020, 21, 5311. [Google Scholar] [CrossRef] [PubMed]
- Rao, C.V.; Yamada, H.Y. Genomic Instability and Colon Carcinogenesis: From the Perspective of Genes. Front. Oncol. 2013, 3, 130. [Google Scholar] [CrossRef] [Green Version]
- Guo, M.; Wang, S.M. Genome Instability-Derived Genes Are Novel Prognostic Biomarkers for Triple-Negative Breast Cancer. Front. Cell Dev. Biol. 2021, 9, 701073. [Google Scholar] [CrossRef] [PubMed]
- Griffis, E.R.; Stuurman, N.; Vale, R.D. Spindly, a Novel Protein Essential for Silencing the Spindle Assembly Checkpoint, Recruits Dynein to the Kinetochore. J. Cell Biol. 2007, 177, 1005–1015. [Google Scholar] [CrossRef] [Green Version]
- Conte, C.; Baird, M.A.; Davidson, M.W.; Griffis, E.R. Spindly Is Required for Rapid Migration of Human Cells. Biol. Open 2018, 7, bio033233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klimaszewska-Wiśniewska, A.; Buchholz, K.; Neska-Długosz, I.; Durślewicz, J.; Grzanka, D.; Zabrzyński, J.; Sopońska, P.; Grzanka, A.; Gagat, M. Expression of Genomic Instability-Related Molecules: Cyclin F, RRM2 and SPDL1 and Their Prognostic Significance in Pancreatic Adenocarcinoma. Cancers 2021, 13, 859. [Google Scholar] [CrossRef]
- Silva, P.M.A.; Delgado, M.L.; Ribeiro, N.; Florindo, C.; Tavares, Á.A.; Ribeiro, D.; Lopes, C.; do Amaral, B.; Bousbaa, H.; Monteiro, L.S. Spindly and Bub3 Expression in Oral Cancer: Prognostic and Therapeutic Implications. Oral Dis. 2019, 25, 1291–1301. [Google Scholar] [CrossRef]
- Tian, X.; Wang, N. Upregulation of ASPM, BUB1B and SPDL1 in Tumor Tissues Predicts Poor Survival in Patients with Pancreatic Ductal Adenocarcinoma. Oncol. Lett. 2020, 19, 3307–3315. [Google Scholar] [CrossRef] [Green Version]
- Kodama, T.; Marian, T.A.; Lee, H.; Kodama, M.; Li, J.; Parmacek, M.S.; Jenkins, N.A.; Copeland, N.G.; Wei, Z. MRTFB Suppresses Colorectal Cancer Development through Regulating SPDL1 and MCAM. Proc. Natl. Acad. Sci. USA 2019, 116, 23625–23635. [Google Scholar] [CrossRef]
- Zheng, J.; Li, T.-K.; Bao, Y.; Zhang, S.-X. Expression and clinical significance of Spindly and Bub3 in oral squamous cell carcinoma. Shanghai J. Stomatol. 2020, 29, 528–532. [Google Scholar] [PubMed]
- Carter, S.L.; Eklund, A.C.; Kohane, I.S.; Harris, L.N.; Szallasi, Z. A Signature of Chromosomal Instability Inferred from Gene Expression Profiles Predicts Clinical Outcome in Multiple Human Cancers. Nat. Genet. 2006, 38, 1043–1048. [Google Scholar] [CrossRef] [PubMed]
- Kawakami, H.; Zaanan, A.; Sinicrope, F.A. Microsatellite Instability Testing and Its Role in the Management of Colorectal Cancer. Curr. Treat. Options Oncol. 2015, 16, 30. [Google Scholar] [CrossRef] [PubMed]
- Andor, N.; Maley, C.C.; Ji, H.P. Genomic Instability in Cancer: Teetering on the Limit of Tolerance. Cancer Res. 2017, 77, 2179–2185. [Google Scholar] [CrossRef] [Green Version]
- Altman, D.G.; McShane, L.M.; Sauerbrei, W.; Taube, S.E. Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK): Explanation and Elaboration. PLoS Med. 2012, 9, e1001216. [Google Scholar] [CrossRef] [Green Version]
- Compton, C.C. Optimal Pathologic Staging: Defining Stage II Disease. Clin. Cancer Res. 2007, 13, 6862s–6870s. [Google Scholar] [CrossRef] [Green Version]
- Greaves, P.; Clear, A.; Coutinho, R.; Wilson, A.; Matthews, J.; Owen, A.; Shanyinde, M.; Lister, T.A.; Calaminici, M.; Gribben, J.G. Expression of FOXP3, CD68, and CD20 at Diagnosis in the Microenvironment of Classical Hodgkin Lymphoma Is Predictive of Outcome. J. Clin. Oncol. 2013, 31, 256–262. [Google Scholar] [CrossRef] [Green Version]
- Romagosa, C.; Simonetti, S.; López-Vicente, L.; Mazo, A.; Lleonart, M.E.; Castellvi, J.; Ramon y Cajal, S. P16(Ink4a) Overexpression in Cancer: A Tumor Suppressor Gene Associated with Senescence and High-Grade Tumors. Oncogene 2011, 30, 2087–2097. [Google Scholar] [CrossRef] [Green Version]
- Koskela, J.T.; Häppölä, P.; Liu, A.; FinnGen; Partanen, J.; Genovese, G.; Artomov, M.; Myllymäki, M.N.M.; Kanai, M.; Zhou, W.; et al. Genetic Variant in SPDL1 Reveals Novel Mechanism Linking Pulmonary Fibrosis Risk and Cancer Protection; Genetic and Genomic Medicine. medRxiv. 2021. Available online: https://www.medrxiv.org/content/10.1101/2021.05.07.21255988v1.full (accessed on 2 December 2021).
- Dhindsa, R.S.; Mattsson, J.; Nag, A.; Wang, Q.; Wain, L.V.; Allen, R.; Wigmore, E.M.; Ibanez, K.; Vitsios, D.; Deevi, S.V.V.; et al. Identification of a Missense Variant in SPDL1 Associated with Idiopathic Pulmonary Fibrosis. Commun. Biol. 2021, 4, 392. [Google Scholar] [CrossRef]
- Kajita, K.; Kuwano, Y.; Satake, Y.; Kano, S.; Kurokawa, K.; Akaike, Y.; Masuda, K.; Nishida, K.; Rokutan, K. Ultraconserved Region-Containing Transformer 2β4 Controls Senescence of Colon Cancer Cells. Oncogenesis 2016, 5, e213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Ge, Z.; Liu, C.; Liu, Z.; Björkholm, M.; Jia, J.; Xu, D. CIP2A Is Overexpressed in Gastric Cancer and Its Depletion Leads to Impaired Clonogenicity, Senescence, or Differentiation of Tumor Cells. Clin. Cancer Res. 2008, 14, 3722–3728. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, J.; Su, H.-K.; Yu, Z.-H.; Xi, S.-Y.; Guo, C.-C.; Hu, Z.-Y.; Qu, Y.; Cai, H.-P.; Zhao, Y.-Y.; Zhao, H.-F.; et al. Skp2 Modulates Proliferation, Senescence and Tumorigenesis of Glioma. Cancer Cell Int. 2020, 20, 71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sacristan, C.; Ahmad, M.U.D.; Keller, J.; Fermie, J.; Groenewold, V.; Tromer, E.; Fish, A.; Melero, R.; Carazo, J.M.; Klumperman, J.; et al. Dynamic Kinetochore Size Regulation Promotes Microtubule Capture and Chromosome Biorientation in Mitosis. Nat. Cell Biol. 2018, 20, 800–810. [Google Scholar] [CrossRef]
- Barisic, M.; Sohm, B.; Mikolcevic, P.; Wandke, C.; Rauch, V.; Ringer, T.; Hess, M.; Bonn, G.; Geley, S. Spindly/CCDC99 Is Required for Efficient Chromosome Congression and Mitotic Checkpoint Regulation. Mol. Biol. Cell 2010, 21, 1968–1981. [Google Scholar] [CrossRef] [Green Version]
- Chan, Y.W.; Fava, L.L.; Uldschmid, A.; Schmitz, M.H.A.; Gerlich, D.W.; Nigg, E.A.; Santamaria, A. Mitotic Control of Kinetochore-Associated Dynein and Spindle Orientation by Human Spindly. J. Cell Biol. 2009, 185, 859–874. [Google Scholar] [CrossRef] [Green Version]
- Gama, J.B.; Pereira, C.; Simões, P.A.; Celestino, R.; Reis, R.M.; Barbosa, D.J.; Pires, H.R.; Carvalho, C.; Amorim, J.; Carvalho, A.X.; et al. Molecular Mechanism of Dynein Recruitment to Kinetochores by the Rod-Zw10-Zwilch Complex and Spindly. J. Cell Biol. 2017, 216, 943–960. [Google Scholar] [CrossRef]
- de Cárcer, G.; Venkateswaran, S.V.; Salgueiro, L.; El Bakkali, A.; Somogyi, K.; Rowald, K.; Montañés, P.; Sanclemente, M.; Escobar, B.; de Martino, A.; et al. Plk1 Overexpression Induces Chromosomal Instability and Suppresses Tumor Development. Nat. Commun. 2018, 9, 3012. [Google Scholar] [CrossRef] [Green Version]
- Kops, G.J.P.L.; Foltz, D.R.; Cleveland, D.W. Lethality to Human Cancer Cells through Massive Chromosome Loss by Inhibition of the Mitotic Checkpoint. Proc. Natl. Acad. Sci. USA 2004, 101, 8699–8704. [Google Scholar] [CrossRef] [Green Version]
- Roylance, R.; Endesfelder, D.; Gorman, P.; Burrell, R.A.; Sander, J.; Tomlinson, I.; Hanby, A.M.; Speirs, V.; Richardson, A.L.; Birkbak, N.J.; et al. Relationship of Extreme Chromosomal Instability with Long-Term Survival in a Retrospective Analysis of Primary Breast Cancer. Cancer Epidemiol. Prev. Biomark. 2011, 20, 2183–2194. [Google Scholar] [CrossRef] [Green Version]
- Chakraborty, U.; Dinh, T.A.; Alani, E. Genomic Instability Promoted by Overexpression of Mismatch Repair Factors in Yeast: A Model for Understanding Cancer Progression. Genetics 2018, 209, 439–456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilczak, W.; Rashed, S.; Hube-Magg, C.; Kluth, M.; Simon, R.; Büscheck, F.; Clauditz, T.S.; Grupp, K.; Minner, S.; Tsourlakis, M.C.; et al. Up-Regulation of Mismatch Repair Genes MSH6, PMS2 and MLH1 Parallels Development of Genetic Instability and Is Linked to Tumor Aggressiveness and Early PSA Recurrence in Prostate Cancer. Carcinogenesis 2017, 38, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Norris, A.M.; Woodruff, R.D.; D’Agostino, R.B.; Clodfelter, J.E.; Scarpinato, K.D. Elevated Levels of the Mismatch Repair Protein PMS2 Are Associated with Prostate Cancer. Prostate 2007, 67, 214–225. [Google Scholar] [CrossRef] [PubMed]
- Krol, K.; Brozda, I.; Skoneczny, M.; Bretner, M.; Bretne, M.; Skoneczna, A. A Genomic Screen Revealing the Importance of Vesicular Trafficking Pathways in Genome Maintenance and Protection against Genotoxic Stress in Diploid Saccharomyces Cerevisiae Cells. PLoS ONE 2015, 10, e0120702. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Foiani, M.; Bartek, J. Golgi Feels DNA’s Pain. Cell 2014, 156, 392–393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neska-Długosz, I.; Buchholz, K.; Durślewicz, J.; Gagat, M.; Grzanka, D.; Tojek, K.; Klimaszewska-Wiśniewska, A. Prognostic Impact and Functional Annotations of KIF11 and KIF14 Expression in Patients with Colorectal Cancer. Int. J. Mol. Sci. 2021, 22, 9732. [Google Scholar] [CrossRef] [PubMed]
- Gzil, A.; Zarębska, I.; Jaworski, D.; Antosik, P.; Durślewicz, J.; Maciejewska, J.; Domanowska, E.; Skoczylas-Makowska, N.; Ahmadi, N.; Grzanka, D.; et al. The Prognostic Value of Leucine-Rich Repeat-Containing G-Protein (Lgr5) and Its Impact on Clinicopathological Features of Colorectal Cancer. J. Cancer Res. Clin. Oncol. 2020, 146, 2547–2557. [Google Scholar] [CrossRef]
- Ogłuszka, M.; Orzechowska, M.; Jędroszka, D.; Witas, P.; Bednarek, A.K. Evaluate Cutpoints: Adaptable Continuous Data Distribution System for Determining Survival in Kaplan-Meier Estimator. Comput. Methods Programs Biomed. 2019. [Google Scholar] [CrossRef]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.V.S.K.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef]
- Jassal, B.; Matthews, L.; Viteri, G.; Gong, C.; Lorente, P.; Fabregat, A.; Sidiropoulos, K.; Cook, J.; Gillespie, M.; Haw, R.; et al. The Reactome Pathway Knowledgebase. Nucleic Acids Res. 2020, 48, D498–D503. [Google Scholar] [CrossRef]

| Clinicopathological Feature | n (%) n = 86 | SPDL1 Expression | p Value | |
|---|---|---|---|---|
| Low n = 44 | High n = 42 | |||
| Age (years) | ||||
| ≤65 | 38 (44.19) | 20 (52.63) | 18 (47.37) | 0.83 |
| >65 | 48 (55.81) | 24 (50.00) | 24 (50.00) | |
| Gender | ||||
| Male | 49 (56.98) | 23 (46.94) | 26 (53.06) | 0.39 |
| Female | 37 (43.02) | 21 (56.76) | 16 (43.24) | |
| Grading | ||||
| G2 | 76 (91.57) | 39 (51.32) | 37 (48.68) | 0.71 |
| G3 | 7 (8.43) | 3 (42.86) | 4 (57.14) | |
| pT status | ||||
| T2 | 13 (15.12) | 6 (46.15) | 7 (53.85) | 0.70 |
| T3 | 60 (69.77) | 30 (50.00) | 30 (50.00) | |
| T4 | 13 (15.12) | 8 (61.54) | 5 (38.46) | |
| pN status | ||||
| N0 | 33 (40.74) | 16 (48.48) | 17 (51.52) | 0.66 |
| N1–N2 | 48 (59.26) | 26 (54.17) | 22 (45.83) | |
| pM status | ||||
| M0 | 42 (52.50) | 20 (47.62) | 22 (52.38) | 0.51 |
| M1 | 38 (47.50) | 21 (55.26) | 17 (44.74) | |
| TNM stage | ||||
| I | 5 (6.25) | 2 (40.00) | 3 (60.00) | 0.31 |
| II | 17 (21.25) | 7 (41.18) | 10 (58.82) | |
| III | 20 (25.00) | 11 (55.00) | 9 (45.00) | |
| IV | 38 (47.50) | 21 (55.26) | 17 (44.74) | |
| VI | ||||
| Absent | 24 (60.00) | 10 (41.67) | 14 (58.33) | 0.52 |
| Present | 16 (40.00) | 9 (56.25) | 7 (43.75) | |
| PNI | ||||
| Absent | 25 (89.29) | 10 (40.00) | 15 (60.00) | 0.09 |
| Present | 3 (10.71) | 3 (100.00) | 0 (0.00) | |
| Resection margin | ||||
| R0 | 65 (75.58) | 33 (50.77) | 32 (49.23) | >0.99 |
| R1−R2 | 21 (24.42) | 11 (52.38) | 10 (47.62) | |
| Primary tumor location | ||||
| Right colon | 30 (34.88) | 16 (53.33) | 14 (46.67) | 0.54 |
| Left colon | 28 (32.56) | 12 (42.86) | 16 (57.14) | |
| Rectum | 28 (32.56) | 16 (57.14) | 12 (42.86) | |
| Variable | Univariate Analysis | Multivariate Analysis | ||||||
|---|---|---|---|---|---|---|---|---|
| HR | 95% CI | p | HR | 95% CI | p | |||
| Lower | Upper | Lower | Upper | |||||
| SPDL1 | 0.45 | 0.25 | 0.82 | 0.009 | 0.42 | 0.23 | 0.78 | 0.006 |
| age | 1.09 | 0.61 | 1.93 | 0.77 | 1.42 | 0.73 | 2.74 | 0.30 |
| gender | 0.99 | 0.55 | 1.75 | 0.96 | 1.32 | 0.71 | 2.45 | 0.39 |
| grade | 2.58 | 1.00 | 6.69 | 0.05 | 3.87 | 1.36 | 11.01 | 0.01 |
| pT | 2.30 | 0.91 | 5.83 | 0.08 | 1.88 | 0.72 | 4.93 | 0.20 |
| pN | 1.70 | 0.93 | 3.11 | 0.09 | 1.16 | 0.58 | 2.29 | 0.68 |
| pM | 2.81 | 1.55 | 5.08 | 0.001 | 3.32 | 1.73 | 6.37 | 0.0003 |
| TNM stage | 2.93 | 1.37 | 6.27 | 0.006 | - | - | - | - |
| resection margin | 1.86 | 0.98 | 3.53 | 0.06 | 2.37 | 1.13 | 4.98 | 0.02 |
| tumor location | ||||||||
| rectum | Ref. | Ref. | ||||||
| right colon | 1.29 | 0.66 | 2.52 | 0.46 | 2.10 | 1.00 | 4.44 | 0.05 |
| left colon | 0.99 | 0.49 | 2.01 | 0.98 | 1.55 | 0.71 | 3.40 | 0.28 |
| Clinicopathological Feature | n (%) n = 275 | SPDL1 Expression | p Value | |
|---|---|---|---|---|
| Low n = 234 | High n = 41 | |||
| Age (years) | ||||
| ≤65 | 129 (46.91) | 106 (82.17) | 23 (17.83) | 0.42 |
| >65 | 146 (53.09) | 128 (87.67) | 18 (12.33) | |
| Gender | ||||
| Male | 150 (54.55 | 125 (83.33) | 25 (16.67) | 0.40 |
| Female | 125 (45.45) | 109 (87.20) | 16 (12.80) | |
| pT status | ||||
| T1 | 6 (2.18) | 6 (100.00) | 0 (0.00) | 0.67 |
| T2 | 43 (15.64) | 39 (90.70) | 4 (9.30) | |
| T3 | 188 (68.36) | 154 (81.91) | 34 (18.09) | |
| T4 | 38 (13.82) | 35 (92.11) | 3 (7.89) | |
| pN status | ||||
| N0 | 160 (58.18) | 137 (85.63) | 23 (14.38) | 0.86 |
| N1–N2 | 115 (41.82) | 97 (84.35) | 18 (15.65) | |
| pM status | ||||
| M0 | 232 (86.25) | 196 (84.48) | 36 (15.52) | >0.99 |
| M1 | 37 (13.75) | 32 (86.49) | 5 (13.51) | |
| TNM stage | ||||
| I | 44 (16.36) | 40 (90.91) | 4 (9.09) | 0.60 |
| II | 106 (39.41) | 88 (83.02) | 18 (16.98) | |
| III | 82 (30.48) | 67 (82.72) | 14 (17.28) | |
| IV | 37 (13.75) | 32 (86.49) | 5 (13.51) | |
| Variable | Univariate Analysis | Multivariate Analysis | ||||||
|---|---|---|---|---|---|---|---|---|
| HR | 95% CI | p | HR | 95% CI | p | |||
| Lower | Upper | Lower | Upper | |||||
| SPDL1 | 0.41 | 0.18 | 0.96 | 0.04 | 0.37 | 0.16 | 0.85 | 0.02 |
| age | 1.68 | 1.01 | 2.81 | 0.047 | 2.25 | 1.30 | 3.88 | 0.004 |
| gender | 1.42 | 0.86 | 2.33 | 0.17 | 1.13 | 0.68 | 1.88 | 0.65 |
| pT | 3.23 | 1.17 | 8.89 | 0.02 | 2.23 | 0.78 | 6.39 | 0.14 |
| pN | 2.45 | 1.50 | 4.02 | 0.0004 | 1.90 | 1.09 | 3.31 | 0.02 |
| pM | 3.71 | 2.16 | 6.36 | <0.0001 | 3.23 | 1.75 | 5.94 | 0.0002 |
| TNM stage | 2.97 | 1.78 | 4.96 | <0.0001 | - | - | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Klimaszewska-Wiśniewska, A.; Buchholz, K.; Durślewicz, J.; Villodre, E.S.; Gagat, M.; Grzanka, D. SPDL1 Is an Independent Predictor of Patient Outcome in Colorectal Cancer. Int. J. Mol. Sci. 2022, 23, 1819. https://doi.org/10.3390/ijms23031819
Klimaszewska-Wiśniewska A, Buchholz K, Durślewicz J, Villodre ES, Gagat M, Grzanka D. SPDL1 Is an Independent Predictor of Patient Outcome in Colorectal Cancer. International Journal of Molecular Sciences. 2022; 23(3):1819. https://doi.org/10.3390/ijms23031819
Chicago/Turabian StyleKlimaszewska-Wiśniewska, Anna, Karolina Buchholz, Justyna Durślewicz, Emilly Schlee Villodre, Maciej Gagat, and Dariusz Grzanka. 2022. "SPDL1 Is an Independent Predictor of Patient Outcome in Colorectal Cancer" International Journal of Molecular Sciences 23, no. 3: 1819. https://doi.org/10.3390/ijms23031819
APA StyleKlimaszewska-Wiśniewska, A., Buchholz, K., Durślewicz, J., Villodre, E. S., Gagat, M., & Grzanka, D. (2022). SPDL1 Is an Independent Predictor of Patient Outcome in Colorectal Cancer. International Journal of Molecular Sciences, 23(3), 1819. https://doi.org/10.3390/ijms23031819







