Blood Bacteria-Free DNA in Septic Mice Enhances LPS-Induced Inflammation in Mice through Macrophage Response
Abstract
1. Introduction
2. Results
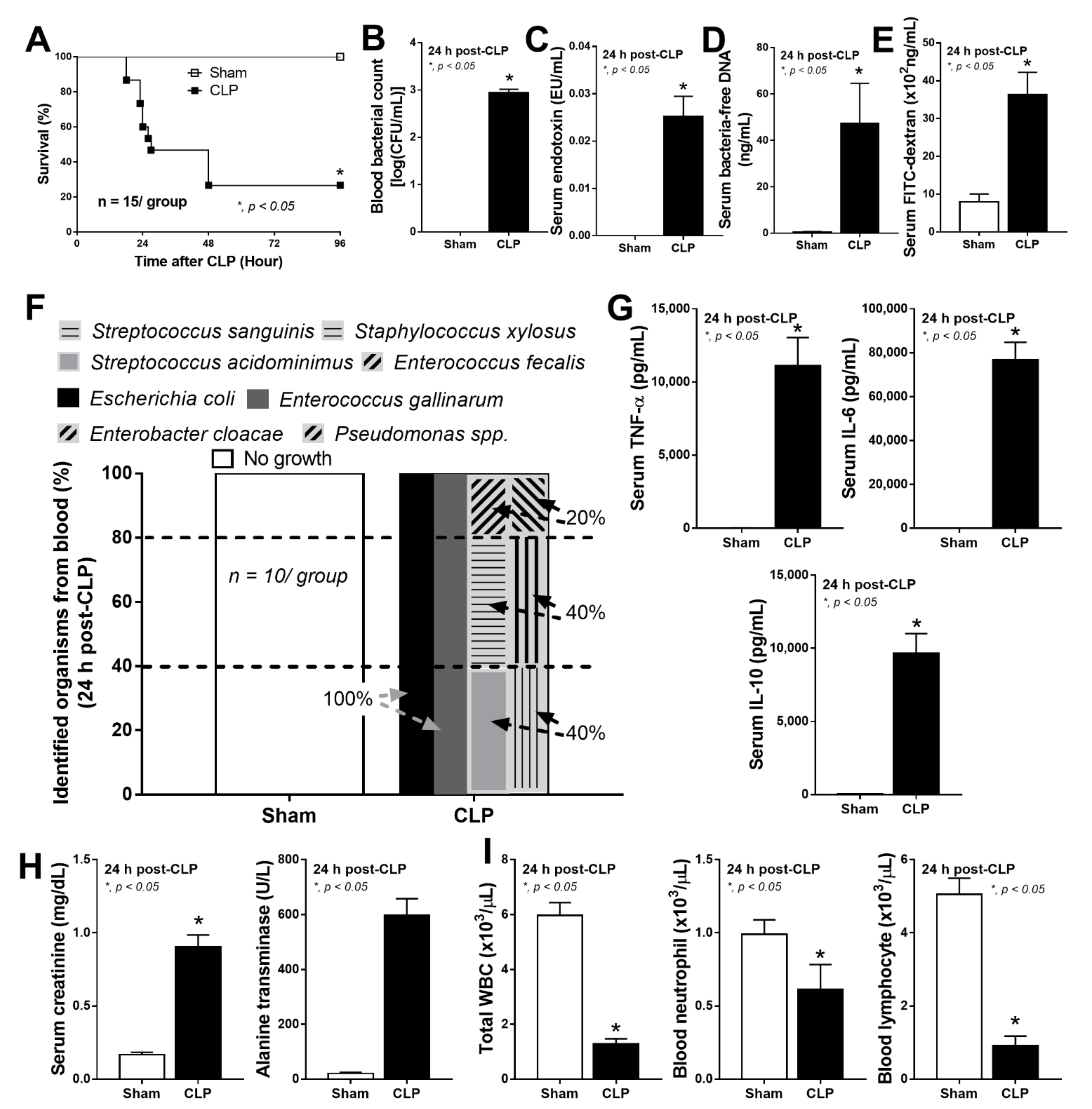
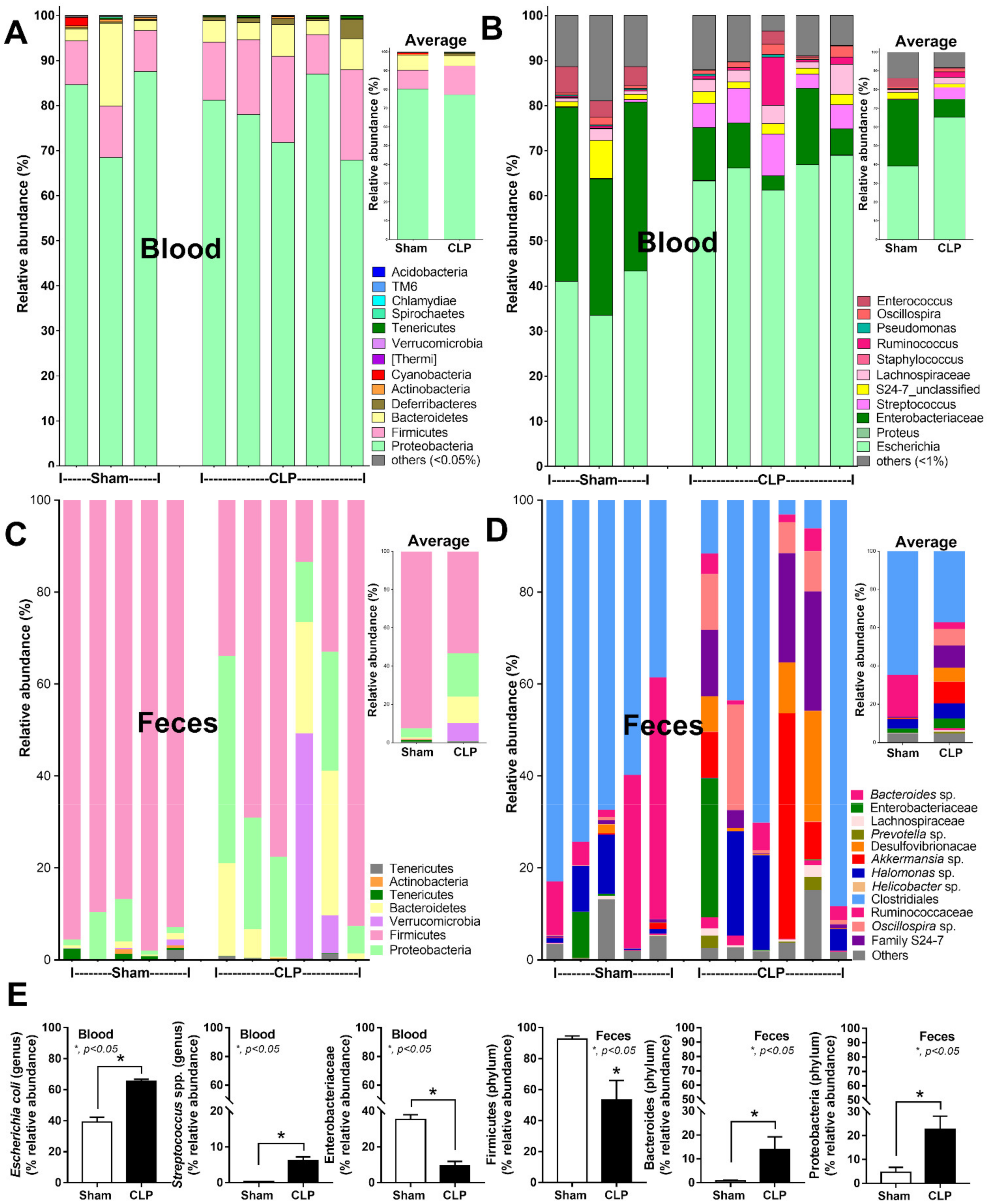
2.1. Bacteria-Free DNA in the Blood of Mice with Cecal Ligation and Puncture Sepsis Was Possibly Derived from the Breakdown of Blood Bacteria and Gut Translocation
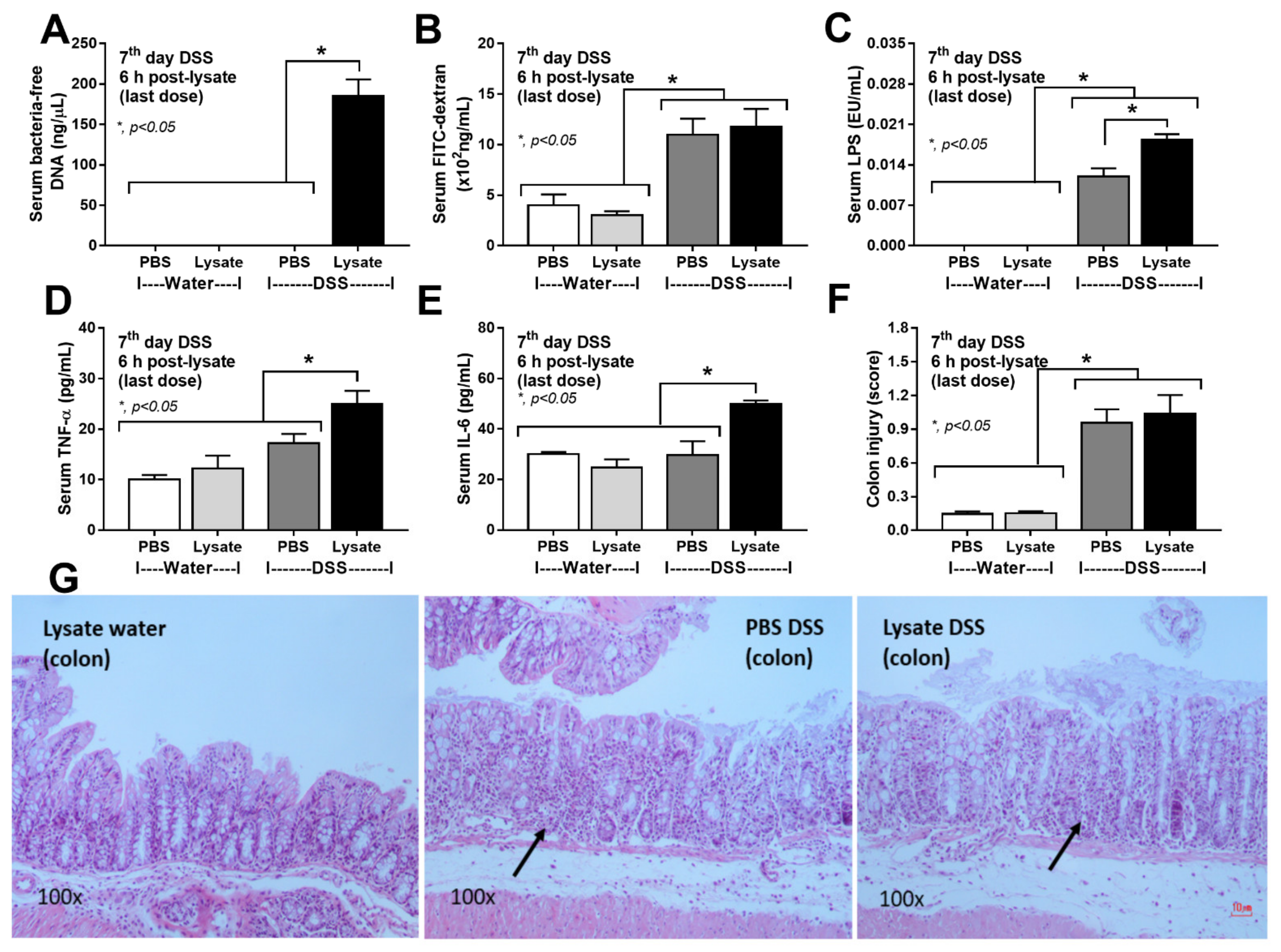
2.2. Bacteria-Free DNA in the Blood of DSS Mice with Oral Gavage by Bacterial Lysate Was Transferred from the Gut Contents
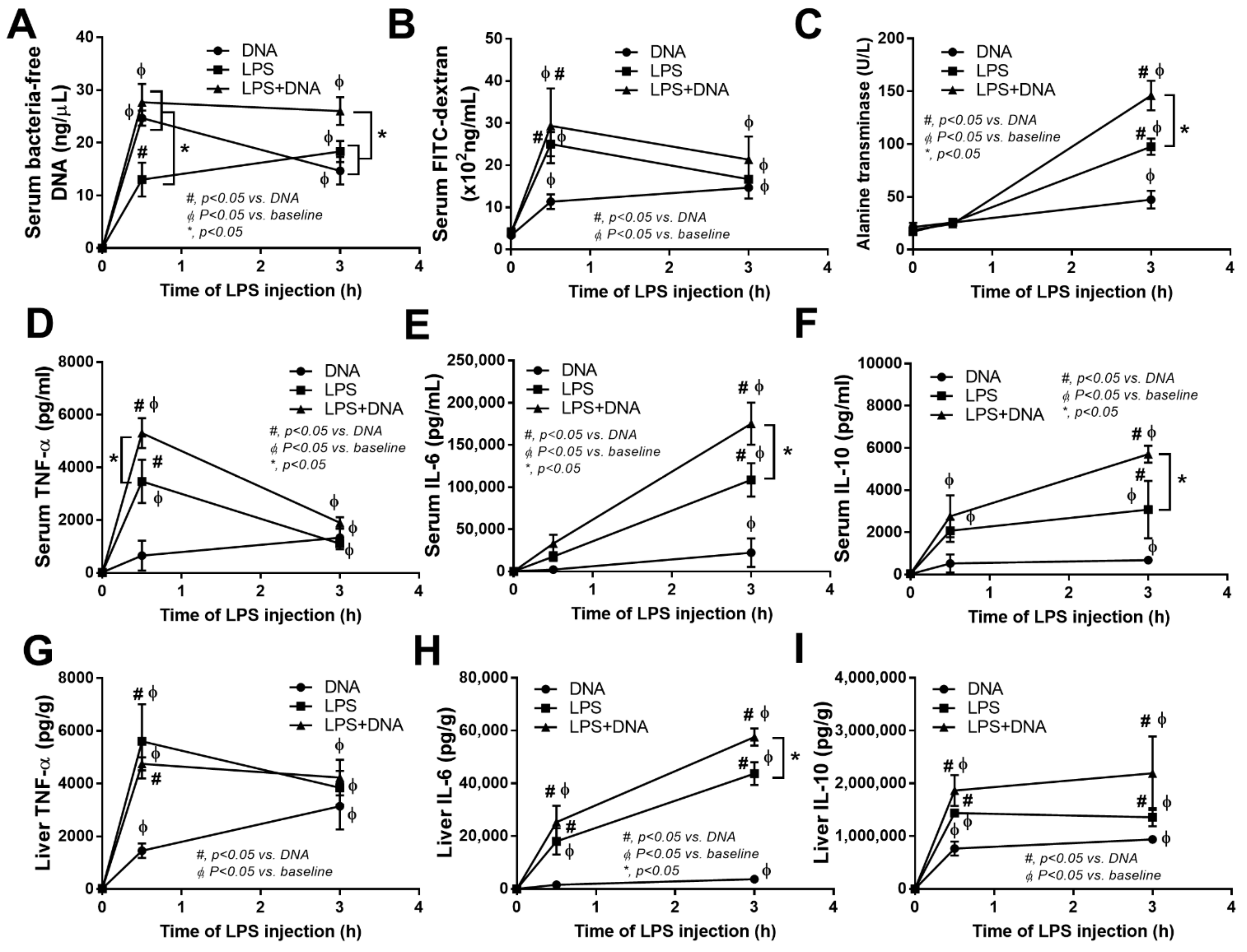
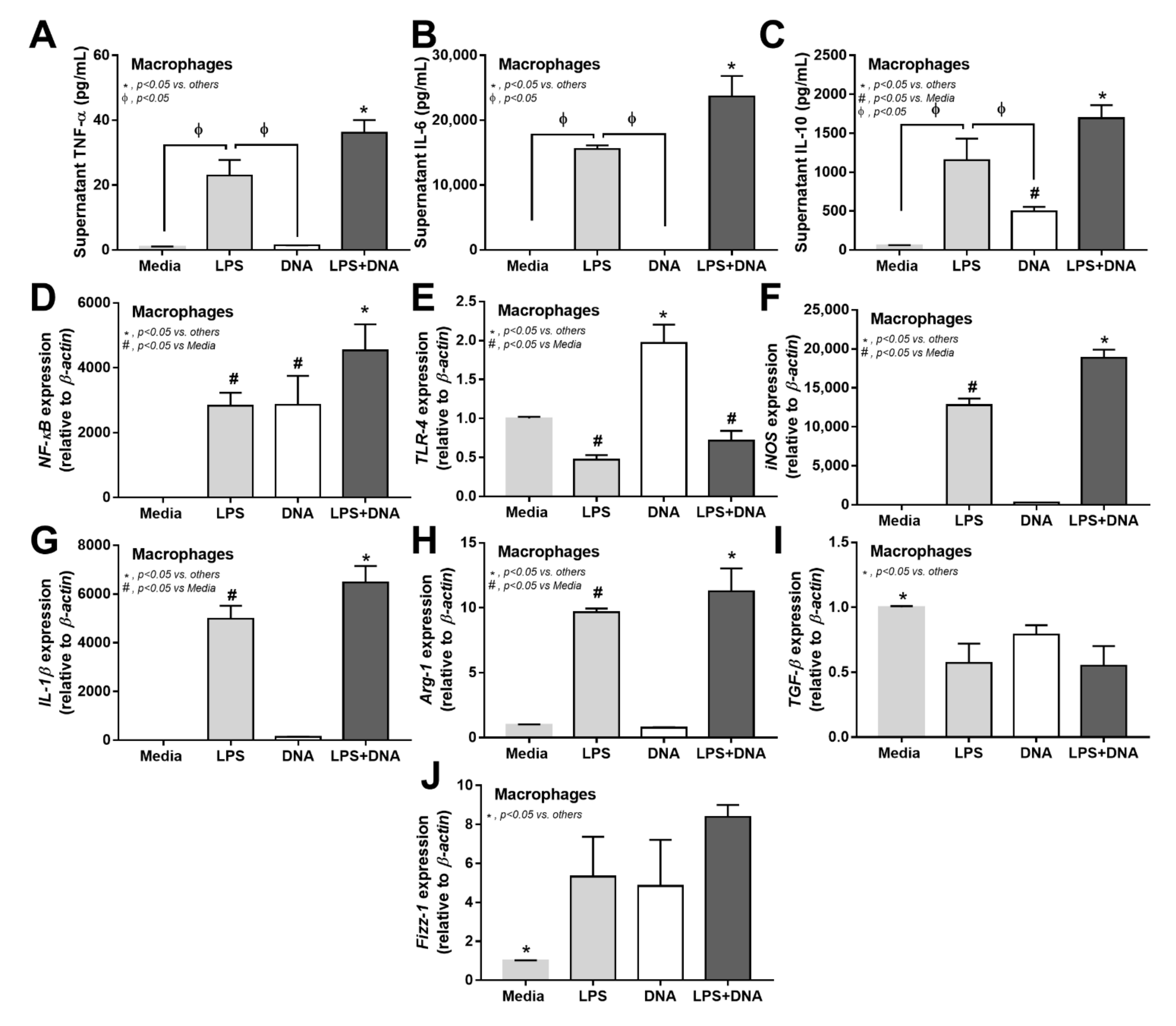
2.3. Additive Inflammatory Effect of Bacteria-Free DNA on LPS Responses in LPS Injection Mouse Model
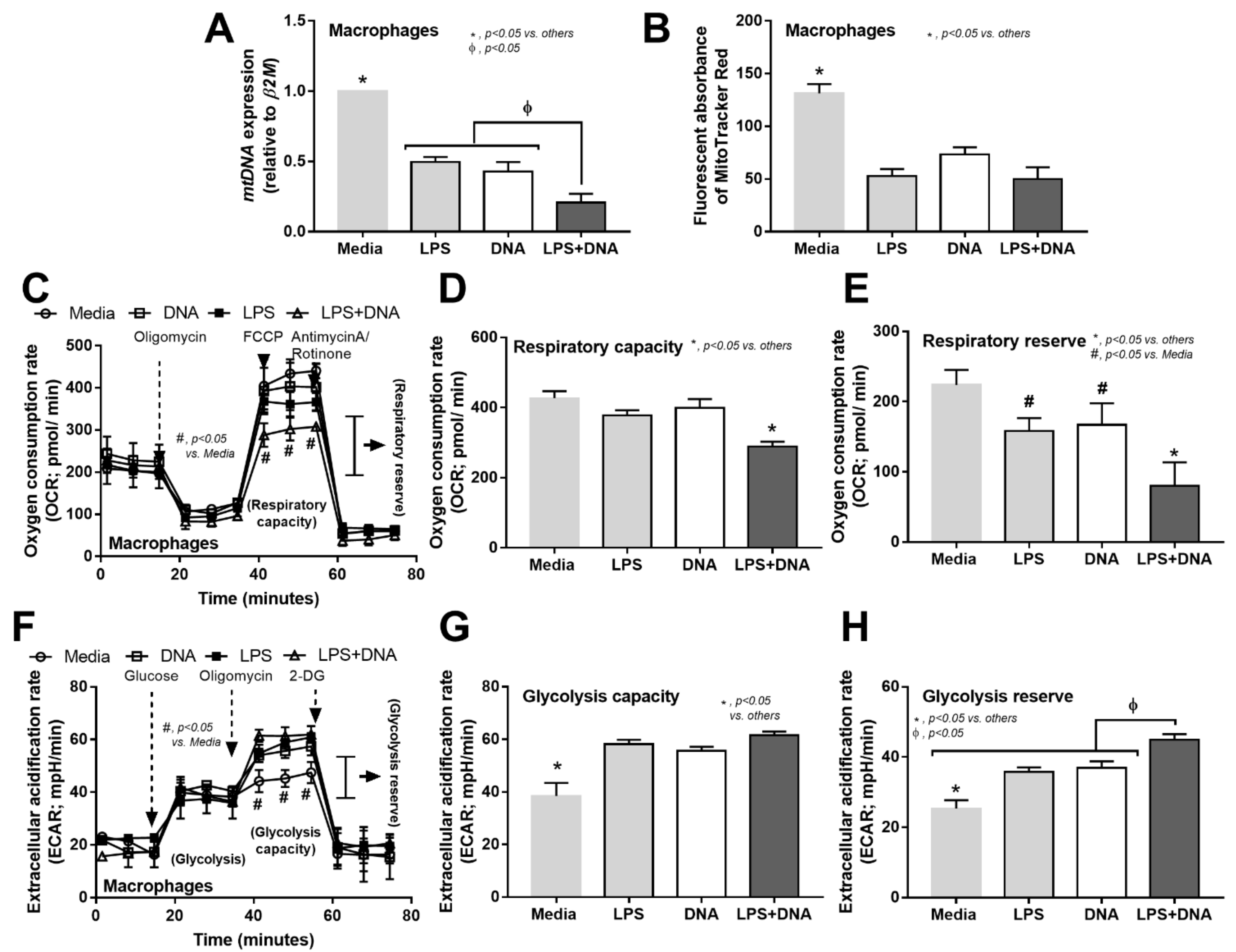
2.4. Additive Inflammatory Effect of Bacteria-Free DNA on LPS Responses, a Possible Impact of Cell Energy Alteration in Macrophages
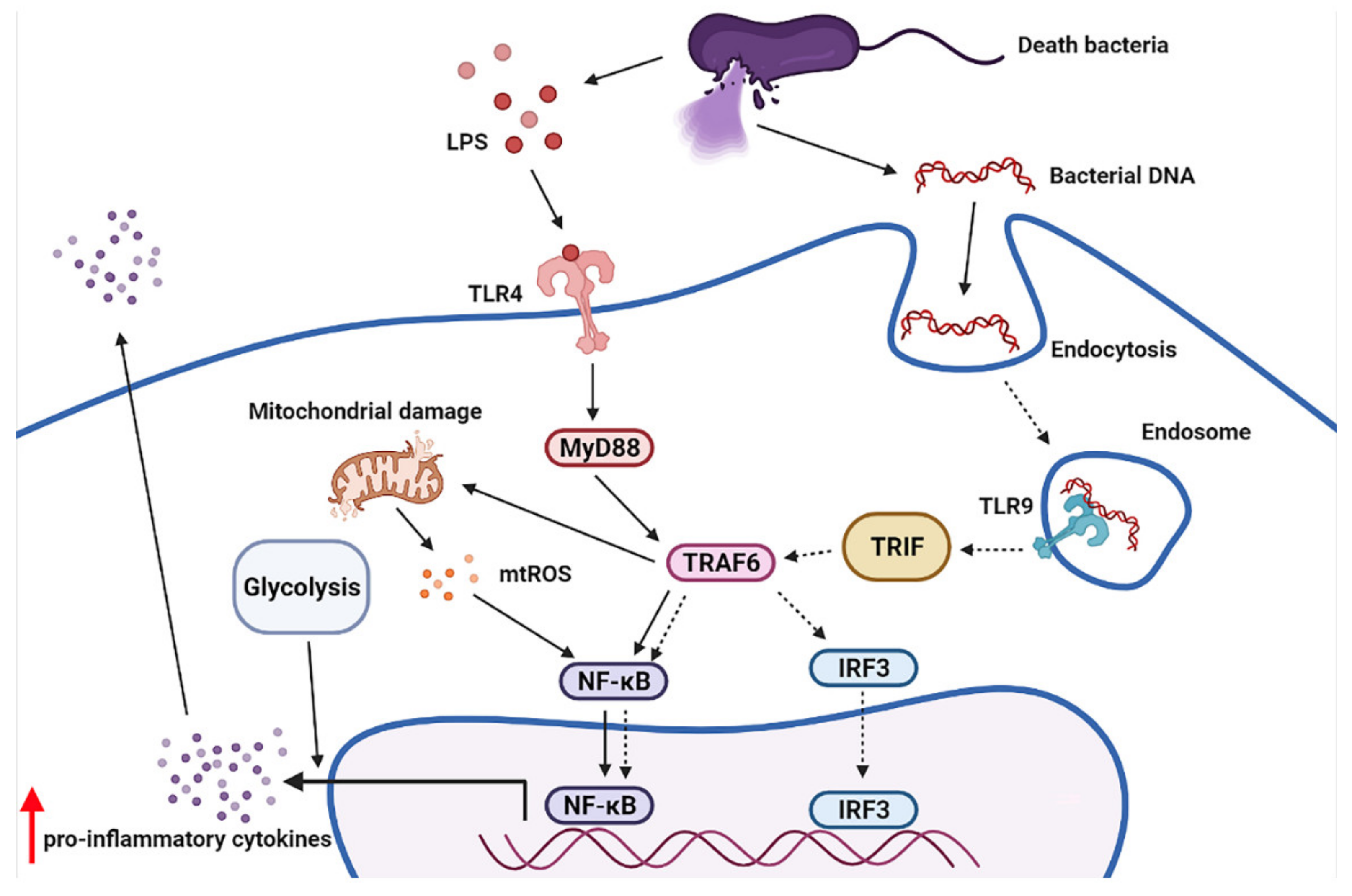
3. Discussion
4. Materials and Methods
4.1. Animal
4.2. Cecal Ligation and Puncture Model and Lipopolysaccharide with and without Bacterial DNA Injection Model
4.3. Dextran Sulfate Solution Model with Bacterial Lysate Administration
4.4. Blood Sample Analysis and Gut Permeability Measurement
4.5. Histology
4.6. Microbiome Analysis of Feces and Blood
4.7. Macrophage Experiments and Inhibition of Endosomal Acidification
4.8. Mitochondrial Evaluation and Extracellular Flux Analysis
4.9. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Singer, M.; Deutschman, C.S.; Seymour, C.W.; Shankar-Hari, M.; Annane, D.; Bauer, M.; Bellomo, R.; Bernard, G.R.; Chiche, J.D.; Coopersmith, C.M.; et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA 2016, 315, 801–810. [Google Scholar] [CrossRef] [PubMed]
- Rajaee, A.; Barnett, R.; Cheadle, W.G. Pathogen- and Danger-Associated Molecular Patterns and the Cytokine Response in Sepsis. Surg. Infect. 2018, 19, 107–116. [Google Scholar] [CrossRef]
- Hotchkiss, R.S.; Moldawer, L.L.; Opal, S.M.; Reinhart, K.; Turnbull, I.R.; Vincent, J.L. Sepsis and septic shock. Nat. Rev. Dis. Primers 2016, 2, 16045. [Google Scholar] [CrossRef] [PubMed]
- Mosser, D.M.; Hamidzadeh, K.; Goncalves, R. Macrophages and the maintenance of homeostasis. Cell Mol. Immunol. 2021, 18, 579–587. [Google Scholar] [CrossRef] [PubMed]
- Kotas, M.E.; Matthay, M.A. Mesenchymal stromal cells and macrophages in sepsis: New insights. Eur. Respir. J. 2018, 51, 1800510. [Google Scholar] [CrossRef]
- O’Dwyer, M.J.; Starczewska, M.H.; Schrenzel, J.; Zacharowski, K.; Ecker, D.J.; Sampath, R.; Brealey, D.; Singer, M.; Libert, N.; Wilks, M.; et al. The detection of microbial DNA but not cultured bacteria is associated with increased mortality in patients with suspected sepsis—A prospective multi-centre European observational study. Clin. Microbiol. Infect. 2017, 23, 208.e1–208.e6. [Google Scholar] [CrossRef]
- Ziegler, I.; Cajander, S.; Rasmussen, G.; Ennefors, T.; Molling, P.; Stralin, K. High nuc DNA load in whole blood is associated with sepsis, mortality and immune dysregulation in Staphylococcus aureus bacteraemia. Infect. Dis. 2019, 51, 216–226. [Google Scholar] [CrossRef]
- Christaki, E.; Giamarellos-Bourboulis, E.J. The complex pathogenesis of bacteremia: From antimicrobial clearance mechanisms to the genetic background of the host. Virulence 2014, 5, 57–65. [Google Scholar] [CrossRef]
- Gutierrez, J.; Guimaraes, A.O.; Lewin-Koh, N.; Berhanu, A.; Xu, M.; Cao, Y.; Kim, J.; Yan, D.; Chang, J.K.; Dinoso, J.B.; et al. Sustained circulating bacterial deoxyribonucleic acid is associated with complicated staphylococcus aureus bacteremia. In Open Forum Infectious Diseases; Oxford University Press: Cambridge, MA, USA, 2019. [Google Scholar]
- Assimakopoulos, S.F.; Triantos, C.; Thomopoulos, K.; Fligou, F.; Maroulis, I.; Marangos, M.; Gogos, C.A. Gut-origin sepsis in the critically ill patient: Pathophysiology and treatment. Infection 2018, 46, 751–760. [Google Scholar] [CrossRef]
- Deitch, E.A. Gut-origin sepsis: Evolution of a concept. Surgeon 2012, 10, 350–356. [Google Scholar] [CrossRef]
- Fasano, A. All disease begins in the (leaky) gut: Role of zonulin-mediated gut permeability in the pathogenesis of some chronic inflammatory diseases. F1000Research 2020, 9, 69. [Google Scholar] [CrossRef]
- Gargari, G.; Mantegazza, G.; Taverniti, V.; Del Bo, C.; Bernardi, S.; Andres-Lacueva, C.; González-Domínguez, R.; Kroon, P.A.; Winterbone, M.S.; Cherubini, A.; et al. Bacterial DNAemia is associated with serum zonulin levels in older subjects. Sci. Rep. 2021, 11, 11054. [Google Scholar] [CrossRef]
- Sirivongrangson, P.; Kulvichit, W.; Payungporn, S.; Pisitkun, T.; Chindamporn, A.; Peerapornratana, S.; Pisitkun, P.; Chitcharoen, S.; Sawaswong, V.; Worasilchai, N.; et al. Endotoxemia and circulating bacteriome in severe COVID-19 patients. Intensive Care Med. Exp. 2020, 8, 72. [Google Scholar] [CrossRef]
- Lindahl, T. Instability and decay of the primary structure of DNA. Nature 1993, 362, 709–715. [Google Scholar] [CrossRef]
- Nielsen, K.M.; Johnsen, P.J.; Bensasson, D.; Daffonchio, D. Release and persistence of extracellular DNA in the environment. Environ. Biosaf. Res. 2007, 6, 37–53. [Google Scholar] [CrossRef]
- Overballe-Petersen, S.; Harms, K.; Orlando, L.A.; Mayar, J.V.M.; Rasmussen, S.; Dahl, T.W.; Rosing, M.T.; Poole, A.M.; Sicheritz-Ponten, T.; Brunak, S.; et al. Bacterial natural transformation by highly fragmented and damaged DNA. Proc. Natl. Acad. Sci. USA 2013, 110, 19860–19865. [Google Scholar] [CrossRef]
- Willerslev, E.; Hansen, A.J.; Rønn, R.; Brand, T.B.; Barnes, I.; Wiuf, C.; Gilichinsky, D.; Mitchell, D.; Cooper, A. Long-term persistence of bacterial DNA. Curr Biol. 2004, 14, R9–R10. [Google Scholar] [CrossRef]
- Jann, B.; Reske, K.; Jann, K. Hetergeniety of Lipopolysachhride, Analysis of polysaccharide chain length by sodium dodacyl gel electrophoriesis. Eur. J. Biochem. 1975, 60, 239–246. [Google Scholar] [CrossRef]
- Leelahavanichkul, A.; Worasilchai, N.; Wannalerdsakun, S.; Jutivorakool, K.; Somparn, P.; Issara-Amphorn, J.; Tachaboon, S.; Srisawat, N.; Finkelman, M.; Chindamporn, A. Gastrointestinal Leakage Detected by Serum (1-->3)-beta-D-Glucan in Mouse Models and a Pilot Study in Patients with Sepsis. Shock 2016, 46, 506–518. [Google Scholar] [CrossRef]
- Amornphimoltham, P.; Yuen, P.S.T.; Star, R.A.; Leelahavanichkul, A. Gut Leakage of Fungal-Derived Inflammatory Mediators: Part of a Gut-Liver-Kidney Axis in Bacterial Sepsis. Dig Dis Sci. 2019, 64, 2416–2428. [Google Scholar] [CrossRef]
- Schrenzel, J. PCR for the diagnosis of sepsis: Hope or hype? Crit. Care 2011, 15, 111. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Struelens, M.J. Detection of Microbial DNAemia: Does it Matter for Sepsis Management? Intensive Care Med. 2010, 36, 193–195. [Google Scholar] [CrossRef][Green Version]
- Cheng, Z.; Abrams, S.T.; Austin, J.; Toh, J.; Wang, S.S.; Wang, Z.; Yu, Q.; Yu, W.; Toh, C.H.; Wang, G. The Central Role and Possible Mechanisms of Bacterial DNAs in Sepsis Development. Mediat. Inflamm. 2020, 2020, 7418342. [Google Scholar] [CrossRef] [PubMed]
- Gosiewski, T.; Ludwig-Galezowska, A.H.; Huminska, K.; Sroka-Oleksiak, A.; Radkowski, P.; Salamon, D.; Wojciechowicz, J.; Kus-Slowinska, M.; Bulanda, M.; Wolkow, P.P. Comprehensive detection and identification of bacterial DNA in the blood of patients with sepsis and healthy volunteers using next-generation sequencing method-the observation of DNAemia. Eur. J. Clin. Microbiol. Infect. Dis. 2017, 36, 329–336. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Li, S.; Li, W.; Ren, J.; Sun, F.; Liu, R.; Zhou, X.J. Rapid diagnosis and comprehensive bacteria profiling of sepsis based on cell-free DNA. J. Transl. Med. 2020, 18, 5. [Google Scholar] [CrossRef]
- Prins, J.M.; van Deventer, S.J.; Kuijper, E.J.; Speelman, P. Clinical relevance of antibiotic-induced endotoxin release. Antimicrob. Agents Chemother. 1994, 38, 1211–1218. [Google Scholar] [CrossRef] [PubMed]
- Hamaguchi, S.; Akeda, Y.; Yamamoto, N.; Seki, M.; Yamamoto, K.; Oishi, K.; Tomono, K. Origin of Circulating Free DNA in Sepsis: Analysis of the CLP Mouse Model. Mediat. Inflamm. 2015, 2015, 614518. [Google Scholar] [CrossRef]
- Rimmele, T.; Kellum, J.A. Clinical review: Blood purification for sepsis. Crit. Care 2011, 15, 205. [Google Scholar] [CrossRef]
- Lilleri, D.; Gerna, G.; Furione, M.; Bernardo, M.E.; Giorgiani, G.; Telli, S.; Baldanti, F.; Locatelli, F. Use of a DNAemia cut-off for monitoring human cytomegalovirus infection reduces the number of preemptively treated children and young adults receiving hematopoietic stem-cell transplantation compared with qualitative pp65 antigenemia. Blood J. Am. Soc. Hematol. 2007, 110, 2757–2760. [Google Scholar] [CrossRef]
- Liao, W.; Zuo, X.; Lin, G.; Zhou, Y.; Fu, Y.; Cai, S.; Wei, P.P.; Liu, Y.X.; Liu, Y.; Ma, G.; et al. Microbial cell-free DNA in plasma of patients with sepsis: A potential diagnostic methodology. Discov. Med. 2020, 29, 129–137. [Google Scholar]
- Phatadon, S.; Win, K.; Sunchai, P.; Trairak, P.; Ariya, C.; Sadudee, P.; Prapaporn, P.; Suwalak, C.; Vorthon, S.; Navaporn, W.; et al. Endotoxemia and circulating bacteriome in severe COVID-19 patients. Intensive Care Med. Exp. 2020, 8, 1–15. [Google Scholar]
- Huang, M.; Cai, S.; Su, J. The Pathogenesis of Sepsis and Potential Therapeutic Targets. Int. J. Mol. Sci. 2019, 20, 5376. [Google Scholar] [CrossRef] [PubMed]
- Cornélie, S.; Wiel, E.; Lund, N.; Lebuffe, G.; Vendeville, C.; Riveau, G.; Vallet, B.; Ban, E. Cytosine-phosphate-guanine (CpG) motifs are sensitizing agents for lipopolysaccharide in toxic shock model. Intensive Care Med. 2002, 28, 1340–1347. [Google Scholar] [CrossRef] [PubMed]
- Yi, A.-K.; Yoon, J.-G.; Hong, S.-C.; Redford, T.W.; Krieg, A.M. Lipopolysaccharide and CpG DNA synergize for tumor necrosis factor-α production through activation of NF-κB. Int. Immunol. 2001, 13, 1391–1404. [Google Scholar] [CrossRef]
- Sparwasser, T.; Miethke, T.; Lipford, G.; Borschert, K.; Häcker, H.; Heeg, K.; Wagner, H. Bacterial DNA causes septic shock. Nature 1997, 386, 336–337. [Google Scholar] [CrossRef]
- Roberts, T.L.; Dunn, J.A.; Terry, T.D.; Jennings, M.P.; Hume, D.A.; Sweet, M.J.; Stacey, K.J. Differences in macrophage activation by bacterial DNA and CpG-containing oligonucleotides. J. Immunol. 2005, 175, 3569–3576. [Google Scholar] [CrossRef]
- Miyake, K.; Onji, M. Endocytosis-free DNA sensing by cell surface TLR 9 in neutrophils: Rapid defense with autoimmune risks. Eur. J. Immunol. 2013, 43, 2006–2009. [Google Scholar] [CrossRef]
- Behnen, M.; Leschczyk, C.; Möller, S.; Batel, T.; Klinger, M.; Solbach, W.; Laskay, T. Immobilized immune complexes induce neutrophil extracellular trap release by human neutrophil granulocytes via FcgammaRIIIB and Mac-1. J. Immunol. 2014, 193, 1954–1965. [Google Scholar] [CrossRef]
- Margolis, S.R.; Wilson, S.C.; Vance, R.E. Evolutionary Origins of cGAS-STING Signaling. Trends Immunol. 2017, 38, 733–743. [Google Scholar] [CrossRef]
- Zhao, Q.; Wei, Y.; Pandol, S.J.; Li, L.; Habtezion, A. STING Signaling Promotes Inflammation in Experimental Acute Pancreatitis. Gastroenterology 2018, 154, 1822–1835.e2. [Google Scholar] [CrossRef]
- Torralba, D.; Baixauli, F.; Villarroya Beltri, C.; Fernández Delgado, I.; Latorre Pellicer, A. Priming of dendritic cells by DNA containing extracellular vesicles from activated T cells through antigen driven con tacts. Nat. Commun. 2018, 9, 2658. [Google Scholar] [CrossRef]
- Roberts, P.J.; Segal, A.W. The digestion of bacterial macromolecules by phagocytic cells: The effect of mepacrine and ethanol. Immunology 1987, 62, 581–586. [Google Scholar]
- Ablasser, A.; Gulen, M.F. The role of cGAS in innate immunity and beyond. J. Mol. Med. 2016, 94, 1085–1093. [Google Scholar] [CrossRef]
- Celhar, T.; Pereira-Lopes, S.; Thornhill, S.I.; Lee, H.Y.; Dhillon, M.K.; Poidinger, M.; Connolly, J.E.; Lim, L.H.; Biswas, S.K.; Fairhurst, A.M. TLR7 and TLR9 ligands regulate antigen presentation by macrophages. Int. Immunol. 2016, 28, 223–232. [Google Scholar] [CrossRef]
- El-Sayed, O.M.; Dewyer, N.A.; Luke, C.E.; Elfline, M.; Laser, A.; Hogaboam, C.; Kunkel, S.L.; Henke, P.K. Intact Toll-like receptor 9 signaling in neutrophils modulates normal thrombogenesis in mice. J. Vasc. Surg. 2016, 64, 1450–1458.e1. [Google Scholar] [CrossRef]
- Roda, J.M.; Parihar, R.; Carson, W.E. CpG-containing oligodeoxynucleotides act through TLR9 to enhance the NK cell cytokine response to antibody-coated tumor cells. J. Immunol. 2005, 175, 1619–1627. [Google Scholar] [CrossRef]
- Sacramento, L.; Trevelin, S.C.; Nascimento, M.S.; Lima-Jùnior, D.S.; Costa, D.L.; Almeida, R.P.; Cunha, F.Q.; Silva, J.S.; Carregaro, V. Toll-like receptor 9 signaling in dendritic cells regulates neutrophil recruitment to inflammatory foci following Leishmania infantum infection. Infect. Immun. 2015, 83, 4604–4616. [Google Scholar] [CrossRef]
- Gao, J.J.; Xue, Q.; Papasian, C.J.; Morrison, D.C. Bacterial DNA and lipopolysaccharide induce synergistic production of TNF-α through a post-transcriptional mechanism. J. Immunol. 2001, 166, 6855–6860. [Google Scholar] [CrossRef]
- Gao, J.J.; Zuvanich, E.G.; Xue, Q.; Horn, D.L.; Silverstein, R.; Morrison, D.C. Cutting edge: Bacterial DNA and LPS act in synergy in inducing nitric oxide production in RAW 264.7 macrophages. J. Immunol. 1999, 163, 4095–4099. [Google Scholar]
- Doi, K.; Leelahavanichkul, A.; Yuen, P.S.; Star, R.A. Animal models of sepsis and sepsis-induced kidney injury. J. Clin. Investig. 2009, 119, 2868–2878. [Google Scholar] [CrossRef]
- Han, C.; Li, G.; Lim, K.; DeFrances, M.C.; Gandhi, C.R.; Wu, T. Transgenic expression of cyclooxygenase-2 in hepatocytes accelerates endotoxin-induced acute liver failure. J. Immunol. 2008, 181, 8027–8035. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Zanoni, I.; Cullen, T.W.; Goodman, A.L.; Kagan, J.C. Mechanisms of Toll-like receptor 4 endocytosis reveal a common immune-evasion strategy used by pathogenic and commensal bacteria. Immunity 2015, 43, 909–922. [Google Scholar] [CrossRef]
- Nedeva, C. Inflammation and Cell Death of the Innate and Adaptive Immune System during Sepsis. Biomolecules 2021, 11, 1011. [Google Scholar] [CrossRef]
- Jaroonwitchawan, T.; Visitchanakun, P.; Dang, P.C.; Ritprajak, P.; Palaga, T.; Leelahavanichkul, A. Dysregulation of lipid metabolism in macrophages is responsible for severe endotoxin tolerance in FcgRIIB-deficient lupus mice. Front. Immunol. 2020, 11, 959. [Google Scholar] [CrossRef] [PubMed]
- Dang, C.P.; Issara-Amphorn, J.; Charoensappakit, A.; Udompornpitak, K.; Bhunyakarnjanarat, T.; Saisorn, W.; Sae-Khow, K.; Leelahavanichkul, A. BAM15, a Mitochondrial Uncoupling Agent, Attenuates Inflammation in the LPS Injection Mouse Model: An Adjunctive Anti-Inflammation on Macrophages and Hepatocytes. J. Innate Immun. 2021, 19, 359–375. [Google Scholar] [CrossRef] [PubMed]
- Issara-Amphorn, J.; Dang, C.P.; Saisorn, W.; Limbutara, K.; Leelahavanichkul, A. Candida Administration in Bilateral Nephrectomy Mice Elevates Serum (1→ 3)-β-D-glucan That Enhances Systemic Inflammation Through Energy Augmentation in Macrophages. Int. J. Mol. Sci. 2021, 22, 5031. [Google Scholar] [CrossRef] [PubMed]
- Ivory, C.P.; Prystajecky, M.; Jobin, C.; Chadee, K. Toll-like receptor 9-dependent macrophage activation by Entamoeba histolytica DNA. Infect. Immun. 2008, 76, 289–297. [Google Scholar] [CrossRef]
- Kiemer, A.K.; Senaratne, R.H.; Hoppstädter, J.; Diesel, B.; Riley, L.W.; Tabeta, K.; Bauer, S.; Beutler, B.; Zuraw, B.L. Attenuated activation of macrophage TLR9 by DNA from virulent mycobacteria. J. Innate Immun. 2009, 1, 29–45. [Google Scholar] [CrossRef]
- Rother, N.; Yanginlar, C.; Lindeboom, R.G.; Bekkering, S.; van Leent, M.M.; Buijsers, B.; Jonkman, I.; de Graaf, M.; Baltissen, M.; Lamers, L.A.; et al. Hydroxychloroquine Inhibits the Trained Innate Immune Response to Interferons. Cell Rep. Med. 2020, 1, 100146. [Google Scholar] [CrossRef]
- Jang, C.; Choi, J.; Byun, M.; Jue, D. Chloroquin inhibits production of TNFalpha, interleukin-1 and interleukin-6 from lipopolysaccharide-stimulated human monocytes/macrophages by different modes. Rheumatology 2006, 45, 703–710. [Google Scholar] [CrossRef]
- Redmann, M.; Benavides, G.A.; Berryhill, T.F.; Wani, W.Y.; Ouyang, X.; Johnson, M.S.; Ravi, S.; Barnes, S.; Darley-Usmar, V.M.; Zhang, J. Inhibition of autophagy with bafilomycin and chloroquine decreases mitochondrial quality and bioenergetic function in primary neurons. Redox Biol. 2017, 11, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Ochman, H.; Caro, A. Genome Size and Structure, Bacterial; Elsevier: Amsterdam, The Netherlands, 2016. [Google Scholar]
- Williams, D.; Trimble, W.L.; Shilts, M.; Meyer, F.; Ochman, H. Rapid quantification of sequence repeats to resolve the size, structure and contents of bacterial genomes. BMC Genom. 2013, 14, 537. [Google Scholar] [CrossRef] [PubMed]
- Bellot, P.; Frances, R.; Such, J. Pathological bacterial translocation in cirrhosis: Pathophysiology, diagnosis and clinical implications. Liver Int. 2013, 33, 31–39. [Google Scholar] [CrossRef]
- Rodríguez-Laiz, G.P.; Zapater, P.; Melgar, P.; Alcázar, C.; Franco, M.; Giménez, P.; Pascual, S.; Bellot, P.; Palazón, J.M.; Rodríguez, M.; et al. Bacterial DNA translocation contributes to systemic inflammation and to minor changes in the clinical outcome of liver transplantation. Sci. Rep. 2019, 9, 835. [Google Scholar] [CrossRef]
- Remick, D.G.; Newcomb, D.E.; Bolgos, G.L.; Call, D.R. Comparison of the mortality and inflammatory response of two models of sepsis: Lipopolysaccharide vs. cecal ligation and puncture. Shock 2000, 13, 110–116. [Google Scholar] [CrossRef]
- Seemann Zohles, F.; Lupp, A. Comprehensive comparison of three different animal models for systemic inflammation. J. Biomed. Sci. 2017, 24, 60. [Google Scholar] [CrossRef]
- Vaiserman, A.; Romanenko, M.; Piven, L.; Moseiko, V.; Lushchak, O.; Kryzhanovska, N.; Guryanov, V.; Koliada, A. Differences in the gut Firmicutes to Bacteroidetes ratio across age groups in healthy Ukrainian population. BMC Microbiol. 2020, 20, 221. [Google Scholar] [CrossRef]
- Baldelli, V.; Scaldaferri, F.; Putignani, L.; Del Chierico, F. The Role of Enterobacteriaceae in Gut Microbiota Dysbiosis in Inflammatory Bowel Diseases. Microorganisms 2021, 9, 697. [Google Scholar] [CrossRef] [PubMed]
- Camilleri, M. Leaky gut: Mechanisms, measurement and clinical implications in humans. Gut 2019, 68, 1516–1526. [Google Scholar] [CrossRef] [PubMed]
- Panpetch, W.; Somboonna, N.; Palasuk, M.; Hiengrach, P.; Finkelman, M.; Tumwasorn, S.; Leelahavanichkul, A. Oral Candida administration in a Clostridium difficile mouse model worsens disease severity but is attenuated by Bifidobacterium. PLoS ONE 2019, 14, e0210798. [Google Scholar] [CrossRef] [PubMed]
- Udompornpitak, K.; Bhunyakarnjanarat, T.; Charoensappakit, A.; Dang, C.P.; Saisorn, W.; Leelahavanichkul, A. Lipopolysaccharide-Enhanced Responses against Aryl Hydrocarbon Receptor in FcgRIIb-Deficient Macrophages, a Profound Impact of an Environmental Toxin on a Lupus-Like Mouse Model. Int. J. Mol. Sci. 2021, 22, 4199. [Google Scholar] [CrossRef]
- Nighot, M.; Al-Sadi, R.; Guo, S.; Rawat, M.; Nighot, P.; Watterson, M.D.; Ma, T.Y. Lipopolysaccharide-Induced Increase in Intestinal Epithelial Tight Permeability Is Mediated by Toll-Like Receptor 4/Myeloid Differentiation Primary Response 88 (MyD88) Activation of Myosin Light Chain Kinase Expression. Am. J. Pathol. 2017, 187, 2698–2710. [Google Scholar] [CrossRef]
- Visitchanakun, P.; Kaewduangduen, W.; Chareonsappakit, A.; Susantitaphong, P.; Pisitkun, P.; Ritprajak, P.; Townamchai, N.; Leelahavanichkul, A. Interference on Cytosolic DNA Activation Attenuates Sepsis Severity: Experiments on Cyclic GMP–AMP Synthase (cGAS) Deficient Mice. Int. J. Mol. Sci. 2021, 22, 11450. [Google Scholar] [CrossRef]
- Cao, Y.; Zhang, X.; Shang, W.; Xu, J.; Wang, X.; Hu, X.; Ao, Y.; Cheng, H. Proinflammatory Cytokines Stimulate Mitochondrial Superoxide Flashes in Articular Chondrocytes In Vitro and In Situ. PLoS ONE 2013, 8, e66444. [Google Scholar] [CrossRef] [PubMed]
- Naik, E.; Dixit, V.M. Mitochondrial reactive oxygen species drive proinflammatory cytokine production. J. Exp. Med. 2011, 208, 417–420. [Google Scholar] [CrossRef]
- Busani, S.; De Biasi, S.; Nasi, M.; Paolini, A.; Venturelli, S.; Tosi, M.; Girardis, M.; Cossarizza, A. Increased Plasma Levels of Mitochondrial DNA and Normal Inflammasome Gene Expression in Monocytes Characterize Patients with Septic Shock Due to Multidrug Resistant Bacteria. Front. Immunol. 2020, 11, 768. [Google Scholar] [CrossRef]
- Timmermans, K.; Kox, M.; Scheffer, G.J.; Pickkers, P. Plasma Nuclear and Mitochondrial DNA Levels, and Markers of Inflammation, Shock, and Organ Damage in Patients with Septic Shock. Shock 2016, 45, 607–612. [Google Scholar] [CrossRef]
- Qiu, P.; Liu, Y.; Zhang, J. Review: The Role and Mechanisms of Macrophage Autophagy in Sepsis. Inflammation 2019, 42, 6–19. [Google Scholar] [CrossRef]
- Soto-Heredero, G.; Gomez de Las Heras, M.M.; Gabande-Rodriguez, E.; Oller, J.; Mittelbrunn, M. Glycolysis—A key player in the inflammatory response. FEBS J. 2020, 287, 3350–3369. [Google Scholar] [CrossRef]
- Pålsson-McDermott, E.M.; O’Neill, L.A. Targeting immunometabolism as an anti-inflammatory strategy. Cell Res. 2020, 30, 300–314. [Google Scholar] [CrossRef]
- Hu, D.; Yang, X.; Xiang, Y.; Li, H.; Yan, H.; Zhou, J.; Caudle, Y.; Zhang, X.; Yin, D. Inhibition of Toll-like receptor 9 attenuates sepsis-induced mortality through suppressing excessive inflammatory response. Cell. Immunol. 2015, 295, 92–98. [Google Scholar] [CrossRef]
- De Madaria, E.; Martinez, J.; Lozano, B.; Sempere, L.; Benlloch, S.; Such, J.; Uceda, F.; Frances, R.; Perez-Mateo, M. Detection and identification of bacterial DNA in serum from patients with acute pancreatitis. Gut 2005, 54, 1293–1297. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yasuda, H.; Leelahavanichkul, A.; Tsunoda, S.; Dear, J.W.; Takahashi, Y.; Ito, S.; Hu, X.; Zhou, H.; Doi, K.; Childs, R.; et al. Chloroquine and inhibition of Toll-like receptor 9 protect from sepsis-induced acute kidney injury. Am. J. Physiol. Renal Physiol. 2008, 294, F1050–F1058. [Google Scholar] [CrossRef]
- Cadegiani, F.A.; Goren, A.; Wambier, C.G.; Mc, J. Early COVID-19 therapy with azithromycin plus nitazoxanide, ivermectin or hydroxychloroquine in outpatient settings significantly improved COVID-19 outcomes compared to known outcomes in untreated patients. New Microbes New Infect. 2021, 43, 100915. [Google Scholar] [CrossRef]
- Galan, L.E.B.; Santos, N.M.D.; Asato, M.S.; Araújo, J.V.; De Lima Moreira, A.; Araújo, A.M.M.; Paiva, A.D.P.; Portella, D.G.S.; Marques, F.S.S.; Silva, G.M.A.; et al. Phase 2 randomized study on chloroquine, hydroxychloroquine or ivermectin in hospitalized patients with severe manifestations of SARS-CoV-2 infection. Pathog. Glob. Health 2021, 115, 235–242. [Google Scholar] [CrossRef]
- Gao, W.; Xiong, Y.; Li, Q.; Yang, H. Inhibition of Toll-Like Receptor Signaling as a Promising Therapy for Inflammatory Diseases: A Journey from Molecular to Nano Therapeutics. Front. Physiol. 2017, 8, 508. [Google Scholar] [CrossRef] [PubMed]
- Boom, R.; Sol, C.J.; Heijtink, R.; Wertheim-van Dillen, P.M.; van der Noordaa, J. Rapid purification of hepatitis B virus DNA from serum. J. Clin. Microbiol. 1991, 29, 1804–1811. [Google Scholar] [CrossRef] [PubMed]
- Hadiji-Abbes, N.; Martin, M.; Benzina, W.; Karray-Hakim, H.; Gergely, C.; Gargouri, A.; Mokdad-Gargouri, R. Extraction and purification of hepatitis B virus-like M particles from a recombinant Saccharomyces cerevisiae strain using alumina powder. J. Virol. Methods 2013, 187, 132–137. [Google Scholar] [CrossRef]
- Tullis, R.H.; Duffin, R.P.; Handley, H.H.; Sodhi, P.; Menon, J.; Joyce, J.A.; Kher, V. Reduction of hepatitis C virus using lectin affinity plasmapheresis in dialysis patients. Blood Purif. 2009, 27, 64–69. [Google Scholar] [CrossRef]
- Lu, Y.C.; Yeh, W.C.; Ohashi, P.S. LPS/TLR4 signal transduction pathway. Cytokine 2008, 42, 145–151. [Google Scholar] [CrossRef]
- Suthers, A.N.; Sarantopoulos, S. TLR7/TLR9-and B cell receptor-signaling crosstalk: Promotion of potentially dangerous B cells. Front. Immunol. 2017, 8, 775. [Google Scholar] [CrossRef]
- Lopez-Armada, M.J.; Riveiro-Naveira, R.R.; Vaamonde-Garcia, C.; Valcarcel-Ares, M.N. Mitochondrial dysfunction and the inflammatory response. Mitochondrion 2013, 13, 106–118. [Google Scholar] [CrossRef]
- Doi, K.; Yuen, P.S.; Eisner, C.; Hu, X.; Leelahavanichkul, A.; Schnermann, J.; Star, R.A. Reduced production of creatinine limits its use as marker of kidney injury in sepsis. J. Am. Soc. Nephrol. 2009, 20, 1217–1221. [Google Scholar] [CrossRef]
- Leelahavanichkul, A.; Bocharov, A.V.; Kurlander, R.; Baranova, I.N.; Vishnyakova, T.G.; Souza, A.C.; Hu, X.; Doi, K.; Vaisman, B.; Amar, M.; et al. Class B scavenger receptor types I and II and CD36 targeting improves sepsis survival and acute outcomes in mice. J. Immunol. 2012, 188, 2749–2758. [Google Scholar] [CrossRef]
- Leelahavanichkul, A.; Huang, Y.; Hu, X.; Zhou, H.; Tsuji, T.; Chen, R.; Kopp, J.B.; Schnermann, J.; Yuen, P.S.; Star, R.A. Chronic kidney disease worsens sepsis and sepsis-induced acute kidney injury by releasing High Mobility Group Box Protein-1. Kidney Int. 2011, 80, 1198–1211. [Google Scholar] [CrossRef]
- Leelahavanichkul, A.; Yasuda, H.; Doi, K.; Hu, X.; Zhou, H.; Yuen, P.S.; Star, R.A. Methyl-2-acetamidoacrylate, an ethyl pyruvate analog, decreases sepsis-induced acute kidney injury in mice. Am. J. Physiol. Renal Physiol. 2008, 295, F1825–F1835. [Google Scholar] [CrossRef]
- Németh, K.; Leelahavanichkul, A.; Yuen, P.S.; Mayer, B.; Parmelee, A.; Robey, P.G.; Leelahavanichkul, K.; Koller, B.H.; Brown, J.M.; Hu, X.; et al. Bone marrow stromal cells attenuate sepsis via prostaglandin E 2–dependent reprogramming of host macrophages to increase their interleukin-10 production. Nat. Med. 2009, 15, 42–49. [Google Scholar] [CrossRef]
- Zafrani, L.; Ince, C.; Yuen, P.S. Microparticles During Sepsis: Target, Canary or Cure? Intensive Care Med. 2013, 39, 1854–1856. [Google Scholar] [CrossRef]
- Denayer, T.; Stöhr, T.; Van Roy, M. Animal models in translational medicine: Validation and prediction. New Horiz. Transl. Med. 2014, 2, 5–11. [Google Scholar] [CrossRef]
- Van der Worp, H.B.; Howells, D.W.; Sena, E.S.; Porritt, M.J.; Rewell, S.; O’Collins, V.; Macleod, M.R. Can animal models of disease reliably inform human studies? PLoS Med. 2010, 7, e1000245. [Google Scholar] [CrossRef]
- Chace, J.H.; Hooker, N.A.; Mildenstein, K.L.; Krieg, A.M.; Cowdery, J.S. Bacterial DNA-induced NK cell IFN-γ production is dependent on macrophage secretion of IL-12. Clin. Immunol. Immunopathol. 1997, 84, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Poth, J.M.; Coch, C.; Busch, N.; Boehm, O.; Schlee, M.; Janke, M.; Zillinger, T.; Schildgen, O.; Barchet, W.; Hartmann, G. Monocyte-mediated inhibition of TLR9-dependent IFN-α induction in plasmacytoid dendritic cells questions bacterial DNA as the active ingredient of bacterial lysates. J. Immunol. 2010, 185, 7367–7373. [Google Scholar] [CrossRef] [PubMed]
- Rizzello, V.; Bonaccorsi, I.; Dongarra, M.L.; Fink, L.N.; Ferlazzo, G. Role of natural killer and dendritic cell crosstalk in immunomodulation by commensal bacteria probiotics. J. Biomed. Biotechnol. 2011, 2011, 473097. [Google Scholar] [CrossRef] [PubMed]
- Visitchanakun, P.; Tangtanatakul, P.; Trithiphen, O.; Soonthornchai, W.; Wongphoom, J.; Tachaboon, S.; Srisawat, N.; Leelahavanichkul, A. Plasma miR-370–3p as a biomarker of sepsis-associated encephalopathy, the transcriptomic profiling analysis of microrna-arrays from mouse brains. Shock 2020, 54, 347–357. [Google Scholar] [CrossRef] [PubMed]
- Saithong, S.; Saisorn, W.; Visitchanakun, P.; Sae-khow, K.; Chiewchengchol, D.; Leelahavanichkul, A. A Synergy Between Endotoxin and (1 -> 3)-Beta-D-Glucan Enhanced Neutrophil Extracellular Traps in Candida Administered Dextran Sulfate Solution Induced Colitis in FcGRIIB-/- Lupus Mice, an Impact of Intestinal Fungi in Lupus. J. Inflamm. Res. 2021, 14, 2333–2352. [Google Scholar] [CrossRef]
- Grant, D.M. Detoxification pathways in the liver. J. Inherit. Metab. Dis. 1991, 14, 421–430. [Google Scholar] [CrossRef]
- Panpetch, W.; Hiengrach, P.; Nilgate, S.; Tumwasorn, S.; Somboonna, N.; Wilantho, A.; Chatthanathon, P.; Prueksapanich, P.; Leelahavanichkul, A. Additional Candida albicans administration enhances the severity of dextran sulfate solution induced colitis mouse model through leaky gut-enhanced systemic inflammation and gut-dysbiosis but attenuated by Lactobacillus rhamnosus L34. Gut Microbes 2020, 11, 465–480. [Google Scholar] [CrossRef]
- Li, Q.R.; Wang, C.Y.; Tang, C.; Zhao, X.F.; He, Q.; Li, J.S. Identification and Characterization of Blood and Neutrophil-Associated Microbiomes in Patients with Severe Acute Pancreatitis Using Next-Generation Sequencing. Front. Cell. Infect. Microbiol. 2018, 8, 5. [Google Scholar] [CrossRef]
- Wongpiyabovorn, J.; Soonthornchai, W.; Wilantho, A.; Palasuk, M.; Payungporn, S.; Sodsai, P.; Poomipak, W.; Weschawalit, S.; Ruchusatsawat, K.; Baillie, G.S.; et al. Effect of tarcolimus on skin microbiome in atopic dermatitis. Allergy 2019, 74, 1400–1406. [Google Scholar] [CrossRef]



| Primers | Forward | Reverse |
|---|---|---|
| Nuclear factor-kB (NF-kB) | 5′-CTTCCTCAGCCATGGTACCTCT-3′ | 5′-CAAGTCTTCATCAGCATCAAACTG-3′ |
| Toll like receptor-4 (TLR-4) | 5′-GGCAGCAGGTGGAATTGTAT-3′ | 5′-AGGCCCCAGAGTTTTGTTCT-3′ |
| Inducible nitric oxide synthase (iNOS) | 5′-ACCCACATCTGGCAGAATGAG-3′ | 5′-AGCCATGACCTTTCGCATTAG-3′ |
| Interleukin-1ß (IL-1ß) | 5′-GAAATGCCACCTTTTGACAGTG-3′ | 5′-TGGATGCTCTCATCAGGACAG-3′ |
| Arginase-1 (Arg-1) | 5′-CTTGGCTTGCTTCGGAACTC-3′ | 5′-GGAGAAGGCGTTTGCTTAGTTC-3′ |
| Transforming Growth Factor-β (TGF-β) | 5′-CAGAGCTGCGCTTGCAGAG-3′ | 5′-GTCAGCAGCCGGTTACCAAG-3′ |
| Resistin-like molecule-α (FIZZ-1) | 5′-GCCAGGTCCTGGAACCTTTC-3′ | 5′-GGAGCAGGGAGATGCAGATGA-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kaewduangduen, W.; Visitchanakun, P.; Saisorn, W.; Phawadee, A.; Manonitnantawat, C.; Chutimaskul, C.; Susantitaphong, P.; Ritprajak, P.; Somboonna, N.; Cheibchalard, T.; et al. Blood Bacteria-Free DNA in Septic Mice Enhances LPS-Induced Inflammation in Mice through Macrophage Response. Int. J. Mol. Sci. 2022, 23, 1907. https://doi.org/10.3390/ijms23031907
Kaewduangduen W, Visitchanakun P, Saisorn W, Phawadee A, Manonitnantawat C, Chutimaskul C, Susantitaphong P, Ritprajak P, Somboonna N, Cheibchalard T, et al. Blood Bacteria-Free DNA in Septic Mice Enhances LPS-Induced Inflammation in Mice through Macrophage Response. International Journal of Molecular Sciences. 2022; 23(3):1907. https://doi.org/10.3390/ijms23031907
Chicago/Turabian StyleKaewduangduen, Warerat, Peerapat Visitchanakun, Wilasinee Saisorn, Ariya Phawadee, Charintorn Manonitnantawat, Chirapas Chutimaskul, Paweena Susantitaphong, Patcharee Ritprajak, Naraporn Somboonna, Thanya Cheibchalard, and et al. 2022. "Blood Bacteria-Free DNA in Septic Mice Enhances LPS-Induced Inflammation in Mice through Macrophage Response" International Journal of Molecular Sciences 23, no. 3: 1907. https://doi.org/10.3390/ijms23031907
APA StyleKaewduangduen, W., Visitchanakun, P., Saisorn, W., Phawadee, A., Manonitnantawat, C., Chutimaskul, C., Susantitaphong, P., Ritprajak, P., Somboonna, N., Cheibchalard, T., Wannigama, D. L., Kueanjinda, P., & Leelahavanichkul, A. (2022). Blood Bacteria-Free DNA in Septic Mice Enhances LPS-Induced Inflammation in Mice through Macrophage Response. International Journal of Molecular Sciences, 23(3), 1907. https://doi.org/10.3390/ijms23031907







