Screening of Differentially Expressed Genes and Localization Analysis of Female Gametophyte at the Free Nuclear Mitosis Stage in Pinus tabuliformis Carr.
Abstract
:1. Introduction
2. Results
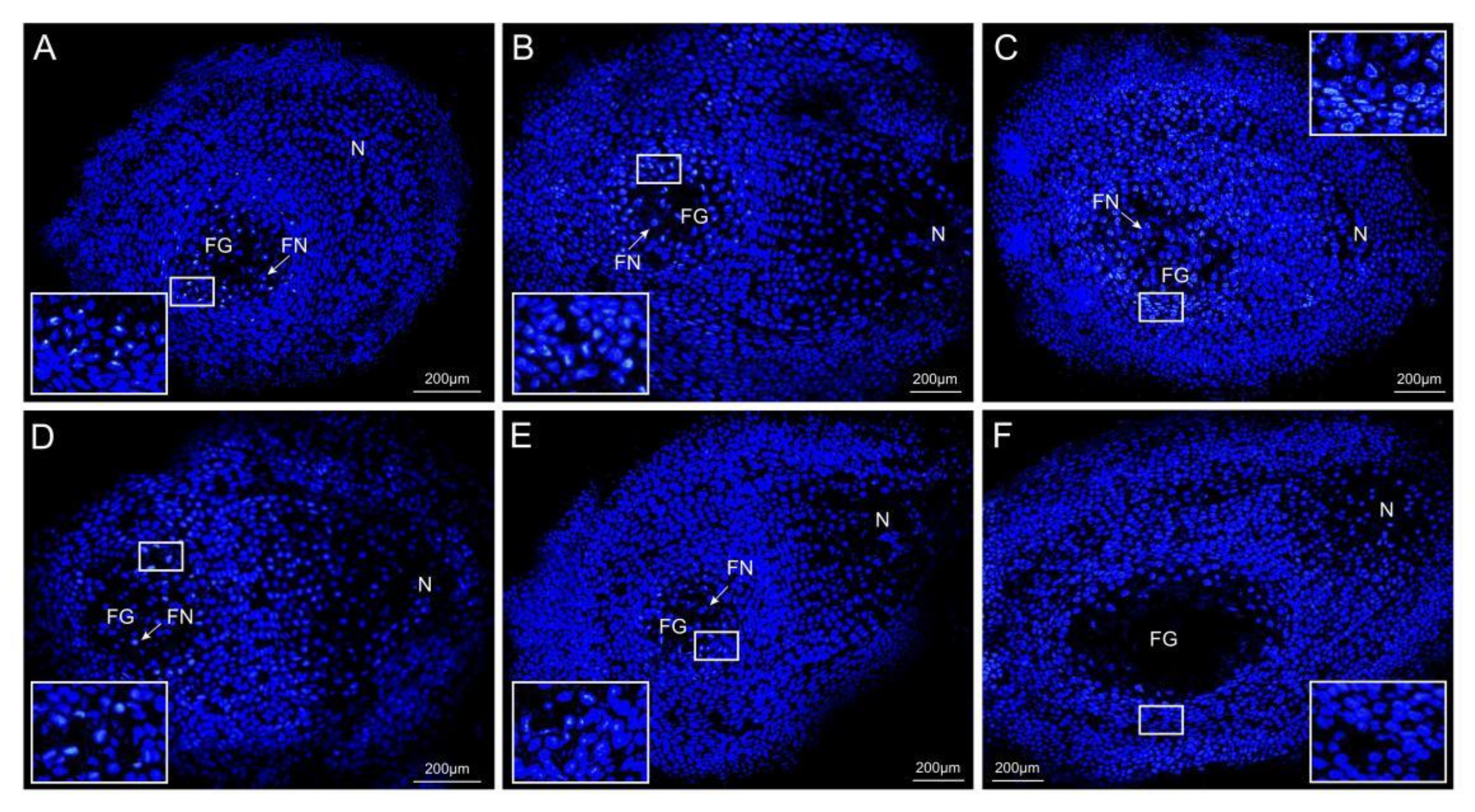
2.1. Ovule Phenotypes of P. tabuliformis
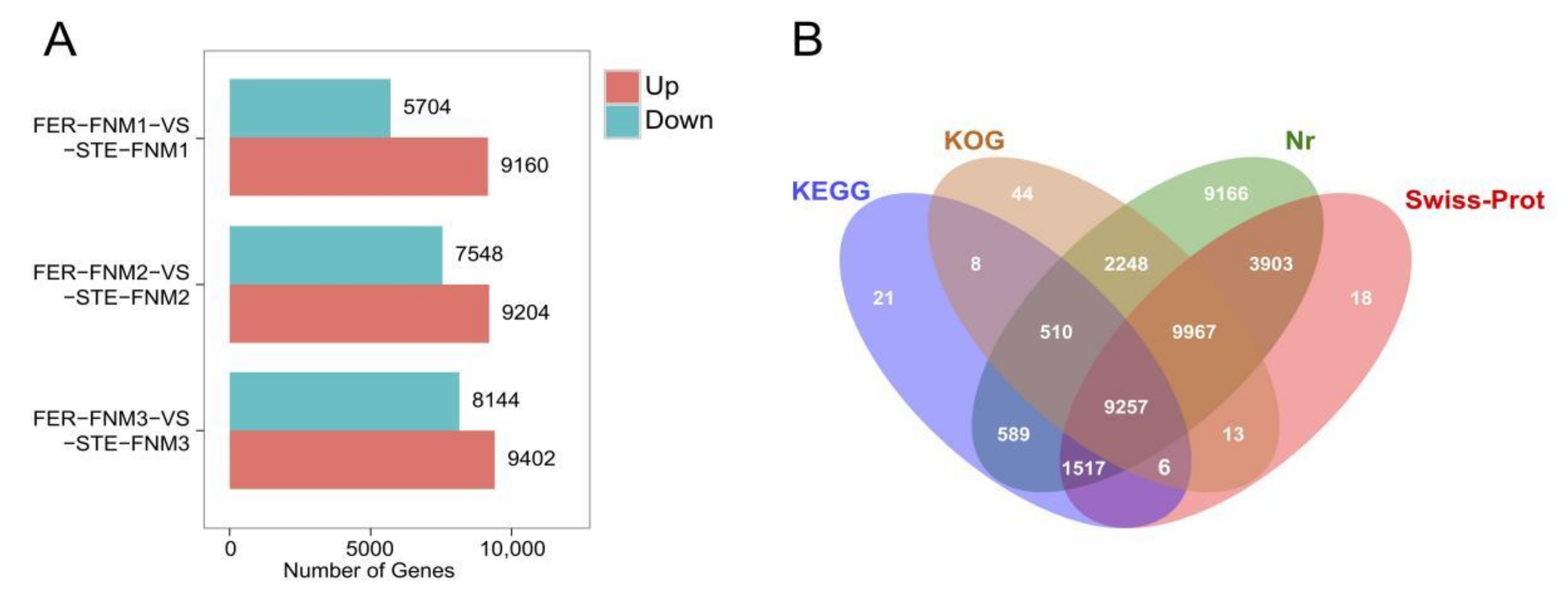
2.2. Sequencing, Read Assembly and Global Data Analysis
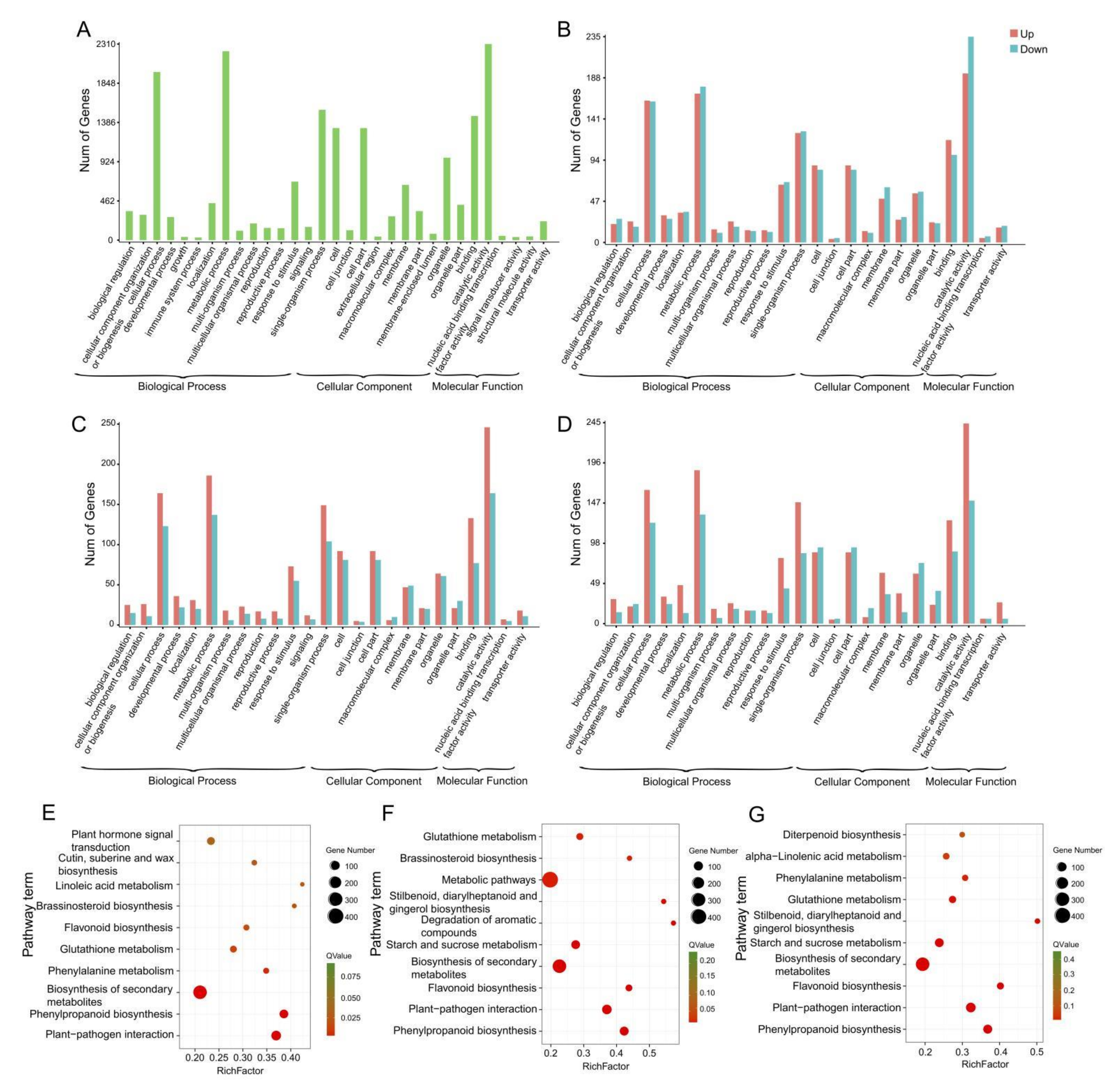
2.3. Gene Annotation and Functional Classification
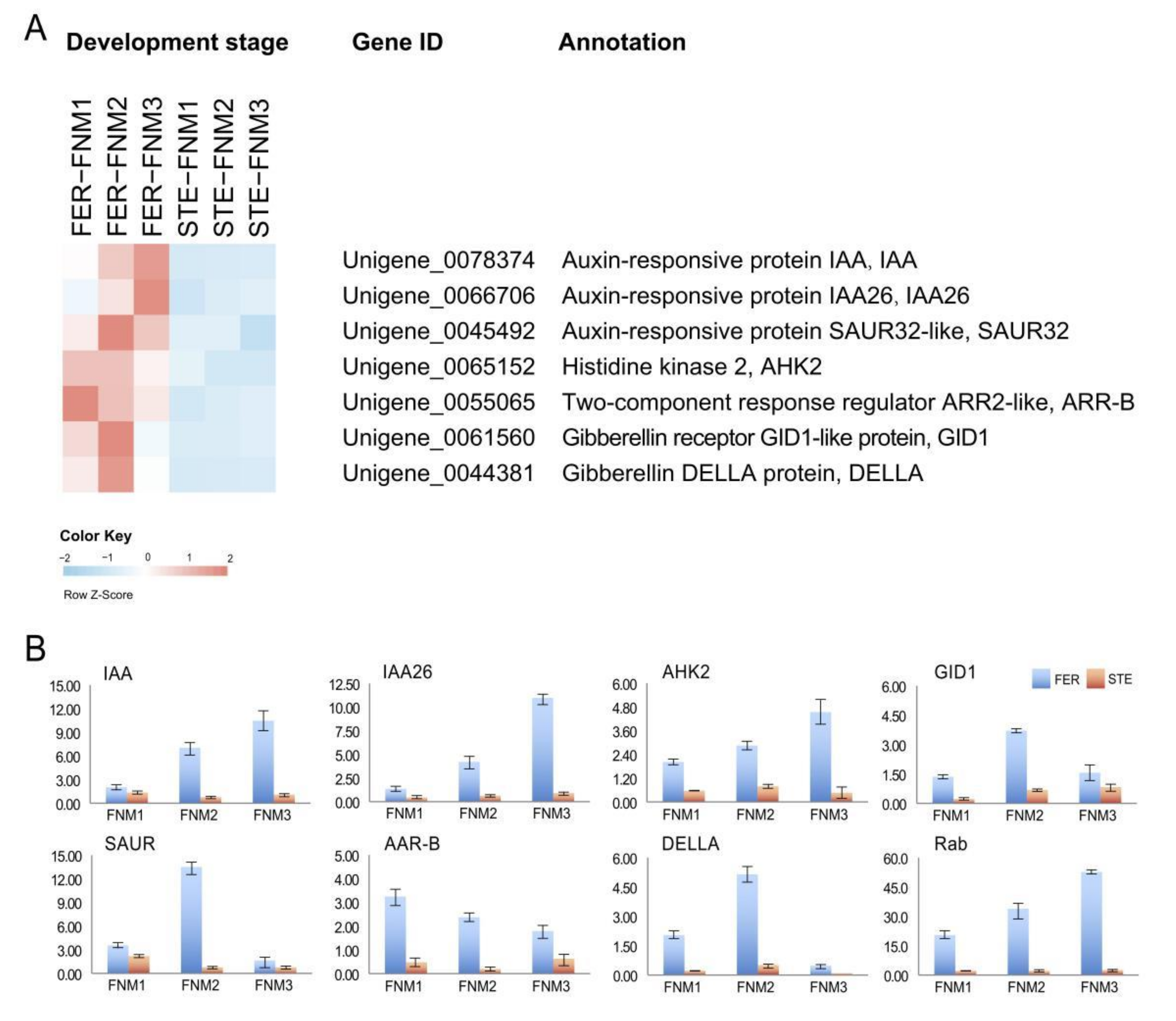
2.4. DEGs Related to Phytohormones in Comparisons between Fertile and Sterile Ovules during FNMM
2.5. Verification of the Gene Expression Profile by qRT-PCR
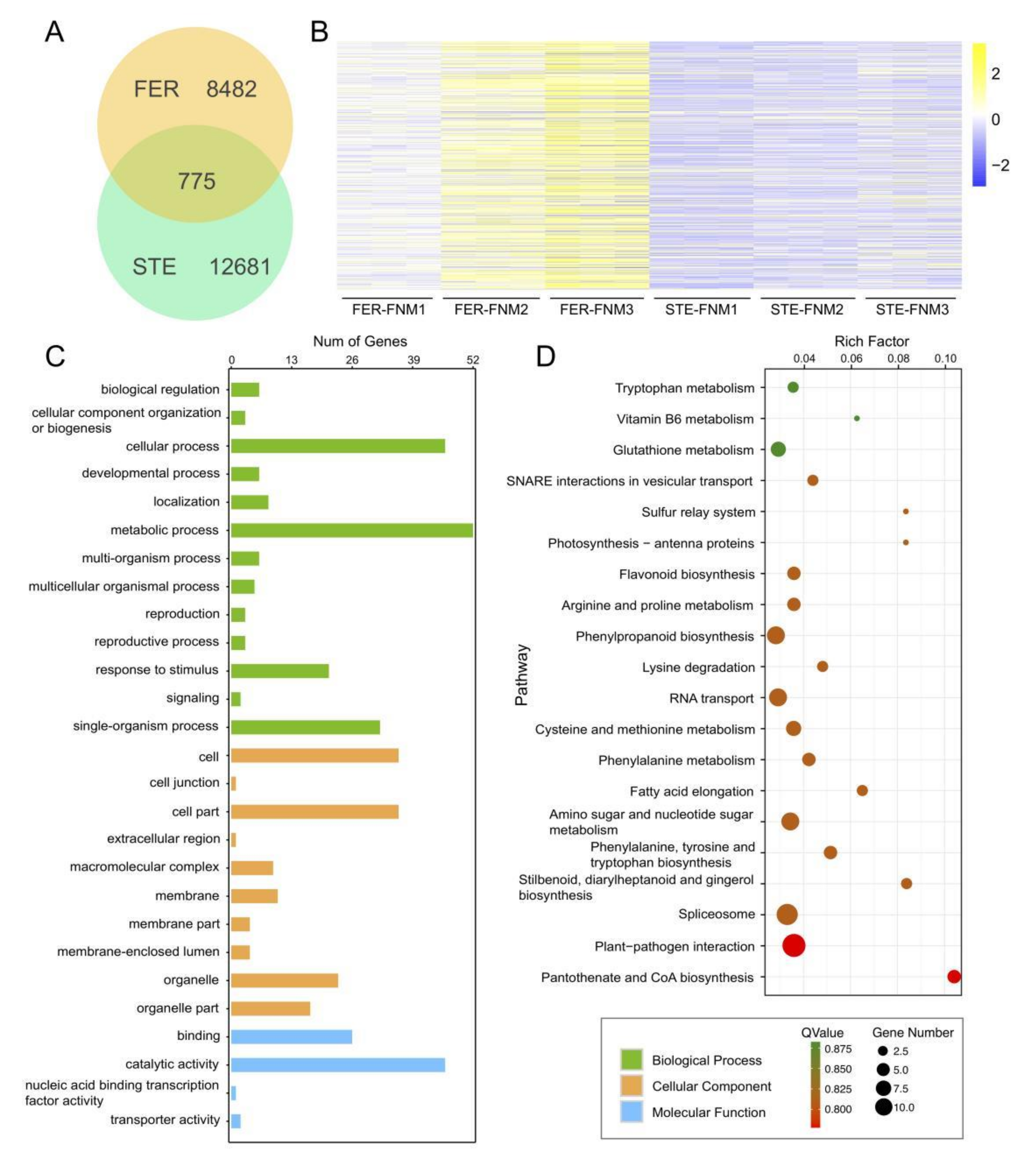
2.6. Expression Patterns of DEGs between STE and FER during the FNM Process
2.7. Spatiotemporal Expression of PtRab in the Ovules
2.8. Development and Characterization of SSR Markers
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. RNA Sample Preparation and RNA-Seq
4.3. De Novo Assembly
4.4. Functional Annotation
4.5. Analysis of Differentially Expressed Genes (DEGs)
4.6. Simple Sequence Repeat (SSR) Marker Prediction
4.7. Fluorescence In Situ Hybridization
4.7.1. Preparation and Labeling of DNA Probes
4.7.2. Preparation of Frozen Sections
4.7.3. Pretreatment of Frozen Sections before the Fluorescence In Situ Hybridization
4.7.4. Fluorescence In Situ Hybridization
4.7.5. Elution and Detection after Hybridization
4.8. Real-Time Quantitative PCR Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hu, Q.; Gao, S.; Li, F. Advances of research on plant female sterility. J. Beijing For. Univ. 2004, 26, 87–91. [Google Scholar]
- Guo, A.; Zheng, C.X. Female gametophyte development. J. Plant Biol. 2013, 56, 345–356. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, J.; Li, H.; Niu, J.; Xue, H.; Liu, B.; Wang, Q.; Luo, X.; Zhang, F.; Zhao, D.; et al. Transcriptomic Analysis Reveals Candidate Genes for Female Sterility in Pomegranate Flowers. Front. Plant Sci. 2017, 8, 1430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Awasthi, A.; Paul, P.; Kumar, S.; Verma, S.K.; Prasad, R.; Dhaliwal, H.S. Abnormal endosperm development causes female sterility in rice insertional mutant OsAPC6. Plant Sci. 2012, 183, 167–174. [Google Scholar] [CrossRef]
- Rosellini, D.; Ferranti, F.; Barone, P.; Veronesi, F. Expression of female sterility in alfalfa (Medicago sativa L.). Sex. Plant Reprod. 2003, 15, 271–279. [Google Scholar] [CrossRef]
- Clark, S.E.; Williams, R.W.; Meyerowitz, E.M. The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 1997, 89, 575–585. [Google Scholar] [CrossRef] [Green Version]
- Alvarez, J.M.; Cortizo, M.; Bueno, N.; Rodríguez, A.; Ordás, R.J. CLAVATA1-LIKE, a leucine-rich-repeat protein receptor kinase gene differentially expressed during adventitious caulogenesis in Pinus pinaster and Pinus pinea. Plant Cell Tissue Organ Cult. PCTOC 2013, 112, 331–342. [Google Scholar] [CrossRef]
- Subbaiah, C.C.; Huber, S.C.; Sachs, M.M.; Rhoads, D. Sucrose synthase: Expanding protein function. Plant Signal Behav. 2007, 2, 28–29. [Google Scholar]
- Kong, J.; Li, Z.; Tan, Y.; Wan, C.; Li, S.; Zhu, Y. Different gene expression patterns of sucrose-starch metabolism during pollen maturation in cytoplasmic male-sterile and male-fertile lines of rice. Physiol. Plant. 2007, 130, 136–147. [Google Scholar] [CrossRef]
- Qiu, Y.; Taylor, A.B.; McManus, H.A. Evolution of the life cycle in land plants. J. Syst. Evol. 2012, 50, 171–194. [Google Scholar] [CrossRef] [Green Version]
- Jin, W.; Gernandt, D.; Wehenkel, C.; Xia, X.; Wei, X.; Wang, X. Phylogenomic and ecological analyses reveal the spatiotemporal evolution of global pines. Proc. Natl. Acad. Sci. USA 2021, 118, e2022302118. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zhang, W.; Gong, Z.; Zheng, C. Morphologic and anatomical observations in the process of ovulate strobilus generation and development in Pinus tabuliformis. J. Beijing For. Univ. 2017, 39, 1–12. [Google Scholar]
- Chen, K.; Abbott, R.J.; Milne, R.I.; Tian, X.; Liu, J. Phylogeography of Pinus tabulaeformis Carr. (Pinaceae), a dominant species of coniferous forest in northern China. Mol. Ecol. 2008, 17, 4276–4288. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Shen, X.; Li, Y. Discovery of female sterility in Pinus tabulaeformis and analysis of its causes. J. Hebei For. Coll. 1992, 7, 93–98. [Google Scholar]
- Bao, R. Primary Study on the Mechanism of Ovule Abortion of Female-Sterile Clone in Pinus Tabulaeformis Carr; Beijing Forestry University: Beijing, China, 2006. [Google Scholar]
- He, Y.; Zhang, C. Relationship between metabolism in developmental ovule and femalesterile in Pinus tabulaeformis Carr. J. Beijing For. Univ. 2007, 29, 88–93. [Google Scholar]
- Ding, S.; Zheng, C.; Bao, R. Analyses of peroxidase isozyme and protein polypeptides in female sterile Pinus tabulaeformis. Acta Bot. Boreal.-Occident. Sin. 2004, 24, 17–20. [Google Scholar]
- Chen, X.; Li, Q.; Li, Y.; Qian, J.; Han, J. Chloroplast genome of Aconitum barbatum var. puberulum (Ranunculaceae) derived from CCS reads using the PacBio RS platform. Front. Plant Sci. 2015, 6, 42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Liu, R.; Wang, J.; Wang, P.; Shen, Y.; Liu, G. The Arabidopsis kinesin gene AtKin-1 plays a role in the nuclear division process during megagametogenesis. Plant Cell Rep. 2014, 33, 819–828. [Google Scholar] [CrossRef]
- López-Almansa, J.C.; Pannell, J.R.; Gil, L. Female sterility in Ulmus minor (Ulmaceae): A hypothesis invoking the cost of sex in a clonal plant. Am. J. Bot. 2003, 90, 603–609. [Google Scholar] [CrossRef]
- Yan, M.; Dai, X.; Li, S.; Yin, D. Sequence Analysis and Comparison of EST-SSRs in Pine, Poplar and Eucalyptus. Genom. Appl. Biol. 2011, 30, 103–109. [Google Scholar]
- Dou, B.; Hou, B.; Xu, H.; Lou, X.; Chi, X.; Yang, J.; Wang, F.; Ni, Z.; Sun, Q. Efficient mapping of a female sterile gene in wheat (Triticum aestivum L.). Genet Res. 2009, 91, 337–343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, D.; Cao, L.; Gao, J. Data mining of simple sequence repeats in Codonopsis pilosula transcriptome. Chin. Tradit. Herb. Drugs 2014, 45, 2390–2394. [Google Scholar]
- Jun, T.; Michel, A.P.; Mian, M.A.R. Development of soybean aphid genomic SSR markers using next generation sequencing. Genome 2011, 54, 360–367. [Google Scholar] [CrossRef]
- Yang, X.; Zhang, K.; Wang, J.; Jia, H.; Li, Y.; Duan, J.; Ma, L. Analysis of Genetic Diversity and Construction of Core Collections of Black Locust Based on SSR Markers. Mol. Plant Breed. 2020, 18, 3086–3097. [Google Scholar]
- Li, Y.; Yang, X.; Zhang, J.; Huang, S.; Xiong, X. Studies on SSR Molecular Markers Based on Transcriptome of Taxus chinensis var. mairei. Acta Hortic. Sin. 2014, 41, 735–745. [Google Scholar]
- Fan, H.; Li, T.; Li, Z.; Lin, Y.; Cai, Y. Characteristics of EST-SSR Distribution in Ginkgo ESTs. Genom. Appl. Biol. 2009, 28, 869–873. [Google Scholar]
- Wu, J.; Cai, C.; Cheng, F.; Cui, H.; Zhou, H. Characterisation and development of EST-SSR markers in tree peony using transcriptome sequences. Mol. Breed. 2014, 34, 1853–1866. [Google Scholar] [CrossRef]
- Fu, W.; Zhao, Z.; Ge, X.; Ding, L.; Li, Z. Anatomy and transcript profiling of gynoecium development in female sterile Brassica napus mediated by one alien chromosome from Orychophragmus violaceus. BMC Genom. 2014, 15, 61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robert, H.S.; Park, C.; Gutièrrez, C.L.; Wójcikowska, B.; Pěnčík, A.; Novák, O.; Chen, J.; Grunewald, W.; Dresselhaus, T.; Friml, J.; et al. Maternal auxin supply contributes to early embryo patterning in Arabidopsis. Nat. Plants 2018, 4, 548–553. [Google Scholar] [CrossRef]
- Pagnussat, G.C.; Alandete-Saez, M.; Bowman, J.L.; Sundaresan, V. Auxin-Dependent Patterning and Gamete Specification in the Arabidopsis Female Gametophyte. Science 2009, 324, 1684–1689. [Google Scholar] [CrossRef] [Green Version]
- Panoli, A.; Martin, M.V.; Alandete-Saez, M.; Simon, M.; Neff, C.; Swarup, R.; Bellido, A.; Yuan, L.; Pagnussat, G.C.; Sundaresan, V. Auxin Import and Local Auxin Biosynthesis Are Required for Mitotic Divisions, Cell Expansion and Cell Specification during Female Gametophyte Development in Arabidopsis thaliana. PLoS ONE 2015, 10, e0126164. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita-Tsujimura, K.; Kakimoto, T. Cytokinin receptors in sporophytes are essential for male and female functions inArabidopsis thaliana. Plant Signal. Behav. 2014, 6, 66–71. [Google Scholar] [CrossRef] [Green Version]
- Cucinotta, M.; Manrique, S.; Cuesta, C.; Benkova, E.; Novak, O.; Colombo, L. CUP-SHAPED COTYLEDON1 (CUC1) and CUC2 regulate cytokinin homeostasis to determine ovule number in Arabidopsis. J. Exp. Bot. 2018, 69, 5169–5176. [Google Scholar] [CrossRef]
- Cheng, C.; Mathews, D.E.; Eric Schaller, G.; Kieber, J.J. Cytokinin-dependent specification of the functional megaspore in the Arabidopsis female gametophyte. Plant J. 2013, 73, 929–940. [Google Scholar] [CrossRef]
- Feng, J.; Wang, C.; Chen, Q.; Chen, H.; Ren, B.; Li, X.; Zuo, J. S-nitrosylation of phosphotransfer proteins represses cytokinin signaling. Nat. Commun. 2013, 4, 1529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agostino, I.B.D.; Kieber, J.J. Molecular Mechanisms of Cytokinin Action; Elsevier Ltd.: London, UK, 1999; Volume 2, pp. 359–364. [Google Scholar]
- Liu, Z.; Dai, X.; Li, J.; Liu, N.; Liu, X.; Li, S.; Xiang, F. The Type-B Cytokinin Response Regulator ARR1 Inhibits Shoot Regeneration in an ARR12-Dependent Manner in Arabidopsis. Plant Cell 2020, 32, 2271–2291. [Google Scholar] [CrossRef] [PubMed]
- Hedden, P.; Sponsel, V. A Century of Gibberellin Research. J. Plant Growth Regul. 2015, 34, 740–760. [Google Scholar] [CrossRef] [Green Version]
- Peng, J.; Carol, P.; Richards, D.E.; King, K.E.; Cowling, R.J.; Murphy, G.P.; Harberd, N.P. The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Gene Dev. 1997, 11, 3194–3205. [Google Scholar] [CrossRef] [Green Version]
- Li, D.; Duan, H.; Liu, H.; Wang, S.; Peng, W.; Yang, X. Progress on gibberellins, gibberellins functional analogues and gibberellins receptor. Chin. J. Pestic. Sci. 2013, 15, 601–608. [Google Scholar]
- Itoh, H.; Sasaki, A.; Ueguchi-Tanaka, M.; Ishiyama, K.; Kobayashi, M.; Hasegawa, Y.; Minami, E.; Ashikari, M.; Matsuoka, M. Dissection of the Phosphorylation of Rice DELLA Protein, SLENDER RICE1. Plant Cell Physiol. 2005, 46, 1392–1399. [Google Scholar] [CrossRef]
- Harberd, N.P. Botany. Relieving DELLA restraint. Science 2003, 299, 1853–1854. [Google Scholar] [CrossRef] [PubMed]
- Harberd, N.P.; Belfield, E.; Yasumura, Y. The Angiosperm Gibberellin-GID1-DELLA Growth Regulatory Mechanism: How an “Inhibitor of an Inhibitor” Enables Flexible Response to Fluctuating Environments. Plant Cell 2009, 21, 1328–1339. [Google Scholar] [CrossRef] [Green Version]
- Kirchhelle, C.; Chow, C.; Foucart, C.; Neto, H.; Stierhof, Y.; Kalde, M.; Walton, C.; Fricker, M.; Smith, R.S.; Jérusalem, A.; et al. The Specification of Geometric Edges by a Plant Rab GTPase Is an Essential Cell-Patterning Principle During Organogenesis in Arabidopsis. Dev. Cell 2016, 36, 386–400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.Y.; Seo, H.S.; Jeong, J.Y.; Lee, S.Y.; Cho, M.J.; Bahk, J.D. Molecular characterization of a rab-related small GTP binding protein cDNA from rice (Oryza sativa L. IR-36). Mol. Cells 1997, 7, 226–230. [Google Scholar] [PubMed]
- Gonçalves, S.; Cairney, J.; Rodríguez, M.P.; Cánovas, F.; Oliveira, M.; Miguel, C. PpRab1, a Rab GTPase from maritime pine is differentially expressed during embryogenesis. Mol. Genet Genom. 2007, 278, 273–282. [Google Scholar] [CrossRef]
- Gutkowska, M.; Wnuk, M.; Nowakowska, J.; Lichocka, M.; Stronkowski, M.M.; Swiezewska, E. Rab geranylgeranyl transferase β subunit is essential for male fertility and tip growth in Arabidopsis. J. Exp. Bot. 2015, 66, 213–224. [Google Scholar] [CrossRef]
- Li, M.; Zheng, C. RAPD analysis in female sterility clone 28 of Pinus tabulae- formis Carr. J. Beijing Forest. Univ. 2002, 24, 35–38. [Google Scholar]
- Zhang, W.; Zheng, C.; Zhao, N.; Li, F. Observation on the formation of ovule tapetum in Pinus tabulaeformis Carr. J. Beijing Forest. Univ. 2015, 37, 53–57. [Google Scholar]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [Green Version]
- Pertea, G.; Huang, X.; Liang, F.; Antonescu, V.; Sultana, R.; Karamycheva, S.; Lee, Y.; White, J.; Cheung, F.; Parvizi, B.; et al. TIGR gene indices clustering tools (TGICL): A software system for fast clustering of large EST datasets. Bioinformatics 2003, 19, 651–652. [Google Scholar] [CrossRef] [Green Version]
- Conesa, A.; Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Talon, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [Green Version]
- Ye, J.; Fang, L.; Zheng, H.; Zhang, Y.; Chen, J.; Zhang, Z.; Wang, J.; Li, S.; Li, R.; Bolund, L.; et al. WEGO: A web tool for plotting GO annotations. Nucleic Acids Res. 2006, 34, W293–W297. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Audic, S.; Claverie, J.M. The significance of digital gene expression profiles. Genome Res. 1997, 7, 986–995. [Google Scholar] [CrossRef] [PubMed]


| Repeat Motif Length | Repeat Number | Total | Percentage | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | >12 | |||
| Di-nucleotide | 795 | 362 | 294 | 193 | 128 | 57 | 15 | 32 | 1876 | 49.95% | ||
| Tri-nucleotide | 821 | 212 | 92 | 22 | 8 | 3 | 0 | 3 | 1161 | 30.91% | ||
| Tetra-nucleotide | 349 | 50 | 1 | 1 | 1 | 402 | 10.7% | |||||
| Penta-nucleotide | 132 | 10 | 1 | 1 | 144 | 3.83% | ||||||
| Hexa-nucleotide | 140 | 28 | 2 | 1 | 2 | 173 | 4.61% | |||||
| Total | 621 | 909 | 1011 | 456 | 319 | 201 | 132 | 57 | 18 | 32 | 3756 | 100% |
| Percentage | 16.53% | 24.2% | 29.47% | 12.14% | 4.49% | 5.51% | 3.61% | 1.52% | 0.48% | 0.85% | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gong, Z.; Hu, H.; Xu, L.; Zhao, Y.; Zheng, C. Screening of Differentially Expressed Genes and Localization Analysis of Female Gametophyte at the Free Nuclear Mitosis Stage in Pinus tabuliformis Carr. Int. J. Mol. Sci. 2022, 23, 1915. https://doi.org/10.3390/ijms23031915
Gong Z, Hu H, Xu L, Zhao Y, Zheng C. Screening of Differentially Expressed Genes and Localization Analysis of Female Gametophyte at the Free Nuclear Mitosis Stage in Pinus tabuliformis Carr. International Journal of Molecular Sciences. 2022; 23(3):1915. https://doi.org/10.3390/ijms23031915
Chicago/Turabian StyleGong, Zaixin, Hailin Hu, Li Xu, Yuanyuan Zhao, and Caixia Zheng. 2022. "Screening of Differentially Expressed Genes and Localization Analysis of Female Gametophyte at the Free Nuclear Mitosis Stage in Pinus tabuliformis Carr." International Journal of Molecular Sciences 23, no. 3: 1915. https://doi.org/10.3390/ijms23031915
APA StyleGong, Z., Hu, H., Xu, L., Zhao, Y., & Zheng, C. (2022). Screening of Differentially Expressed Genes and Localization Analysis of Female Gametophyte at the Free Nuclear Mitosis Stage in Pinus tabuliformis Carr. International Journal of Molecular Sciences, 23(3), 1915. https://doi.org/10.3390/ijms23031915





