An Evaluation of Different 3D Cultivation Models on Expression Profiles of Human Periodontal Ligament Fibroblasts with Compressive Strain
Abstract
:1. Introduction
2. Results
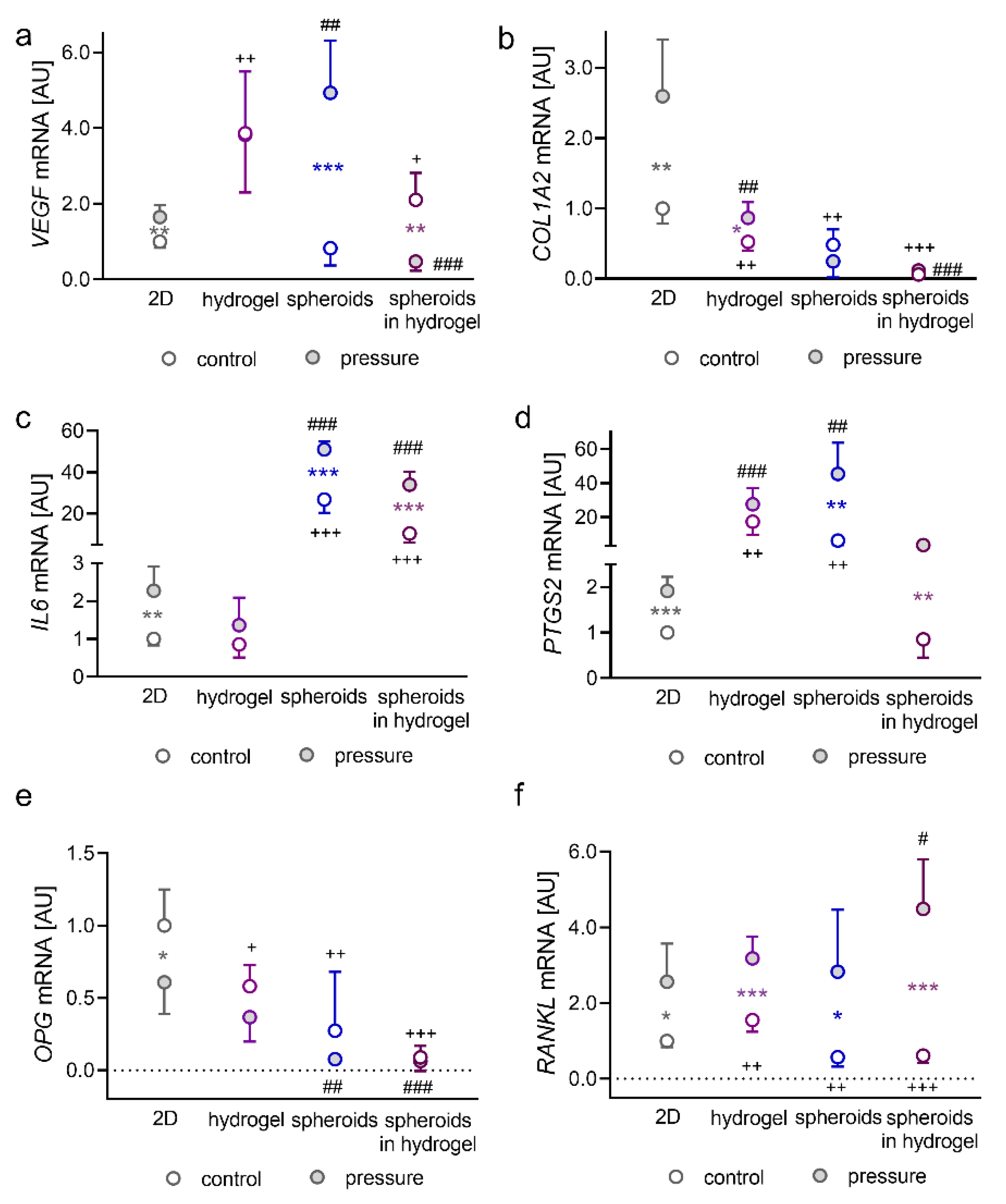
2.1. Impact of Pressure Application on PDLFs Cultivated as Monolayers in Hydrogel, as Spheroids, or as Spheroids Embedded in Hydrogel
2.2. Impact of Pressure Application on PDLFs Cultivated in Different Solid 3D Scaffolds
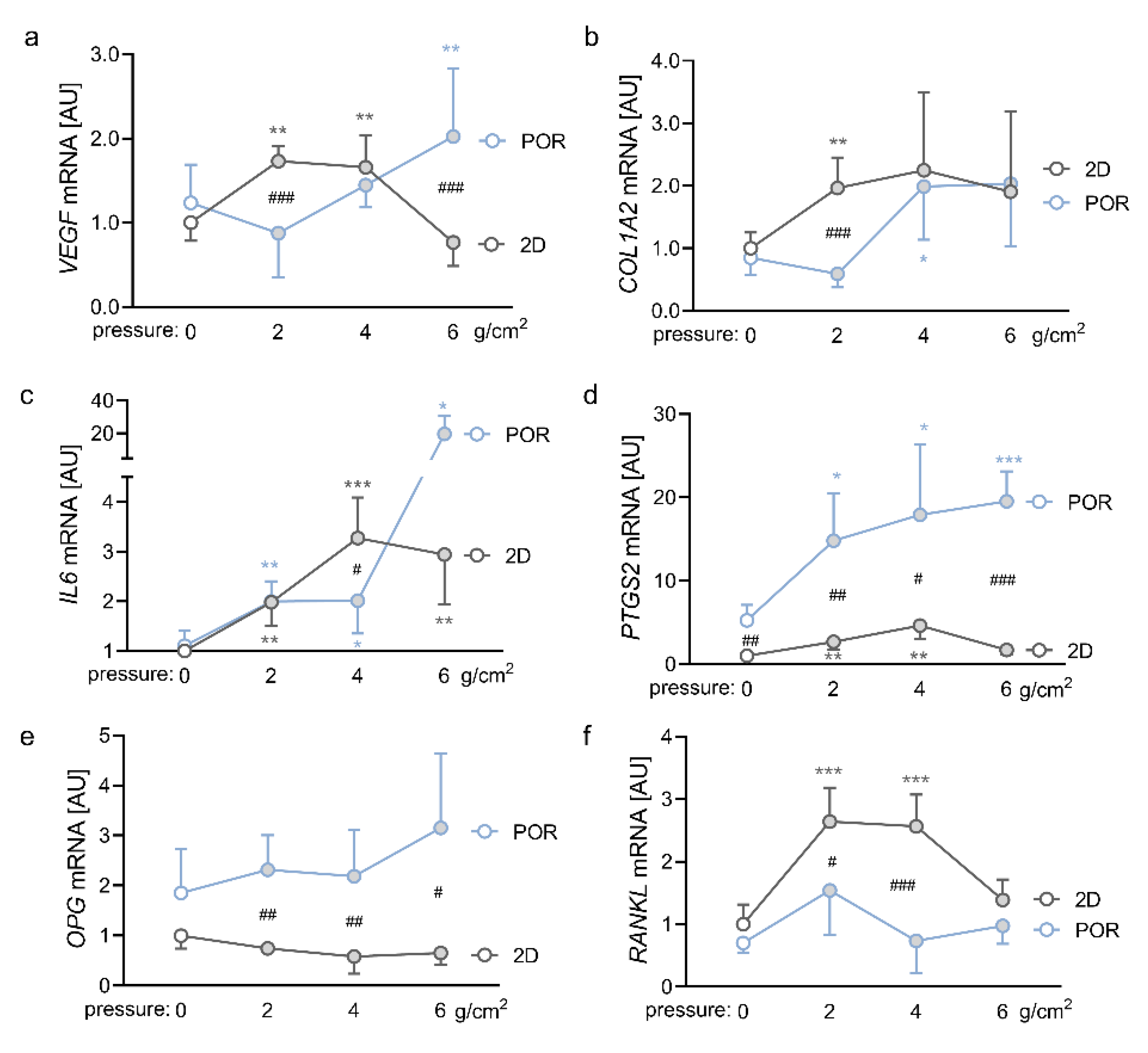
2.3. Impact of Different Magnitudes of Compressive Force on the Expression Profiles of PDLFs in the POR Scaffold
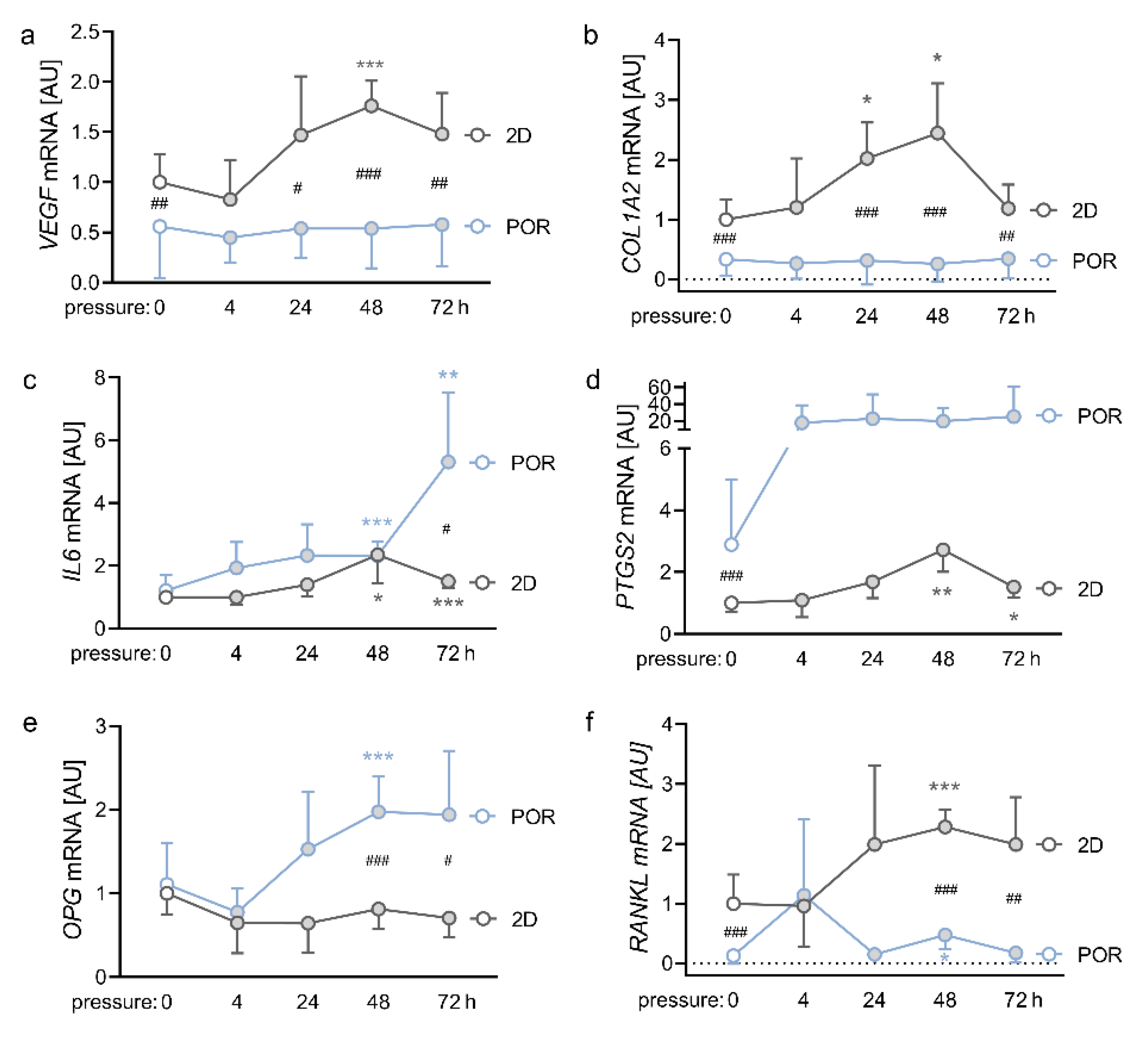
2.4. Impact of Different Compression Times on the Expression Profiles of PDLFs in the POR Scaffold
3. Discussion
4. Materials and Methods
4.1. Isolation and Cultivation of PDLFs
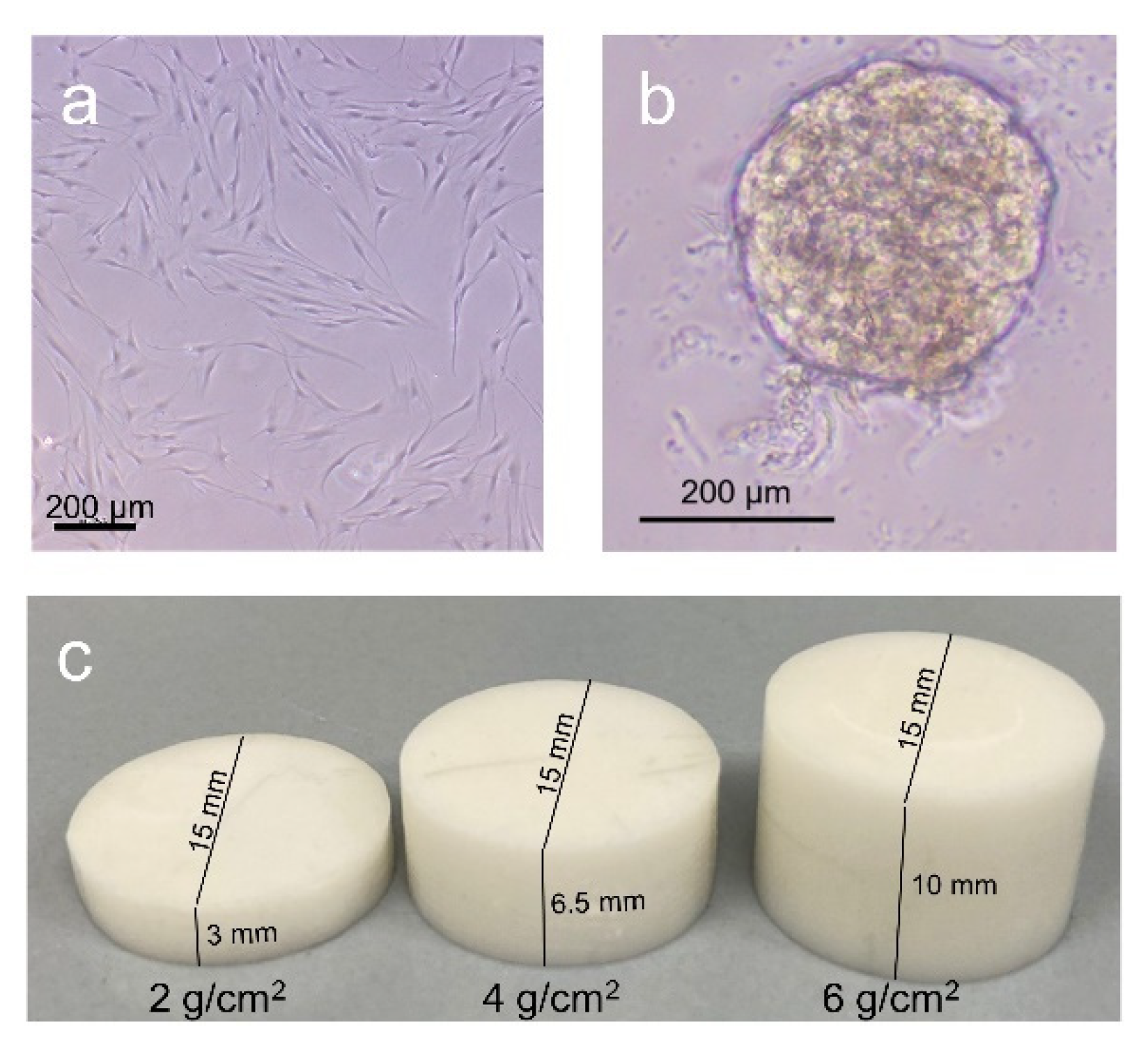
4.2. Cell Culture Experiments with Hydrogel and Spheroids
4.3. Cell Culture Experiments with 3D Scaffolds
4.4. Cell Culture Experiments in POR Scaffolds with Different Magnitudes of Compression
4.5. Cell Culture Experiments in POR Scaffolds with Different Periods of Compression
4.6. RNA Isolation
4.7. cDNA Synthesis
4.8. Quantitative Real-Time PCR (RT–qPCR)
4.9. Statistical Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Proffit, W.R.; Fields, H.W.; Larson, B.E.; Sarver, D.M. Contemporary Orthodontics; Elsevier: Philadelphia, PA, USA, 2019. [Google Scholar]
- Sharma, S.; Narkhede, S.; Sonawane, S.; Gangurde, P. Evaluation of Patient’s Personal Reasons and Experience with Orthodontic Treatment. J. Int. Oral Health 2013, 5, 78–81. [Google Scholar] [PubMed]
- Grimm, S.; Frazão, P.; Antunes, J.L.F.; Castellanos, R.A.; Narvai, P.C. Dental injury among Brazilian schoolchildren in the state of São Paulo. Dent. Traumatol. 2004, 20, 134–138. [Google Scholar] [CrossRef] [PubMed]
- Gaikwad, S.S.; Gheware, A.; Kamatagi, L.; Pasumarthy, S.; Pawar, V.; Fatangare, M. Dental caries and its relationship to malocclusion in permanent dentition among 12–15 year old school going children. J. Int. Oral Health 2014, 6, 27–30. [Google Scholar] [PubMed]
- Geiger, A.M. Malocclusion as an etiologic factor in periodontal disease: A retrospective essay. Am. J. Orthod. Dentofac. Orthop. 2001, 120, 112–115. [Google Scholar] [CrossRef] [PubMed]
- Zanella, S.M.; Pereira, S.S.; Barbisan, J.N.; Vieira, L.; Saba-Chujfi, E.; Haas, A.N.; Rösing, C.K. Periodontal disease, tooth loss and coronary heart disease assessed by coronary angiography: A cross-sectional observational study. J. Periodontal Res. 2016, 51, 221–227. [Google Scholar] [CrossRef] [PubMed]
- Sandy, J.R.; Farndale, R.W.; Meikle, M.C. Recent advances in understanding mechanically induced bone remodeling and their relevance to orthodontic theory and practice. Am. J. Orthod. Dentofac. Orthop. 1993, 103, 212–222. [Google Scholar] [CrossRef]
- Meikle, M.C. The tissue, cellular, and molecular regulation of orthodontic tooth movement. 100 years after Carl Sandstedt. Eur. J. Orthod. 2006, 28, 221–240. [Google Scholar] [CrossRef]
- Kanzaki, H.; Chiba, M.; Shimizu, Y.; Mitani, H. Periodontal ligament cells under mechanical stress induce osteoclastogenesis by receptor activator of nuclear factor kappaB ligand up-regulation via prostaglandin E2 synthesis. J. Bone Miner. Res. 2002, 17, 210–220. [Google Scholar] [CrossRef] [Green Version]
- Beertsen, W.; McCulloch, C.A.; Sodek, J. The periodontal ligament: A unique, multifunctional connective tissue. Periodontol. 2000 1997, 13, 20–40. [Google Scholar] [CrossRef]
- Hassell, T.M. Tissues and cells of the periodontium. Periodontol. 2000 1993, 3, 9–38. [Google Scholar] [CrossRef]
- McCulloch, C.A.; Bordin, S. Role of fibroblast subpopulations in periodontal physiology and pathology. J. Periodontal Res. 1991, 26 Pt 1, 144–154. [Google Scholar] [CrossRef]
- Lekic, P.; McCulloch, C. Periodontal ligament cell populations: The central role of fibroblasts in creating a unique tissue. Anat. Rec. 1996, 245, 327–341. [Google Scholar] [CrossRef]
- Krishnan, V.; Davidovitch, Z. Cellular, molecular, and tissue-level reactions to orthodontic force. Am. J. Orthod. Dentofac. Orthop. 2006, 129, 469.E1–469.E32. [Google Scholar] [CrossRef] [PubMed]
- Di Domenico, M.; D’apuzzo, F.; Feola, A.; Cito, L.; Monsurrò, A.; Pierantoni, G.M.; Berrino, L.; de Rosa, A.; Polimeni, A.; Perillo, L. Cytokines and VEGF induction in orthodontic movement in animal models. J. Biomed. Biotechnol. 2012, 2012, 201689. [Google Scholar] [CrossRef] [PubMed]
- Graber, L.W.; Vanarsdall, R.L.; Vig, K.W.L.; Huang, G.J. Orthodontics. Current Principles and Techniques, 6th ed.; Elsevier: St. Louis, MI, USA, 2017. [Google Scholar]
- Baumrind, S.; Buck, D.L. Rate changes in cell replication and protein synthesis in the periodontal ligament incident to tooth movement. Am. J. Orthod. 1970, 57, 109–131. [Google Scholar] [CrossRef]
- Baker, B.M.; Chen, C.S. Deconstructing the third dimension: How 3D culture microenvironments alter cellular cues. J. Cell Sci. 2012, 125 Pt 13, 3015–3024. [Google Scholar] [CrossRef] [Green Version]
- Thippabhotla, S.; Zhong, C.; He, M. 3D cell culture stimulates the secretion of in vivo like extracellular vesicles. Sci. Rep. 2019, 9, 13012. [Google Scholar] [CrossRef] [Green Version]
- Duval, K.; Grover, H.; Han, L.-H.; Mou, Y.; Pegoraro, A.F.; Fredberg, J.; Chen, Z. Modeling Physiological Events in 2D vs. 3D Cell Culture. Physiology (Bethesda) 2017, 32, 266–277. [Google Scholar] [CrossRef]
- Loessner, D.; Stok, K.S.; Lutolf, M.P.; Hutmacher, D.W.; Clements, J.A.; Rizzi, S.C. Bioengineered 3D platform to explore cell-ECM interactions and drug resistance of epithelial ovarian cancer cells. Biomaterials 2010, 31, 8494–8506. [Google Scholar] [CrossRef] [Green Version]
- Zorlutuna, P.; Annabi, N.; Camci-Unal, G.; Nikkhah, M.; Cha, J.M.; Nichol, J.W.; Manbachi, A.; Bae, H.; Chen, S.; Khademhosseini, A. Microfabricated biomaterials for engineering 3D tissues. Adv. Mater. 2012, 24, 1782–1804. [Google Scholar] [CrossRef] [Green Version]
- Brezulier, D.; Pellen-Mussi, P.; Tricot-Doleux, S.; Novella, A.; Sorel, O.; Jeanne, S. Development of a 3D human osteoblast cell culture model for studying mechanobiology in orthodontics. Eur. J. Orthod. 2020, 42, 387–395. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, J.; Heinrich, M.A.; Teixeira, L.M.; Prakash, J. 3D In Vitro Model (R)evolution: Unveiling Tumor-Stroma Interactions. Trends Cancer 2021, 7, 249–264. [Google Scholar] [CrossRef] [PubMed]
- Mabry, K.M.; Payne, S.Z.; Anseth, K.S. Microarray analyses to quantify advantages of 2D and 3D hydrogel culture systems in maintaining the native valvular interstitial cell phenotype. Biomaterials 2016, 74, 31–41. [Google Scholar] [CrossRef] [Green Version]
- Melissaridou, S.; Wiechec, E.; Magan, M.; Jain, M.V.; Chung, M.K.; Farnebo, L.; Roberg, K. The effect of 2D and 3D cell cultures on treatment response, EMT profile and stem cell features in head and neck cancer. Cancer Cell Int. 2019, 19, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bott, K.; Upton, Z.; Schrobback, K.; Ehrbar, M.; Hubbell, J.A.; Lutolf, M.P.; Rizzi, S.C. The effect of matrix characteristics on fibroblast proliferation in 3D gels. Biomaterials 2010, 31, 8454–8464. [Google Scholar] [CrossRef] [Green Version]
- Jensen, C.; Teng, Y. Is It Time to Start Transitioning from 2D to 3D Cell Culture? Front. Mol. Biosci. 2020, 7, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rhee, S. Fibroblasts in three dimensional matrices: Cell migration and matrix remodeling. Exp. Mol. Med. 2009, 41, 858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, M.; Zhang, C.; Yang, Y. Effects of mechanical forces on osteogenesis and osteoclastogenesis in human periodontal ligament fibroblasts: A systematic review of in vitro studies. Bone Joint Res. 2019, 8, 19–31. [Google Scholar] [CrossRef]
- Nikolova, M.P.; Chavali, M.S. Recent advances in biomaterials for 3D scaffolds: A review. Bioact. Mater. 2019, 4, 271–292. [Google Scholar] [CrossRef]
- Knight, E.; Przyborski, S. Advances in 3D cell culture technologies enabling tissue-like structures to be created in vitro. J. Anat. 2015, 227, 746–756. [Google Scholar] [CrossRef] [Green Version]
- Laschke, M.W.; Menger, M.D. Life is 3D: Boosting Spheroid Function for Tissue Engineering. Trends Biotechnol. 2017, 35, 133–144. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-N.; Nasir, A.; Camacho, S.; Berry, D.L.; Schmidt, M.O.; Pearson, G.W.; Riegel, A.T.; Wellstein, A. Monitoring Cancer Cell Invasion and T-Cell Cytotoxicity in 3D Culture. JoVE J. 2020, e61392. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Shen, S.-M.; Zhao, X.-Y.; Chen, G.-Q. Targeted genes and interacting proteins of hypoxia inducible factor-1. Int. J. Biochem. Mol. Biol. 2012, 3, 165–178. [Google Scholar]
- Kuschel, A.; Simon, P.; Tug, S. Functional regulation of HIF-1α under normoxia—Is there more than post-translational regulation? J. Cell. Physiol. 2012, 227, 514–524. [Google Scholar] [CrossRef] [PubMed]
- Li, M.L.; Yi, J.; Yang, Y.; Zhang, X.; Zheng, W.; Li, Y.; Zhao, Z. Compression and hypoxia play independent roles while having combinative effects in the osteoclastogenesis induced by periodontal ligament cells. Angle Orthod. 2016, 86, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Miyagawa, A.; Chiba, M.; Hayashi, H.; Igarashi, K. Compressive force induces VEGF production in periodontal tissues. J. Dent. Res. 2009, 88, 752–756. [Google Scholar] [CrossRef] [PubMed]
- Kook, S.-H.; Hwang, J.-M.; Park, J.-S.; Kim, E.-M.; Heo, J.-S.; Jeon, Y.-M.; Lee, J.-C. Mechanical force induces type I collagen expression in human periodontal ligament fibroblasts through activation of ERK/JNK and AP-1. J. Cell Biochem. 2009, 106, 1060–1067. [Google Scholar] [CrossRef]
- Hakkinen, K.M.; Harunaga, J.S.; Doyle, A.D.; Yamada, K.M. Direct comparisons of the morphology, migration, cell adhesions, and actin cytoskeleton of fibroblasts in four different three-dimensional extracellular matrices. Tissue Eng. Part A 2011, 17, 713–724. [Google Scholar] [CrossRef] [Green Version]
- Oortgiesen, D.A.W.; Yu, N.; Bronckers, A.L.J.J.; Yang, F.; Walboomers, X.F.; Jansen, J.A. A three-dimensional cell culture model to study the mechano-biological behavior in periodontal ligament regeneration. Tissue Eng. Part C Methods 2012, 18, 81–89. [Google Scholar] [CrossRef] [Green Version]
- Berendsen, A.D.; Smit, T.H.; Walboomers, X.F.; Everts, V.; Jansen, J.A.; Bronckers, A.L.J.J. Three-dimensional loading model for periodontal ligament regeneration in vitro. Tissue Eng. Part C Methods 2009, 15, 561–570. [Google Scholar] [CrossRef] [Green Version]
- Vaheri, A.; Enzerink, A.; Räsänen, K.; Salmenperä, P. Nemosis, a novel way of fibroblast activation, in inflammation and cancer. Exp. Cell. Res. 2009, 315, 1633–1638. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, R.M. Cell and environment interactions in tumor microregions: The multicell spheroid model. Science 1988, 240, 177–184. [Google Scholar] [CrossRef] [PubMed]
- Htwe, S.S.; Harrington, H.; Knox, A.; Rose, F.; Aylott, J.; Haycock, J.W.; Ghaemmaghami, A.M. Investigating NF-κB signaling in lung fibroblasts in 2D and 3D culture systems. Respir. Res. 2015, 16, 144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barnes, P.J.; Karin, M. Nuclear factor-kappaB: A pivotal transcription factor in chronic inflammatory diseases. N. Engl. J. Med. 1997, 336, 1066–1071. [Google Scholar] [CrossRef] [PubMed]
- Proff, P.; Römer, P. The molecular mechanism behind bone remodelling: A review. Clin. Oral Investig. 2009, 13, 355–362. [Google Scholar] [CrossRef]
- Tyrovola, J.B.; Spyropoulos, M.N.; Makou, M.; Perrea, D. Root resorption and the OPG/RANKL/RANK system: A mini review. J. Oral Sci. 2008, 50, 367–376. [Google Scholar] [CrossRef] [Green Version]
- Schröder, A. Expression kinetics of human periodontal ligament fibroblasts in the early phases of orthodontic tooth movement. J. Orofac. Orthop. 2018, 79, 337–351. [Google Scholar] [CrossRef]
- Ullrich, N.; Schröder, A.; Jantsch, J.; Spanier, G.; Proff, P.; Kirschneck, C. The role of mechanotransduction versus hypoxia during simulated orthodontic compressive strain-an in vitro study of human periodontal ligament fibroblasts. Int. J. Oral Sci. 2019, 11, 33. [Google Scholar] [CrossRef] [Green Version]
- Ducy, P.; Schinke, T.; Karsenty, G. The osteoblast: A sophisticated fibroblast under central surveillance. Science 2000, 289, 1501–1504. [Google Scholar] [CrossRef]
- Shen, X.-Q.; Geng, Y.-M.; Liu, P.; Huang, X.-Y.; Li, S.-Y.; Liu, C.-D.; Zhou, Z.; Xu, P.-P. Magnitude-dependent response of osteoblasts regulated by compressive stress. Sci. Rep. 2017, 7, 44925. [Google Scholar] [CrossRef] [Green Version]
- Ren, Y.; Maltha, J.C.; Kuijpers-Jagtman, A.M. Optimum force magnitude for orthodontic tooth movement: A systematic literature review. Angle Orthod. 2003, 73, 86–92. [Google Scholar] [CrossRef] [PubMed]
- Schröder, A.; Gubernator, J.; Nazet, U.; Spanier, G.; Jantsch, J.; Proff, P.; Kirschneck, C. Effects of sodium chloride on the gene expression profile of periodontal ligament fibroblasts during tensile strain. J. Orofac. Orthop. 2020, 81, 360–370. [Google Scholar] [CrossRef] [PubMed]
- Garlet, T.P.; Coelho, U.; Silva, J.S.; Garlet, G.P. Cytokine expression pattern in compression and tension sides of the periodontal ligament during orthodontic tooth movement in humans. Eur. J. Oral Sci. 2007, 115, 355–362. [Google Scholar] [CrossRef] [PubMed]
- Kirschneck, C.; Batschkus, S.; Proff, P.; Köstler, J.; Spanier, G.; Schröder, A. Valid gene expression normalization by RT-qPCR in studies on hPDL fibroblasts with focus on orthodontic tooth movement and periodontitis. Sci. Rep. 2017, 7, 14751. [Google Scholar] [CrossRef] [Green Version]
- De La Zerda, A.; Kratochvil, M.J.; Suhar, N.A.; Heilshorn, S.C. Review: Bioengineering strategies to probe T cell mechanobiology. APL Bioeng. 2018, 2, 21501. [Google Scholar] [CrossRef]
- Ma, Y.; Ji, Y.; Huang, G.; Ling, K.; Zhang, X.; Xu, F. Bioprinting 3D cell-laden hydrogel microarray for screening human periodontal ligament stem cell response to extracellular matrix. Biofabrication 2015, 7, 44105. [Google Scholar] [CrossRef]

| Gene Symbol | Gene Name | 5′-Forward Primer-3′ | 5′-Reverse Primer-3′ |
|---|---|---|---|
| COL1A2 | Collagen-type-I-alpha-2 | AGAAACACGTCTGGCTAGGAG | GCATGAAGGCAAGTTGGGTAG |
| IL6 | Interleukin-6 | TGGCAGAAAACAACCTGAACC | CCTCAAACTCCAAAAGACCAGTG |
| OPG | Osteoprotegerin | TGTCTTTGGTCTCCTGCTAACTC | ACGCTCCAGGACTTATACCG |
| PPIB | Peptidylprolyl isomerase A | TTCCATCGTGTAATCAAGGACTTC | GCTCACCGTAGATGCTCTTTC |
| PTGS2 | Prostaglandin endoperoxide synthase 2 | GAGCAGGCAGATGAAATACCAGTC | TGTCACCATAGAGTGCTTCCAAC |
| RANKL | Receptor activator of NFk-B ligand | ATACCCTGATGAAAGGAGGA | GGGGCTCAATCTATATCTCG |
| RPL22 | Ribosomal protein L22 | TGATTGCACCCACCCTGTAG | GGTTCCCAGCTTTTCCGTTC |
| VEGF | Vascular endothelial growth factor | TGCAGACCAAAGAAAGATAGAGC | ACGCTCCAGGACTTATACCG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schröder, A.; Schöniger, R.; Oeldemann, J.; Spanier, G.; Proff, P.; Jantsch, J.; Kirschneck, C.; Ullrich, N. An Evaluation of Different 3D Cultivation Models on Expression Profiles of Human Periodontal Ligament Fibroblasts with Compressive Strain. Int. J. Mol. Sci. 2022, 23, 2029. https://doi.org/10.3390/ijms23042029
Schröder A, Schöniger R, Oeldemann J, Spanier G, Proff P, Jantsch J, Kirschneck C, Ullrich N. An Evaluation of Different 3D Cultivation Models on Expression Profiles of Human Periodontal Ligament Fibroblasts with Compressive Strain. International Journal of Molecular Sciences. 2022; 23(4):2029. https://doi.org/10.3390/ijms23042029
Chicago/Turabian StyleSchröder, Agnes, Ricarda Schöniger, Juliane Oeldemann, Gerrit Spanier, Peter Proff, Jonathan Jantsch, Christian Kirschneck, and Niklas Ullrich. 2022. "An Evaluation of Different 3D Cultivation Models on Expression Profiles of Human Periodontal Ligament Fibroblasts with Compressive Strain" International Journal of Molecular Sciences 23, no. 4: 2029. https://doi.org/10.3390/ijms23042029







