WMR Peptide as Antifungal and Antibiofilm against Albicans and Non-Albicans Candida Species: Shreds of Evidence on the Mechanism of Action
Abstract
:1. Introduction
2. Results
2.1. WMR Activity In Vitro and In Vivo on Planktonic Cells and Biofilm
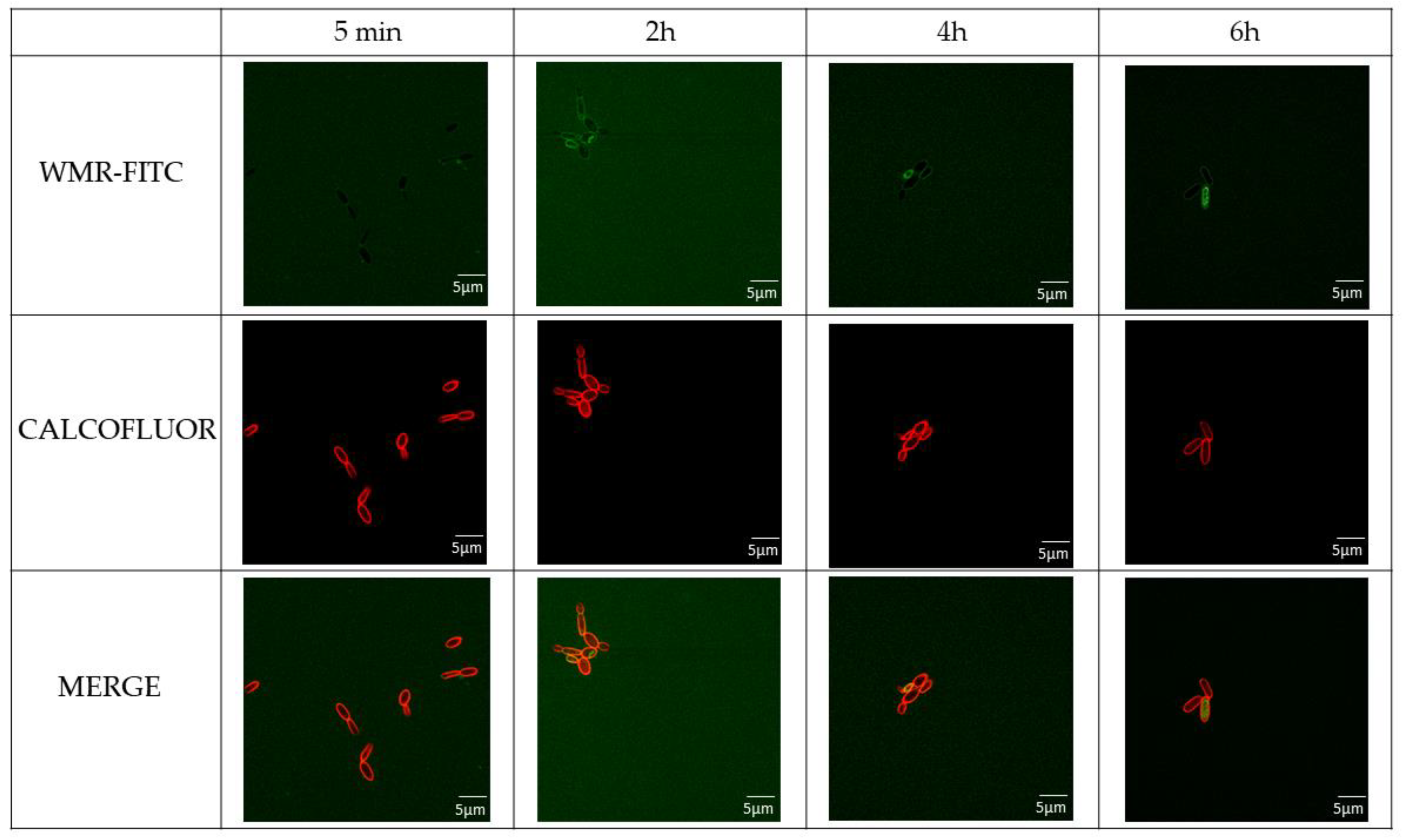
2.2. WMR Mechanism of Activity on Planktonic Cells and Localization of FITC-Labeled WMR
3. Discussion
4. Materials and Methods
4.1. Candida Strains, Media and Growth Conditions
4.2. Peptide Synthesis
4.3. Fluorescein-Labelling of WMR
4.4. Determination of Minimum Inhibitory Concentrations and Minimum Fungicidal Concentration of Planktonic Cells
4.5. Biofilm Formation, Inhibition and Eradication
4.6. Effect of WMR on Gene Expression during Biofilm Inhibition
4.7. Determination of In Vivo Antifungal Effects Using the G. mellonella Infection Model
4.8. Time–Kill Kinetic Analysis
4.9. Checkerboard Assays: Assessment of the In Vitro Synergy Activity of WMR Alone and in Combination with Fluconazole
4.10. Measurement of Intracellular ROS Levels and Mitochondrial Specific ROS Accumulation
4.11. Cell Rescue Assay Using ROS Scavengers
4.12. Rh6G Efflux Assay
4.13. CLSM Analysis
4.14. Localization of FITC-Labeled Peptide in C. parapsilosis
4.15. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cortegiani, A.; Misseri, G.; Fasciana, T.; Giammanco, A.; Giarratano, A.; Chowdhary, A. Epidemiology, clinical characteristics, resistance, and treatment of infections by Candida auris. J. Intensive Care 2018, 6, 69. [Google Scholar] [CrossRef] [PubMed]
- Dadar, M.; Tiwari, R.; Karthik, K.; Chakraborty, S.; Shahali, Y.; Dhama, K. Candida albicans—Biology, molecular characterization, pathogenicity, and advances in diagnosis and control—An update. Microb. Pathog. 2018, 117, 128–138. [Google Scholar] [CrossRef] [PubMed]
- Rajendran, R.; Sherry, L.; Nile, C.J.; Sherriff, A.; Johnson, E.M.; Hanson, M.F.; Williams, C.; Munro, C.A.; Jones, B.J.; Ramage, G. Biofilm formation is a risk factor for mortality in patients with Candida albicans bloodstream infection-Scotland, 2012–2013. Clin. Microbiol. Infect. 2016, 22, 87–93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leite de Andrade, M.C.; Soares de Oliveira, M.A.; Santos, F.; Ximenes Vilela, P.B.; da Silva, M.N.; Macedo, D.P.C.; de Lima Neto, R.G.; Neves, H.J.P.; Brandao, I.S.L.; Chaves, G.M.; et al. A new approach by optical coherence tomography for elucidating biofilm formation by emergent Candida species. PLoS ONE 2017, 12, e0188020. [Google Scholar] [CrossRef] [Green Version]
- Whaley, S.G.; Berkow, E.L.; Rybak, J.M.; Nishimoto, A.T.; Barker, K.S.; Rogers, P.D. Azole Antifungal Resistance in Candida albicans and Emerging Non-albicans Candida Species. Front. Microbiol. 2017, 7, 2173. [Google Scholar] [CrossRef] [Green Version]
- Hendrickson, J.A.; Hu, C.; Aitken, S.L.; Beyda, N. Antifungal Resistance: A Concerning Trend for the Present and Future. Curr. Infect. Dis. Rep. 2019, 21, 47. [Google Scholar] [CrossRef]
- Bhattacharya, S.A.-O.; Sae-Tia, S.; Fries, B.A.-O. Candidiasis and Mechanisms of Antifungal Resistance. Antibiotics 2020, 9, 312. [Google Scholar] [CrossRef]
- Du, H.A.-O.; Bing, J.A.-O.; Hu, T.A.-O.; Ennis, C.A.-O.; Nobile, C.A.-O.; Huang, G.A.-O. Candida auris: Epidemiology, biology, antifungal resistance, and virulence. PLOS Pathog. 2020, 16, e1008921. [Google Scholar] [CrossRef]
- Falanga, A.; Lombardi, L.; Franci, G.; Vitiello, M.; Iovene, M.R.; Morelli, G.; Galdiero, M.; Galdiero, S. Marine Antimicrobial Peptides: Nature Provides Templates for the Design of Novel Compounds against Pathogenic Bacteria. Int. J. Mol. Sci. 2016, 17, 785. [Google Scholar] [CrossRef] [Green Version]
- Lombardi, L.; Falanga, A.; Del Genio, V.; Galdiero, S. A New Hope: Self-Assembling Peptides with Antimicrobial Activity. Pharmaceutics 2019, 11, 166. [Google Scholar] [CrossRef] [Green Version]
- Rima, M.; Rima, M.; Fajloun, Z.; Sabatier, J.M.; Bechinger, B.; Naas, T. Antimicrobial Peptides: A Potent Alternative to Antibiotics. Antibiotics 2021, 10, 95. [Google Scholar] [CrossRef]
- Huan, Y.; Kong, Q.; Mou, H.; Yi, H. Antimicrobial Peptides: Classification, Design, Application and Research Progress in Multiple Fields. Front. Microbiol. 2020, 11, 582779. [Google Scholar] [CrossRef]
- Galdiero, S.; Falanga, A.; Berisio, R.; Grieco, P.; Morelli, G.; Galdiero, M. Antimicrobial peptides as an opportunity against bacterial diseases. Curr. Med. Chem. 2015, 22, 1665–1677. [Google Scholar] [CrossRef]
- Scudiero, O.; Galdiero, S.; Nigro, E.; Del Vecchio, L.; Di Noto, R.; Cantisani, M.; Colavita, I.; Galdiero, M.; Cassiman, J.J.; Daniele, A.; et al. Chimeric beta-defensin analogs, including the novel 3NI analog, display salt-resistant antimicrobial activity and lack toxicity in human epithelial cell lines. Antimicrob. Agents Chemother. 2013, 57, 1701–1708. [Google Scholar] [CrossRef] [Green Version]
- Scudiero, O.; Nigro, E.; Cantisani, M.; Colavita, I.; Leone, M.; Mercurio, F.A.; Galdiero, M.; Pessi, A.; Daniele, A.; Salvatore, F.; et al. Design and activity of a cyclic mini-β-defensin analog: A novel antimicrobial tool. Int. J. Nanomed. 2015, 10, 6523–6539. [Google Scholar] [CrossRef] [Green Version]
- Ashrafuzzaman, M. The Antimicrobial Peptide Gramicidin S Enhances Membrane Adsorption and Ion Pore Formation Potency of Chemotherapy Drugs in Lipid Bilayers. Membranes 2021, 11, 247. [Google Scholar] [CrossRef]
- Priyadarshini, D.; Ivica, J.; Separovic, F.; de Planque, M.R.R. Characterisation of cell membrane interaction mechanisms of antimicrobial peptides by electrical bilayer recording. Biophys. Chem. 2021, 281, 106721. [Google Scholar] [CrossRef]
- Di Somma, A.A.-O.; Moretta, A.; Canè, C.; Cirillo, A.; Duilio, A. Antimicrobial and Antibiofilm Peptides. Biomolecules 2020, 10, 652. [Google Scholar] [CrossRef] [Green Version]
- Subramanian, S.; Ross, N.W.; MacKinnon, S.L. Myxinidin, a novel antimicrobial peptide from the epidermal mucus of hagfish, Myxine glutinosa L. Mar. Biotechnol. 2009, 11, 748–757. [Google Scholar] [CrossRef]
- Cantisani, M.; Leone, M.; Mignogna, E.; Kampanaraki, K.; Falanga, A.; Morelli, G.; Galdiero, M.; Galdiero, S. Structure-activity relations of myxinidin, an antibacterial peptide derived from the epidermal mucus of hagfish. Antimicrob. Agents Chemother. 2013, 57, 5665–5673. [Google Scholar] [CrossRef] [Green Version]
- Cantisani, M.; Finamore, E.; Mignogna, E.; Falanga, A.; Nicoletti, G.F.; Pedone, C.; Morelli, G.; Leone, M.; Galdiero, M.; Galdiero, S. Structural insights into and activity analysis of the antimicrobial peptide myxinidin. Antimicrob. Agents Chemother. 2014, 58, 5280–5290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lombardi, L.; Stellato, M.I.; Oliva, R.; Falanga, A.; Galdiero, M.; Petraccone, L.; D’Errico, G.; De Santis, A.; Galdiero, S.; Del Vecchio, P. Antimicrobial peptides at work: Interaction of myxinidin and its mutant WMR with lipid bilayers mimicking the P. aeruginosa and E. coli membranes. Sci. Rep. 2017, 7, 44425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lombardi, L.; Shi, Y.; Falanga, A.; Galdiero, E.; de Alteriis, E.; Franci, G.; Chourpa, I.; Azevedo, H.S.; Galdiero, S. Enhancing the Potency of Antimicrobial Peptides through Molecular Engineering and Self-Assembly. Biomacromolecules 2019, 20, 1362–1374. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Xie, Z.; Tan, X.; Guo, R.; Song, Y.; Xie, X.; Wang, R.; Li, L.; Wang, M.; Zhang, Y.A.-O. Temporin-Like Peptides Show Antimicrobial and Anti-Biofilm Activities against Streptococcus mutans with Reduced Hemolysis. Molecules 2020, 25, 5724. [Google Scholar] [CrossRef]
- Bellavita, R.A.-O.; Casciaro, B.; Di Maro, S.A.-O.; Brancaccio, D.; Carotenuto, A.A.-O.; Falanga, A.; Cappiello, F.; Buommino, E.; Galdiero, S.; Novellino, E.A.-O.; et al. First-in-Class Cyclic Temporin L Analogue: Design, Synthesis, and Antimicrobial Assessment. J. Med. Chem. 2021, 64, 11675–11694. [Google Scholar] [CrossRef]
- Chen, J.; Hu, N.; Xu, H.; Liu, Q.; Yu, X.; Zhang, Y.; Huang, Y.; Tan, J.; Huang, X.; Zeng, L. Molecular Epidemiology, Antifungal Susceptibility, and Virulence Evaluation of Candida Isolates Causing Invasive Infection in a Tertiary Care Teaching Hospital. Front. Cell. Infect. Microbiol. 2021, 11, 721439. [Google Scholar] [CrossRef]
- Pammi, M.; Holland, L.; Butler, G.; Gacser, A.; Bliss, J.M. Candida parapsilosis is a significant neonatal pathogen: A systematic review and meta-analysis. Pediatr. Infect. Dis. J. 2013, 32, e206–e216. [Google Scholar] [CrossRef] [Green Version]
- Trofa, D.; Gácser, A.; Nosanchuk, J.D. Candida parapsilosis, an emerging fungal pathogen. Clin. Microbiol. Rev. 2008, 21, 606–625. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Ren, H.; Wang, D.; Zhang, M.; Sun, S.; Zhao, Y. The synergistic antifungal effects of gypenosides combined with fluconazole against resistant Candida albicans via inhibiting the drug efflux and biofilm formation. Biomed. Pharmacother. 2020, 130, 110580. [Google Scholar] [CrossRef]
- Łukowska-Chojnacka, E.; Mierzejewska, J.; Milner-Krawczyk, M.; Bondaryk, M.; Staniszewska, M. Synthesis of novel tetrazole derivatives and evaluation of their antifungal activity. Bioorganic Med. Chem. 2016, 24, 6058–6065. [Google Scholar] [CrossRef]
- Kovács, R.; Majoros, L. Fungal Quorum-Sensing Molecules: A Review of Their Antifungal Effect against Candida Biofilms. J. Fungi 2020, 6, 99. [Google Scholar] [CrossRef]
- Day, A.M.; Quinn, J. Stress-Activated Protein Kinases in Human Fungal Pathogens. Front. Cell. Infect. Microbiol. 2019, 9, 261. [Google Scholar] [CrossRef] [Green Version]
- Merlino, F.; Tomassi, S.; Yousif, A.M.; Messere, A.; Marinelli, L.; Grieco, P.; Novellino, E.; Cosconati, S.; Di Maro, S. Boosting Fmoc Solid-Phase Peptide Synthesis by Ultrasonication. Org. Lett. 2019, 21, 6378–6382. [Google Scholar] [CrossRef]
- Wendt, M.; Bellavita, R.; Gerber, A.; Efrém, N.L.; van Ramshorst, T.; Pearce, N.M.; Davey, P.R.J.; Everard, I.; Vazquez-Chantada, M.; Chiarparin, E.; et al. Bicyclic β-Sheet Mimetics that Target the Transcriptional Coactivator β-Catenin and Inhibit Wnt Signaling. Angew. Chem. (Int. Ed. Engl.) 2021, 60, 13937–13944. [Google Scholar] [CrossRef]
- Wayne, P. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts, 3rd ed.; M27-A3; Clinical and Laboratory Standards Institute (CLSI): Wayne, PA, USA, 2008. [Google Scholar]
- Stepanovic, S.; Vukovic, D.; Dakic, I.; Savic, B.; Svabic-Vlahovic, M. A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J. Microbiol. Methods 2000, 40, 175–179. [Google Scholar] [CrossRef]
- Stepanović, S.; Vuković, D.; Hola, V.; Di Bonaventura, G.; Djukić, S.; Cirković, I.; Ruzicka, F. Quantification of biofilm in microtiter plates: Overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. APMIS Acta Pathol. Microbiol. Et Immunol. Scand. 2007, 115, 891–899. [Google Scholar] [CrossRef]
- Di Onofrio, V.; Gesuele, R.; Maione, A.; Liguori, G.; Liguori, R.; Guida, M.; Nigro, R.; Galdiero, E. Prevention of Pseudomonas aeruginosa Biofilm Formation on Soft Contact Lenses by Allium sativum Fermented Extract (BGE) and Cannabinol Oil Extract (CBD). Antibiotics 2019, 8, 258. [Google Scholar] [CrossRef] [Green Version]
- Riesgo, A.; Pérez-Porro, A.R.; Carmona, S.; Leys, S.P.; Giribet, G. Optimization of preservation and storage time of sponge tissues to obtain quality mRNA for next-generation sequencing. Mol. Ecol. Resour. 2012, 12, 312–322. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Horgan, G.W.; Dempfle, L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002, 30, e36. [Google Scholar] [CrossRef]
- Maione, A.; de Alteriis, E.; Carraturo, F.; Galdiero, S.; Falanga, A.; Guida, M.; Di Cosmo, A.; Maselli, V.; Galdiero, E. The Membranotropic Peptide gH625 to Combat Mixed Candida albicans/Klebsiella pneumoniae Biofilm: Correlation between In Vitro Anti-Biofilm Activity and In Vivo Antimicrobial Protection. J. Fungi 2021, 7, 26. [Google Scholar] [CrossRef]
- Frenkel, M.; Mandelblat, M.; Alastruey-Izquierdo, A.; Mendlovic, S.; Semis, R.; Segal, E. Pathogenicity of Candida albicans isolates from bloodstream and mucosal candidiasis assessed in mice and Galleria mellonella. J. De Mycol. Med. 2016, 26, 1–8. [Google Scholar] [CrossRef]
- Li, X.; Zhao, Y.; Huang, X.; Yu, C.; Yang, Y.; Sun, S. Ambroxol Hydrochloride Combined with Fluconazole Reverses the Resistance of Candida albicans to Fluconazole. Front. Cell. Infect. Microbiol. 2017, 7, 124. [Google Scholar] [CrossRef]
- Galdiero, E.; Salvatore, M.M.; Maione, A.; Carraturo, F.; Galdiero, S.; Falanga, A.; Andolfi, A.; Salvatore, F.; Guida, M. Impact of the Peptide WMR-K on Dual-Species Biofilm Candida albicans/Klebsiella pneumoniae and on the Untargeted Metabolomic Profile. Pathogens 2021, 10, 214. [Google Scholar] [CrossRef]
- Tan, J.; Jiang, S.; Tan, L.; Shi, H.; Yang, L.; Sun, Y.; Wang, X. Antifungal Activity of Minocycline and Azoles Against Fluconazole-90Resistant Candida Species. Front. Microbiol. 2021, 12, 649026. [Google Scholar] [CrossRef]
- Chang, C.K.; Kao, M.C.; Lan, C.Y. Antimicrobial Activity of the Peptide LfcinB15 against Candida albicans. J. Fungi 2021, 7. [Google Scholar] [CrossRef]
- Soto, S.M. Role of efflux pumps in the antibiotic resistance of bacteria embedded in a biofilm. Virulence 2013, 4, 223–229. [Google Scholar] [CrossRef] [Green Version]
- Heydorn, A.; Nielsen, A.T.; Hentzer, M.; Sternberg, C.; Givskov, M.; Ersbøll, B.K.; Molin, S. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 2000, 146 Pt 10, 2395–2407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
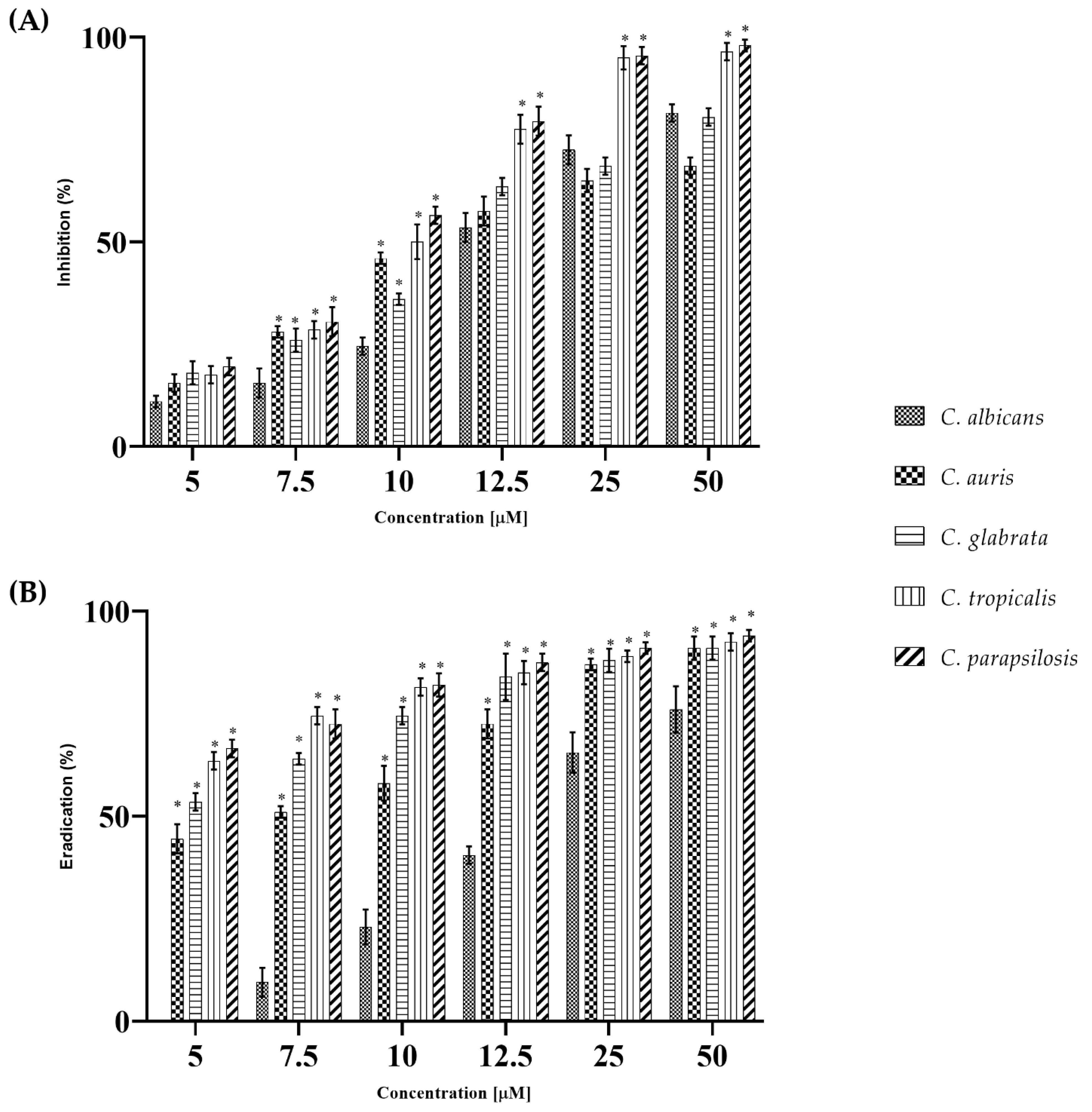
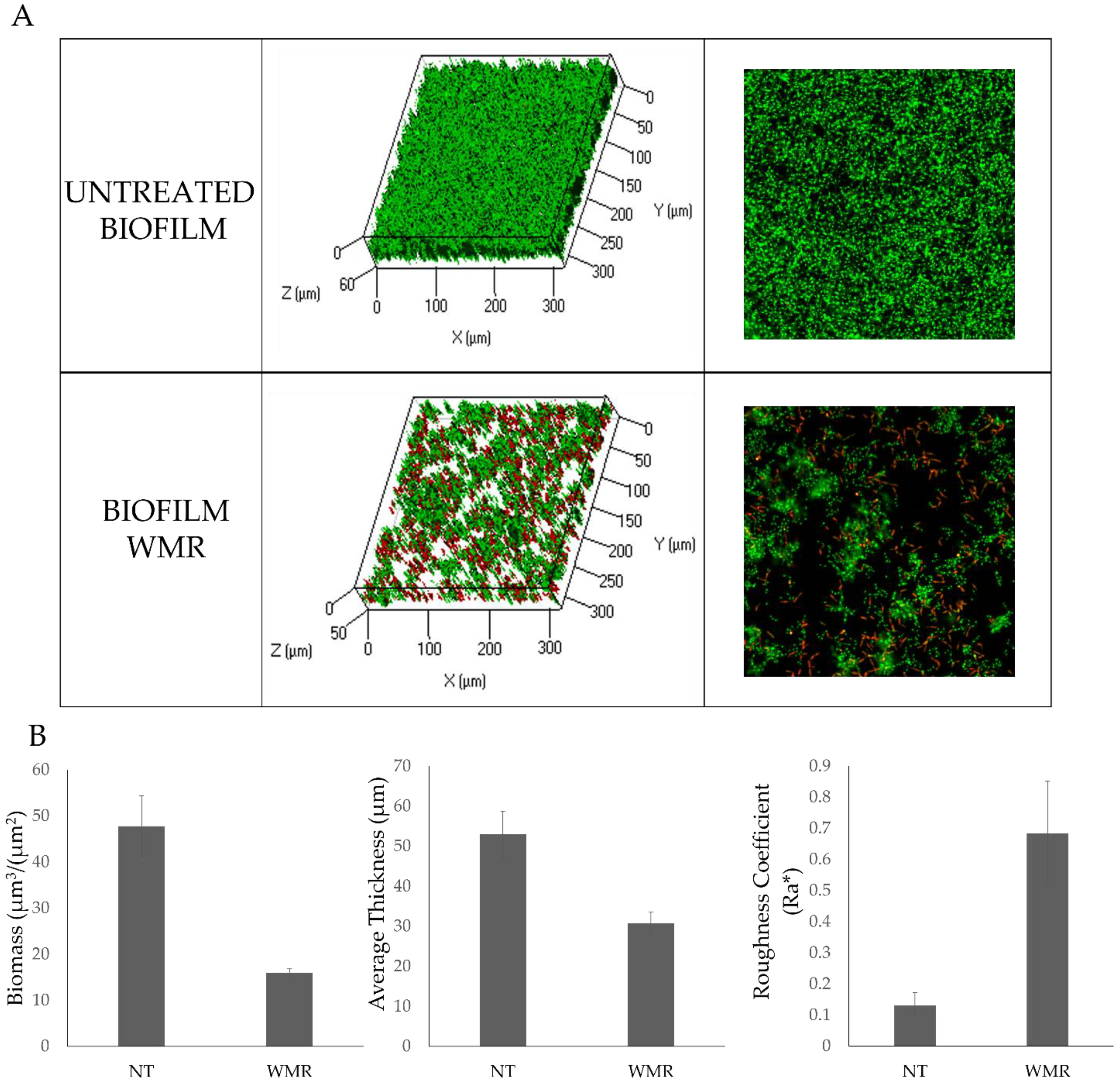



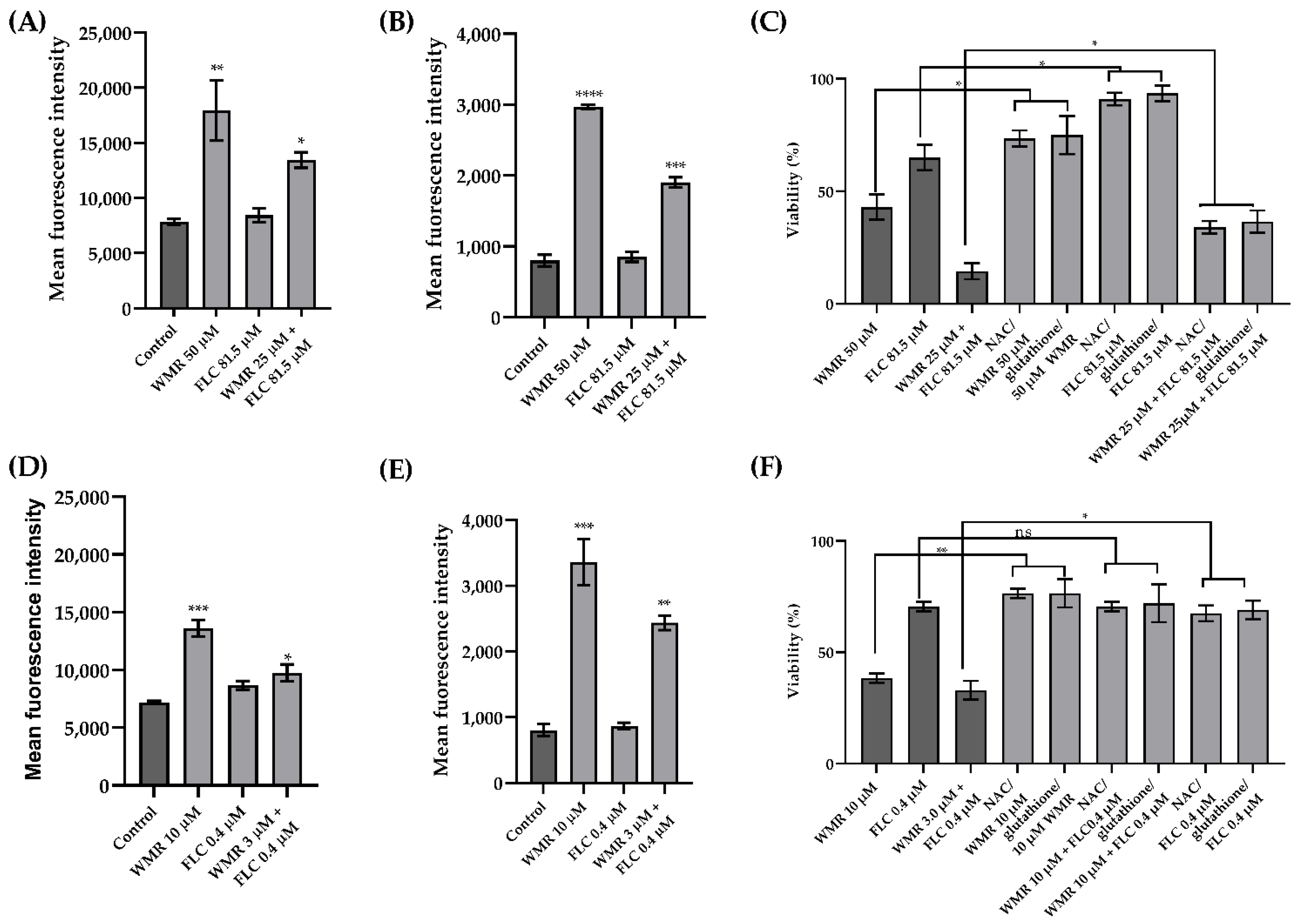
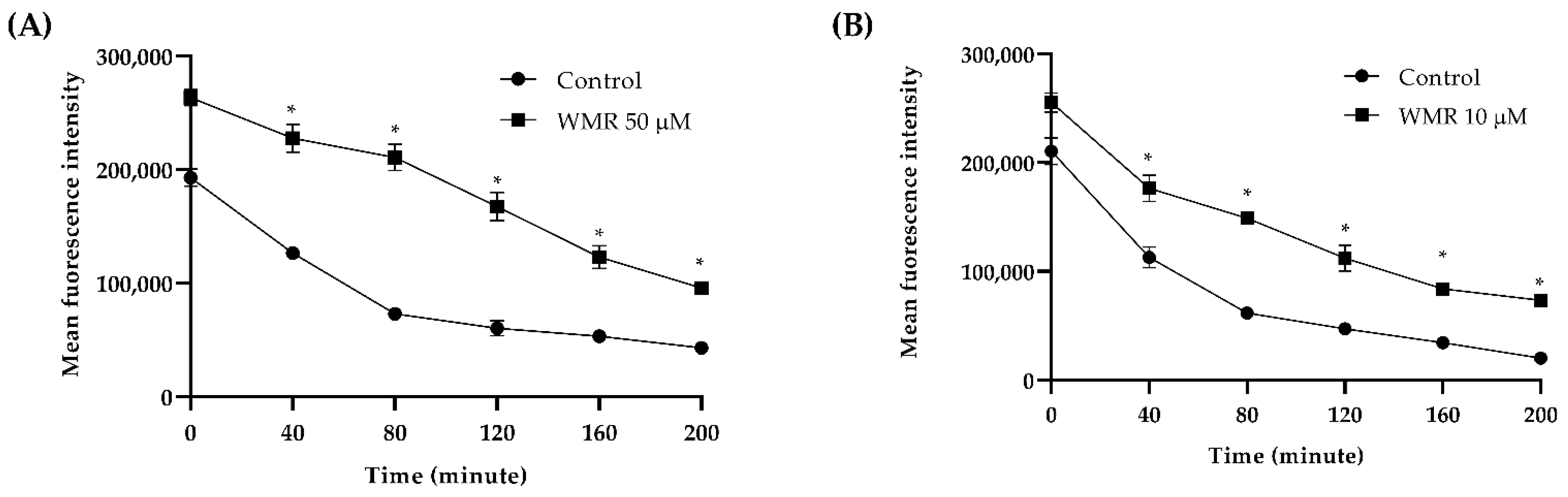
| Strains | WMR (µM) | FLC (µM) | ||
|---|---|---|---|---|
| MIC | MFC | MIC | MFC | |
| C. albicans | >50.0 | >50.0 | >163.0 | >163.0 |
| C. auris | >50.0 | >50.0 | 163.0 | >163.0 |
| C. glabrata | >50.0 | >50.0 | >163.0 | >163.0 |
| C. tropicalis | 25.0 | 50.0 | 81.5 | >163.0 |
| C. parapsilosis | 25.0 | 25.0 | 16.3 | 32.6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maione, A.; Bellavita, R.; de Alteriis, E.; Galdiero, S.; Albarano, L.; La Pietra, A.; Guida, M.; Parrilli, E.; D’Angelo, C.; Galdiero, E.; et al. WMR Peptide as Antifungal and Antibiofilm against Albicans and Non-Albicans Candida Species: Shreds of Evidence on the Mechanism of Action. Int. J. Mol. Sci. 2022, 23, 2151. https://doi.org/10.3390/ijms23042151
Maione A, Bellavita R, de Alteriis E, Galdiero S, Albarano L, La Pietra A, Guida M, Parrilli E, D’Angelo C, Galdiero E, et al. WMR Peptide as Antifungal and Antibiofilm against Albicans and Non-Albicans Candida Species: Shreds of Evidence on the Mechanism of Action. International Journal of Molecular Sciences. 2022; 23(4):2151. https://doi.org/10.3390/ijms23042151
Chicago/Turabian StyleMaione, Angela, Rosa Bellavita, Elisabetta de Alteriis, Stefania Galdiero, Luisa Albarano, Alessandra La Pietra, Marco Guida, Ermenegilda Parrilli, Caterina D’Angelo, Emilia Galdiero, and et al. 2022. "WMR Peptide as Antifungal and Antibiofilm against Albicans and Non-Albicans Candida Species: Shreds of Evidence on the Mechanism of Action" International Journal of Molecular Sciences 23, no. 4: 2151. https://doi.org/10.3390/ijms23042151













