Transcriptome and Metabolite Conjoint Analysis Reveals the Seed Dormancy Release Process in Callery Pear
Abstract
:1. Introduction
2. Results
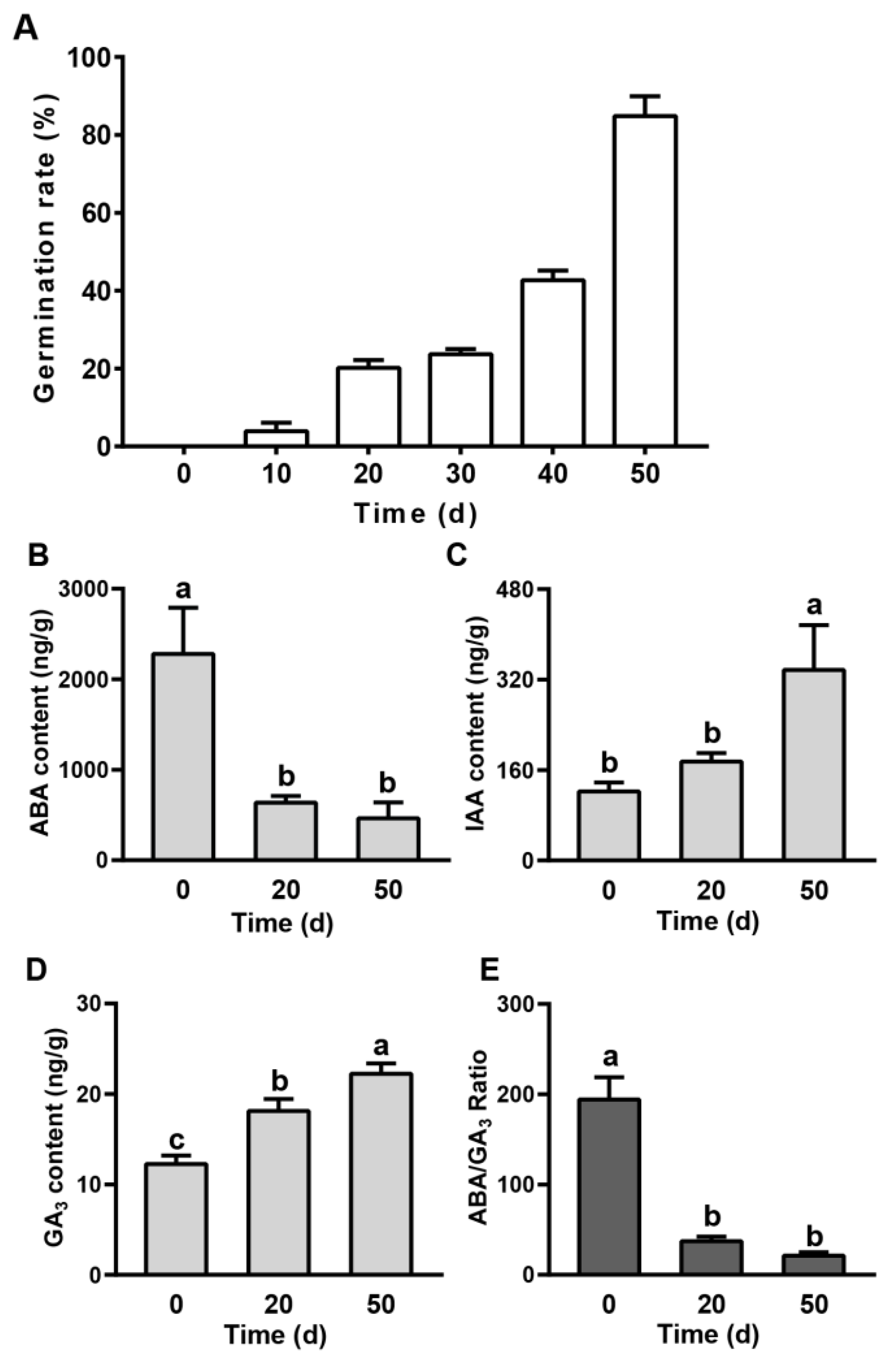
2.1. Evaluation of the Seed Dormancy Status and Phytohormones in Pear Seed during Dormancy Release Progression
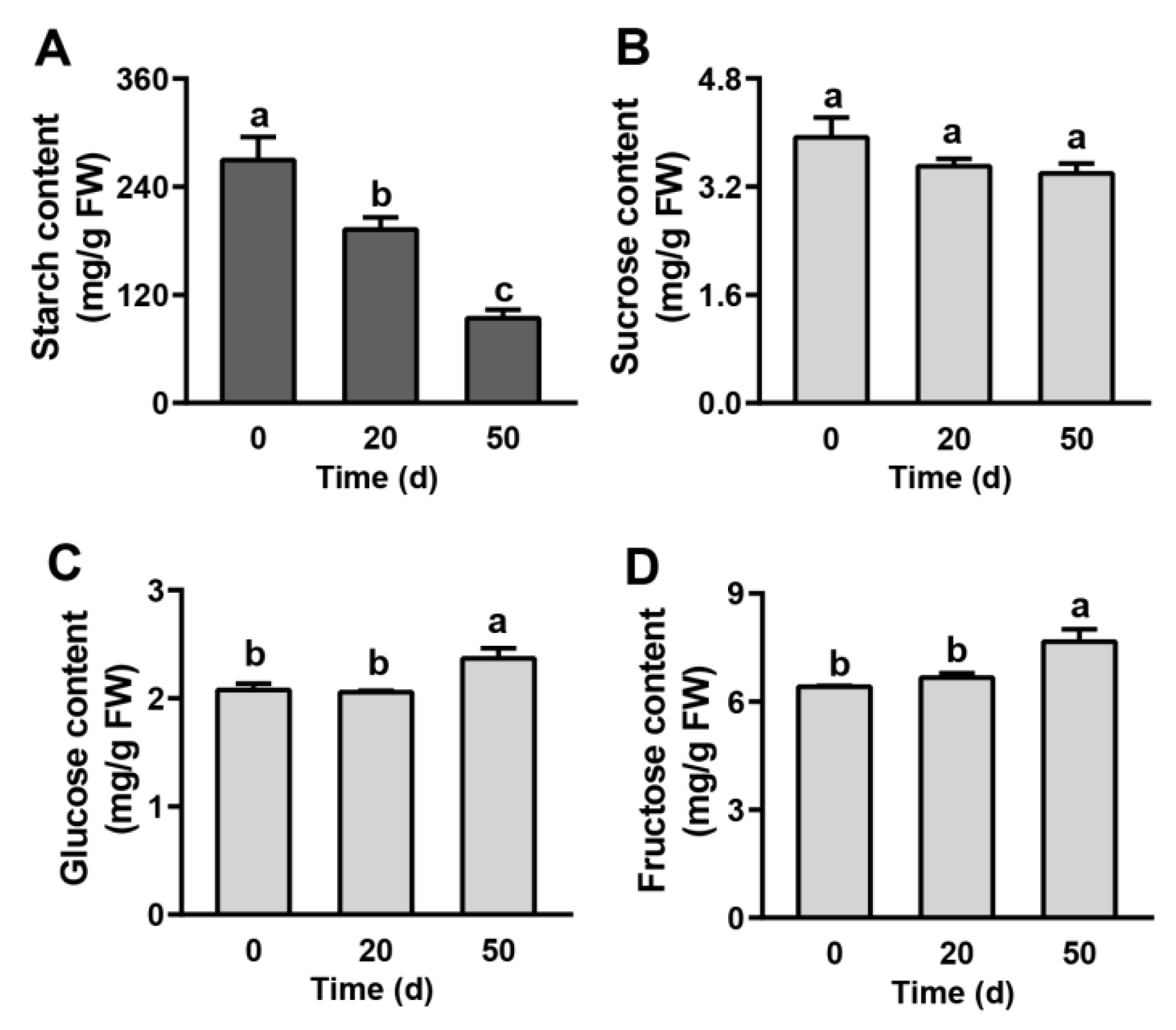
2.2. Carbohydrate Contents in Seeds during Dormancy Release Progression
2.3. Transcriptome Analysis of Seeds during Dormancy Release Progression
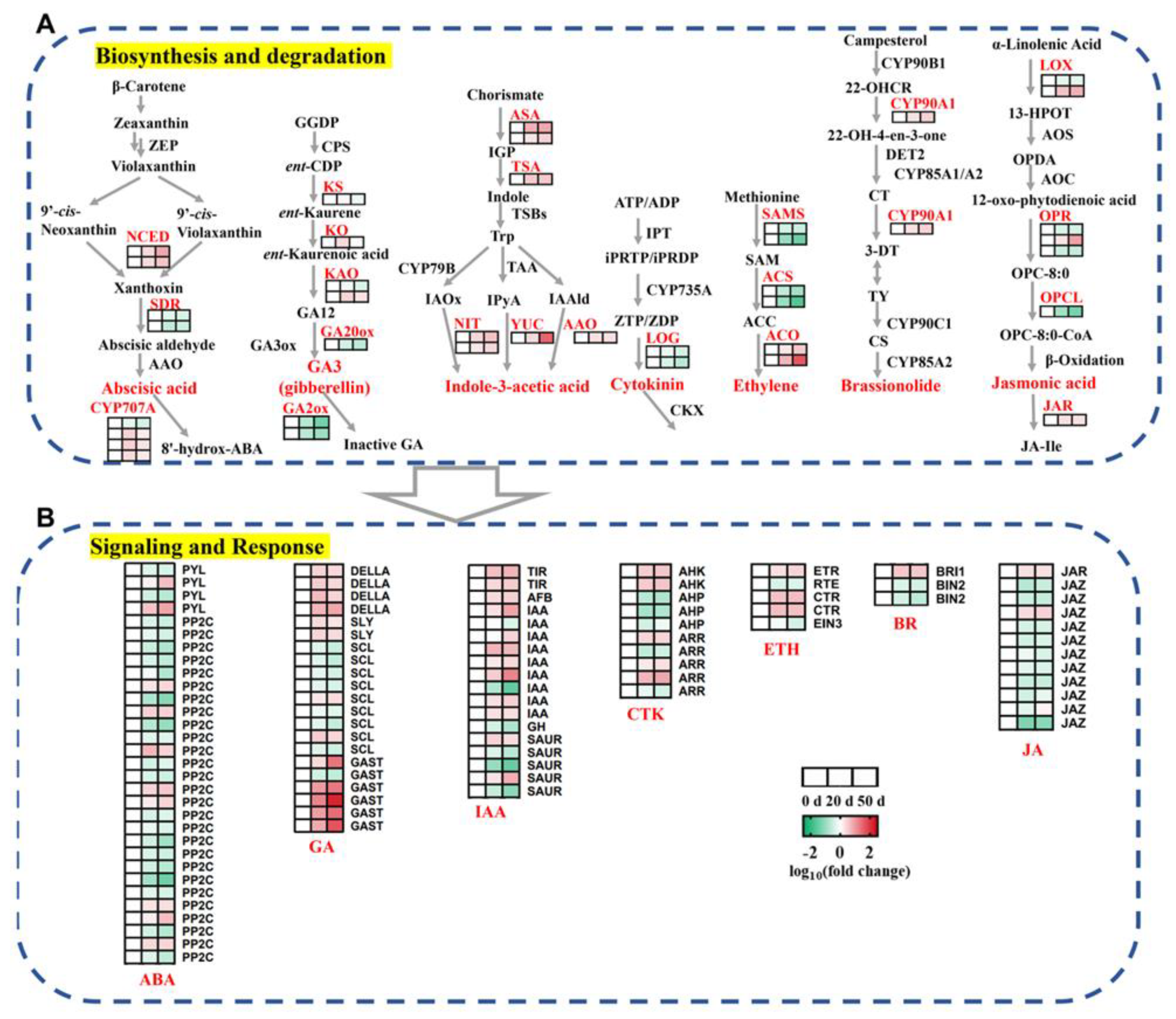
2.4. DEGs Involved in the Biosynthesis, Metabolism, and Signaling Transduction of Plant Hormones during Seed Dormancy Release Progression
2.5. DEGs Involved in Carbohydrate Biosynthesis and Metabolism during Seed Dormancy Release Progression
2.6. DEGs Involved in Lipid Synthesis and Catabolism during Seed Dormancy Release Progression
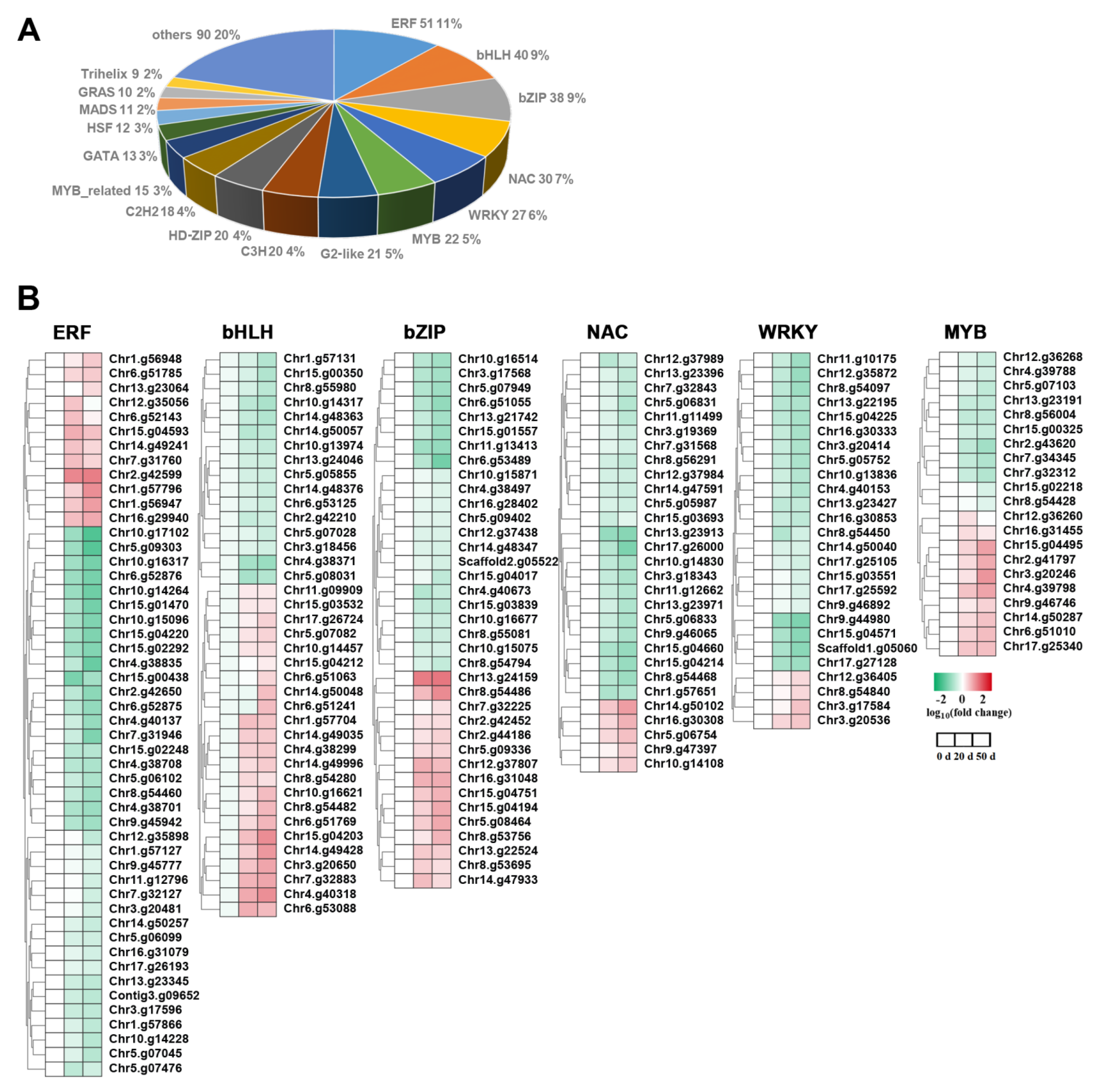
2.7. TFs Were Differentially Expressed in Callery Pear Seed during Dormancy Release Progression
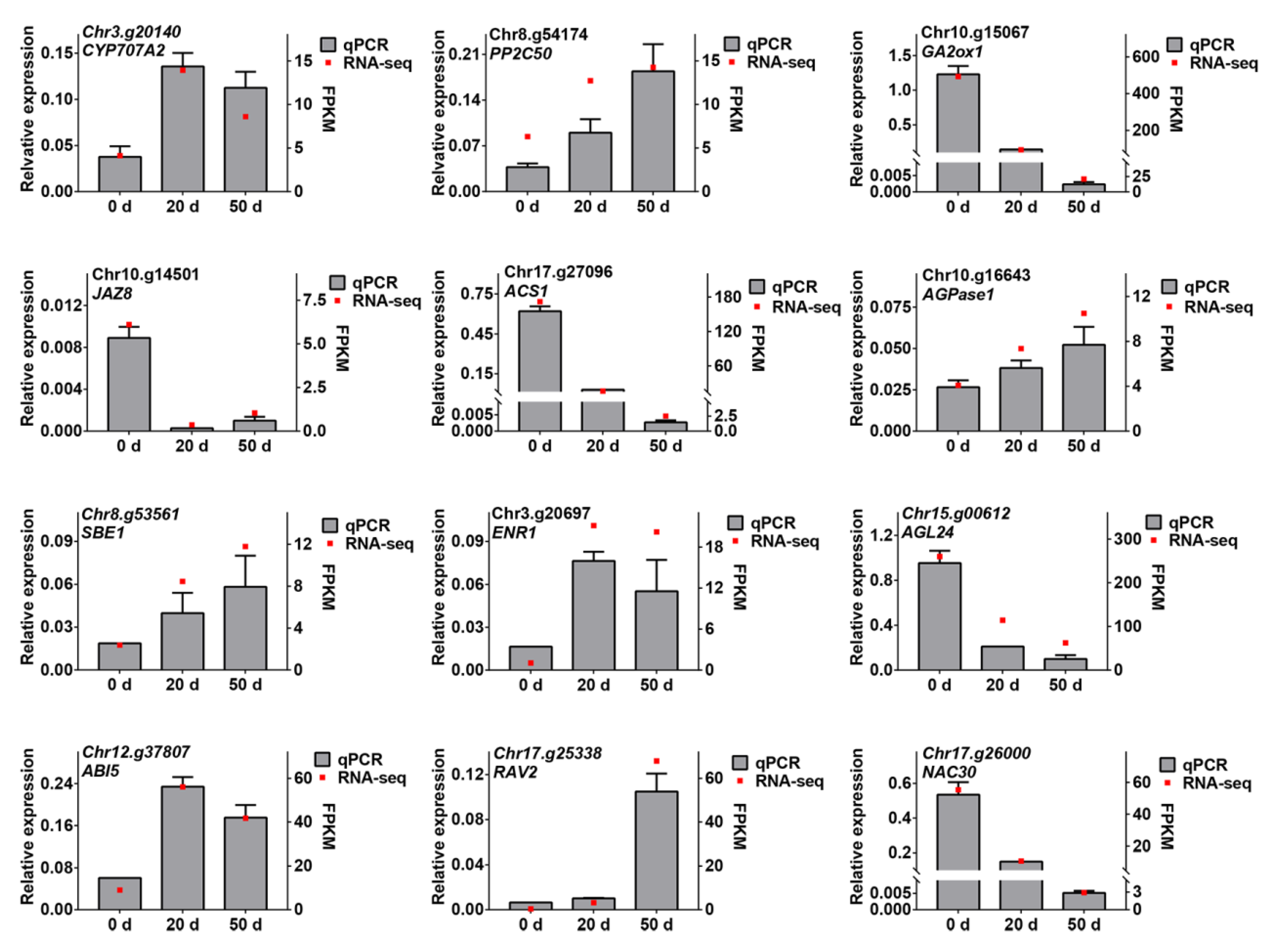
2.8. Validation of RNA-Seq by qRT-PCR Analysis
3. Discussion
3.1. Metabolism and Regulation of Plant Hormones in Callery Pear Seed Dormancy Release Progression
3.2. Metabolism and Regulation of Storage Reserves in Callery Pear Seed Dormancy Release Progression
3.3. Transcription Factors Play a Pivotal Role in Callery Pear Seed Dormancy Release Progression
4. Materials and Methods
4.1. Plant Materials and Treatments
4.2. Measurements of Hormone and Sugar Contents
4.3. RNA Extraction and Transcriptome Sequencing
4.4. Differential Expression Analysis
4.5. Quantitative Real-Time PCR (qRT-PCR) Validation
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chao, W.S.; Doğramacı, M.; Horvath, D.P.; Foley, M.E.; Anderson, J.V. Dormancy Induction and Release in Buds and Seeds; Springer International Publishing: Berlin/Heidelberg, Germany, 2015; pp. 235–256. [Google Scholar]
- Bao, J.P.; Zhang, S.L. Changes in germination, storage materials and antioxidant enzyme activities in pear (Pyrus betulaefolia Bge. and Pyrus calleryana Dcne.) stock seeds during cold stratification. Seed Sci. Technol. 2011, 39, 655–659. [Google Scholar] [CrossRef]
- Baskin, J.M.; Baskin, C.C. A classification system for seed dormancy. Seed Sci. Res. 2004, 14, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Dai, M.; Zhang, S.; Shi, Z. Exploring the hormonal and molecular regulation of sand pear (Pyrus pyrifolia) seed dormancy. Seed Sci. Res. 2013, 23, 15–25. [Google Scholar] [CrossRef]
- Bao, J.P.; Zhang, S.L. Effects of seed coat, chemicals and hormones on breaking dormancy in pear rootstock seeds (Pyrus betulaefolia Bge. and Pyrus calleryana Dcne.). Seed Sci. Technol. 2010, 38, 348–357. [Google Scholar] [CrossRef]
- Wang, D.; Gao, Z.; Du, P.; Xiao, W.; Tan, Q.; Chen, X.; Li, L.; Gao, D. Expression of ABA Metabolism-Related Genes Suggests Similarities and Differences Between Seed Dormancy and Bud Dormancy of Peach (Prunus persica). Front Plant Sci. 2016, 6, 1248. [Google Scholar] [CrossRef]
- Qi, K.J.; Wu, X.; Gao, X.; Li, L.F.; Sun, X.J.; Xie, Z.H.; Gu, C.; Zhang, S.L. Metabolome and transcriptome analyses unravel the inhibition of embryo germination by abscisic acid in pear. Sci. Hortic. 2022, 292, 110652. [Google Scholar]
- Kucera, B.; Cohn, M.A.; Leubner-Metzger, G. Plant hormone interactions during seed dormancy release and germination. Seed Sci. Res. 2005, 15, 281–307. [Google Scholar] [CrossRef]
- Chang, E.; Deng, N.; Zhang, J.; Liu, J.; Chen, L.; Zhao, X.; Abbas, M.; Jiang, Z.; Shi, S. Proteome-Level Analysis of Metabolism- and Stress-Related Proteins during Seed Dormancy and Germination in Gnetum parvifolium. J. Agric. Food Chem. 2018, 66, 3019–3029. [Google Scholar] [CrossRef] [PubMed]
- Seo, M.; Hanada, A.; Kuwahara, A.; Endo, A.; Okamoto, M.; Yamauchi, Y.; North, H.; Marion-Poll, A.; Sun, T.P.; Koshiba, T.; et al. Regulation of hormone metabolism in Arabidopsis seeds: Phytochrome regulation of abscisic acid metabolism and abscisic acid regulation of gibberellin metabolism. Plant J. 2006, 48, 354–366. [Google Scholar] [CrossRef] [PubMed]
- Finkelstein, R.; Reeves, W.; Ariizumi, T.; Steber, C. Molecular Aspects of Seed Dormancy. Annu. Rev. Plant Biol. 2008, 59, 387–415. [Google Scholar] [CrossRef] [Green Version]
- Sugliani, M.; Brambilla, V.; Clerkx, E.J.M.; Koornneef, M.; Soppe, W.J.J. The conserved splicing factor SUA controls alternative splicing of the developmental regulator ABI3 in Arabidopsis. Plant Cell 2010, 22, 1936–1946. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piskurewicz, U.; Jikumaru, Y.; Kinoshita, N.; Nambara, E.; Kamiya, Y.; Lopez-Molina, L. The gibberellic acid signaling repressor RGL2 inhibits Arabidopsis seed germination by stimulating abscisic acid synthesis and ABI5 activity. Plant Cell 2008, 20, 2729–2745. [Google Scholar] [CrossRef] [Green Version]
- Cantoro, R.; Crocco, C.D.; Benech-Arnold, R.L.; Rodriguez, M.V. In vitro binding of Sorghum bicolor transcription factors ABI4 and ABI5 to a conserved region of a GA 2-OXIDASE promoter: Possible role of this interaction in the expression of seed dormancy. J. Exp. Bot. 2013, 64, 5721–5735. [Google Scholar] [PubMed] [Green Version]
- Shu, K.; Zhang, H.; Wang, S.; Chen, M.; Wu, Y.; Tang, S.; Liu, C.; Feng, Y.; Cao, X.; Xie, Q. ABI4 regulates primary seed dormancy by regulating the biogenesis of abscisic acid and gibberellins in Arabidopsis. PLoS Genet. 2013, 9, e1003577. [Google Scholar]
- Soriano, D.; Orozcosegovia, A.; Márquezguzmán, J.; Kitajima, K.; Buen, A.G.; Huante, P. Seed reserve composition in 19 tree species of a tropical deciduous forest in Mexico and its relationship to seed germination and seedling growth. Ann. Bot. 2011, 107, 939–951. [Google Scholar] [CrossRef]
- Rosental, L.; Nonogaki, H.; Fait, A. Activation and regulation of primary metabolism during seed germination. Seed Sci. Res. 2014, 24, 1–15. [Google Scholar] [CrossRef]
- Sakr, S.; Wang, M.; Dedaldechamp, F.; Perez-Garcia, M.-D.; Oge, L.; Hamama, L.; Atanassova, R. The Sugar-Signaling Hub: Overview of Regulators and Interaction with the Hormonal and Metabolic Network. Int. J. Mol. Sci. 2018, 19, 2506. [Google Scholar] [CrossRef] [Green Version]
- Zeeman, S.C.; Kossmann, J.; Smith, A.M. Starch: Its Metabolism, Evolution, and Biotechnological Modification in Plants. Annu. Rev. Plant Biol. 2010, 61, 209–234. [Google Scholar] [CrossRef] [Green Version]
- Ruan, Y.L. Sucrose metabolism: Gateway to diverse carbon use and sugar signaling. Annu. Rev. Plant Biol. 2014, 65, 33–67. [Google Scholar] [CrossRef]
- Macneill, G.J.; Sahar, M.; Minow, M.; Patterson, J.A.; Tetlow, I.J.; Emes, M.J. Starch as a source, starch as a sink: The bifunctional role of starch in carbon allocation. J. Exp. Bot. 2017, 16, 4433–4453. [Google Scholar]
- Liu, X.; Huang, X.; Kong, X.X.; Zhang, J.; Wang, C.L. Sucrose synthase is involved in the carbohydrate metabolism-based regulation of seed dormancy release in Pyrus calleryana decne. J. Hortic. Sci. Biotechnol. 2020, 95, 1–10. [Google Scholar] [CrossRef]
- Nee, G.; Xiang, Y.; Soppe, W.J. The release of dormancy, a wake-up call for seeds to germinate. Curr. Opin. Plant Biol. 2017, 35, 8–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gai, S.; Zhang, Y.; Liu, C.; Zhang, Y.; Zheng, G. Transcript profiling of Paoenia ostii during artificial chilling induced dormancy release identifies activation of GA pathway and carbohydrate metabolism. PLoS ONE 2013, 8, e55297. [Google Scholar]
- Shi, Y.; Ding, Y.; Yang, S. Molecular regulation of CBF signaling in cold acclimation. Trends Plant Sci. 2018, 23, 623–637. [Google Scholar]
- Finch-Savage, W.E.; Leubner-Metzger, G. Seed dormancy and the control of germination. New Phytol. 2006, 171, 501–523. [Google Scholar] [CrossRef]
- Al-Bukhari, F.M.; Qrunfleh, M.M.; Al-Eisawi, D.M. The propagation of Pyrus syriaca by seeds and stem cuttings. Acta Hortic. 2002, 596, 419–424. [Google Scholar] [CrossRef]
- Matilla, A.J. Seed Dormancy: Molecular Control of Its Induction and Alleviation. Plants 2020, 9, 1402. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.Y.; Kuo, S.R.; Chien, C.T. Roles of gibberellins and abscisic acid in dormancy and germination of red bayberry (Myrica rubra) seeds. Tree Physiol. 2008, 28, 1431–1439. [Google Scholar] [CrossRef] [PubMed]
- Walker, M.; Perez, M.; Steinbrecher, T.; Gawthrop, F.; Pavlovic, I.; Novak, O.; Tarkowska, D.; Strnad, M.; Marone, F.; Nakabayashi, K.; et al. Molecular mechanisms and hormonal regulation underpinning morphological dormancy: A case study using Apium graveolens (Apiaceae). Plant J. 2021, 108, 1020–1036. [Google Scholar] [CrossRef]
- Jia, Z.C.; Zhao, B.B.; Liu, S.; Lu, Z.G.; Chang, B.; Jiang, H.R.; Cui, H.; He, Q.S.; Li, W.X.; Jin, B.; et al. Embryo transcriptome and miRNA analyses reveal the regulatory network of seed dormancy in Ginkgo biloba. Tree Physiol. 2021, 41, 571–588. [Google Scholar] [CrossRef] [PubMed]
- Kushiro, T.; Okamoto, M.; Nakabayashi, K.; Yamagishi, K.; Kitamura, S.; Asami, T.; Hirai, N.; Koshiba, T.; Kamiya, Y.; Nambara, E. The Arabidopsis cytochrome P450 CYP707A encodes ABA 8′-hydroxylases: Key enzymes in ABA catabolism. EMBO J. 2004, 23, 1647–1656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okamoto, M.; Kuwahara, A.; Seo, M.; Kushiro, T.; Asami, T.; Hirai, N.; Kamiya, Y.; Koshiba, T.; Nambara, E. CYP707A1 and CYP707A2, which encode ABA 8’-hydroxylases, are indispensable for a proper control of seed dormancy and germination in Arabidopsis. Plant Physiol. 2006, 141, 97–107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, G.H.; Liu, Y.G.; Ye, N.H.; Liu, R.; Zhang, J.H. Involvement of the abscisic acid catabolic gene CYP707A2 in the glucose-induced delay in seed germination and post-germination growth of Arabidopsis. Physiol. Plant 2011, 143, 375–384. [Google Scholar] [CrossRef]
- Shu, K.; Liu, X.; Xie, Q.; He, Z. Two faces of one seed: Hormonal regulation of dormancy and germination. Mol. Plant 2016, 9, 34–45. [Google Scholar] [PubMed] [Green Version]
- Yang, B.; Cheng, J.P.; Wang, J.K.; Cheng, Y.H.; He, Y.Q.; Zhang, H.S.; Wang, Z.F. Physiological characteristics of cold stratification on seed dormancy release in rice. Plant Growth Regul. 2019, 89, 131–141. [Google Scholar]
- Hubbard, K.E.; Nishimura, N.; Hitomi, K.; Getzoff, E.D.; Schroeder, J.I. Early abscisic acid signal transduction mechanisms: Newly discovered components and newly emerging questions. Genes Dev. 2010, 24, 1695–1708. [Google Scholar] [CrossRef] [Green Version]
- Merlot, S.; Gosti, F.; Guerrier, D.; Vavasseur, A.; Giraudat, J. The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signaling pathway. Plant J. 2001, 25, 295–303. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Nishimura, N.; Kitahata, N.; Kuromori, T.; Ito, T.; Asami, T.; Shinozaki, K.; Hirayama, T. ABA-hypersensitive germination3 encodes a protein phosphatase 2C (AtPP2CA) that strongly regulates abscisic acid signaling during germination among Arabidopsis protein phosphatase 2Cs. Plant Physiol. 2006, 140, 115–126. [Google Scholar] [CrossRef] [Green Version]
- Nishimura, N.; Yoshida, T.; Kitahata, N.; Asami, T.; Shinozaki, K.; Hirayama, T. ABA-Hypersensitive Germination1 encodes a protein phosphatase 2C, an essential component of abscisic acid signaling in Arabidopsis seed. Plant J. 2007, 50, 935–949. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.; Lee, Y.; Park, J.; Lee, N.; Choi, G. HONSU, a protein phosphatase 2C, regulates seed dormancy by inhibiting ABA signaling in Arabidopsis. Plant Cell Physiol. 2013, 54, 555–572. [Google Scholar] [PubMed] [Green Version]
- Schwechheimer, C. Gibberellin signaling in plants—the extended version. Front Plant Sci. 2012, 2, 107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colebrook, E.H.; Thomas, S.G.; Phillips, A.L.; Hedden, P. The role of gibberellin signalling in plant responses to abiotic stress. J. Exp. Biol. 2014, 217, 67–75. [Google Scholar]
- Dai, C.; Xue, H.W. Rice early flowering1, a CKI, phosphorylates DELLA protein SLR1 to negatively regulate gibberellin signalling. EMBO J. 2010, 29, 1916–1927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.; Cheng, H.; King, K.E.; Wang, W.; He, Y.; Hussain, A.; Lo, J.; Harberd, N.; Peng, J.R. Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genes Dev. 2002, 5, 646–658. [Google Scholar] [CrossRef] [Green Version]
- Tyler, L.; Thomas, S.G.; Hu, J.; Dill, A.; Alonso, J.M.; Ecker, J.R.; Sun, T.P. DELLA proteins and gibberellin-regulated seed germination and floral development in Arabidopsis. Plant Physiol. 2004, 135, 1008–1019. [Google Scholar] [CrossRef] [Green Version]
- Lv, L.; Huo, X.; Wen, L.; Gao, Z.; Khalil-ur-Rehman, M. Isolation and role of PmRGL2 in GA-mediated floral bud dormancy release in Japanese Apricot (Prunus mume Siebold et Zucc.). Front. Plant Sci. 2018, 9, 27. [Google Scholar] [PubMed]
- Ramaih, S.; Guedira, M.; Paulsen, G.M. Relationship of indoleacetic acid and tryptophan to dormancy and preharvest sprouting of wheat. Funct. Plant Biol. 2003, 30, 939–945. [Google Scholar] [CrossRef] [PubMed]
- Bai, B.; Novak, O.; Ljung, K.; Hanson, J.; Bentsink, L. Combined transcriptome and translatome analyses reveal a role for tryptophan-dependent auxin biosynthesis in the control of DOG1-dependent seed dormancy. New Phytol. 2018, 217, 1077–1085. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pellizzaro, A.; Neveu, M.; Lalanne, D.; Vu, B.L.; Kanno, Y.; Seo, M.; Leprince, O.; Buitink, J. A role for auxin signaling in the acquisition of longevity during seed maturation. New Phytol. 2020, 225, 284–296. [Google Scholar] [CrossRef] [Green Version]
- Preston, J.; Tatematsu, K.; Kanno, Y.; Hobo, T.; Kimura, M.; Jikumaru, Y.; Yano, R.; Kamiya, Y.; Nambara, E. Temporal expression patterns of hormone metabolism genes during imbibition of Arabidopsis thaliana seeds: A comparative study on dormant and non-dormant accessions. Plant Cell Physiol. 2009, 50, 1786–1800. [Google Scholar] [PubMed] [Green Version]
- Ayele, B.T.; Ozga, J.A.; Wickramarathna, A.D.; Reinecke, D.M. Gibberellin metabolism and transport during germination and young seedling growth of pea (Pisum sativum L.). J. Plant Growth Regul. 2012, 31, 235–252. [Google Scholar]
- Liu, A.; Gao, F.; Kanno, Y.; Jordan, M.C.; Kamiya, Y.; Seo, M.; Ayele, B.T. Regulation of wheat seed dormancy by after-ripening is mediated by specific transcriptional switches that induce changes in seed hormone metabolism and signaling. PLoS ONE 2013, 8, e56570. [Google Scholar]
- Miransari, M.; Smith, D.L. Plant hormones and seed germination. Environ. Exp. Bot. 2014, 99, 110–121. [Google Scholar]
- Corbineau, F.; Xia, Q.; Bailly, C.; El-Maarouf-Bouteau, H. Ethylene, a key factor in the regulation of seed dormancy. Front. Plant Sci. 2014, 5, 539. [Google Scholar] [PubMed] [Green Version]
- Sun, M.; Tuan, P.A.; Izydorczyk, M.S.; Ayele, B.T. Ethylene regulates post-germination seedling growth in wheat through spatial and temporal modulation of ABA/GA balance. J. Exp. Bot. 2020, 71, 1985–2004. [Google Scholar] [CrossRef]
- Fait, A.; Angelovici, R.; Less, H.; Ohad, I.; Urbanczyk-Wochniak, E.; Fernie, A.R.; Galili, G. Arabidopsis seed development and germination is associated with temporally distinct metabolic switches. Plant Physiol. 2006, 142, 839–854. [Google Scholar] [PubMed] [Green Version]
- Xu, P.L.; Tang, G.Y.; Cui, W.P.; Chen, G.X.; Ma, C.L.; Zhu, J.Q.; Li, P.X.; Shan, L.; Liu, Z.J.; Wan, S.B. Transcriptional Differences in Peanut (Arachis hypogaea L.) Seeds at the Freshly Harvested, After-ripening and Newly Germinated Seed Stages: Insights into the Regulatory Networks of Seed Dormancy Release and Germination. PLoS ONE 2020, 15, e0219413. [Google Scholar] [CrossRef] [PubMed]
- Rui, Y.K.; Wang, W.Y.; Fazana, R.; Liu, Q. Fatty acids composition of apple and pear seed oils. Int. J. Food Prop. 2009, 12, 774–779. [Google Scholar]
- Weitbrecht, K.; Muller, K.; Leubner-Metzger, G. First off the mark: Early seed germination. J. Exp. Bot. 2011, 62, 3289–3309. [Google Scholar]
- Zhu, L.; Zhao, X.Z.; Xu, Y.; Wang, Q.; Wang, H.Y.; Wu, D.Z.; Jiang, L.X. Effect of germination potential on storage lipids and transcriptome changes in premature developing seeds of oilseed rape (Brassica napus L.). Theor. Appl. Genet. 2020, 133, 2839–2852. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, S.L.; Charlton, W.L.; Baker, A.; Graham, I.A. Germination and storage reserve mobilization are regulated independently in Arabidopsis. Plant J. 2002, 31, 639–647. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.W.; Chiang, C.M.; Tseng, T.H.; Yu, S.M. Interaction between rice MYBGA and the gibberellin response element controls tissue-specific sugar sensitivity of α-amylase genes. Plant Cell 2006, 18, 2326–2340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, M.X.; Du, X.; Zhu, Y.; Wang, Z.; Hua, S.J.; Li, Z.L.; Guo, W.L.; Zhang, G.P.; Peng, J.R.; Jiang, L.X. Seed Fatty Acid Reducer acts downstream of gibberellin signalling pathway to lower seed fatty acid storage in Arabidopsis. Plant Cell Environ. 2012, 35, 2155–2169. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Ye, N.; Zhang, J. Glucose-induced delay of seed germination in rice is mediated by the suppression of ABA catabolism rather than an enhancement of ABA biosynthesis. Plant Cell Physiol. 2009, 50, 644–651. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, H.; Zhao, Y.; Feng, Z.; Li, Q.; Yang, H.Q.; Luan, S.; Li, J.; He, Z.H. Auxin controls seed dormancy through stimulation of abscisic acid signaling by inducing ARF-mediated ABI3 activation in Arabidopsis. Proc. Natl. Acad. Sci. USA 2013, 110, 15485–15490. [Google Scholar] [CrossRef] [Green Version]
- Licausi, F.; Ohme-Takagi, M.; Perata, P. APETALA2/Ethylene Responsive Factor (AP2/ERF) transcription factors: Mediators of stress responses and developmental programs. New Phytol. 2013, 199, 639–649. [Google Scholar] [PubMed]
- Leida, C.; Conesa, A.; Llacer, G.; Badenes, M.L.; Rios, G. Histone modifications and expression of DAM6 gene in peach are modulated during bud dormancy release in a cultivar-dependent manner. New Phytol. 2012, 193, 67–80. [Google Scholar] [CrossRef] [PubMed]
- Niu, Q.F.; Li, J.Z.; Cai, D.Y.; Qian, M.J.; Jia, H.M.; Bai, S.L.; Hussain, S.; Liu, G.Q.; Teng, Y.W.; Zheng, X.Y. Dormancy-associated MADS-box genes and microRNAs jointly control dormancy transition in pear (Pyrus pyrifolia white pear group) flower bud. J. Exp. Bot. 2016, 67, 239–257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wind, J.J.; Peviani, A.; Snel, B.; Hanson, J.; Smeekens, S.C. ABI4: Versatile activator and repressor. Trends Plant Sci. 2013, 18, 125–132. [Google Scholar] [PubMed]
- Longo, C.; Holness, S.; De Angelis, V.; Lepri, A.; Occhigrossi, S.; Ruta, V.; Vittorioso, P. From the Outside to the Inside: New Insights on the Main Factors That Guide Seed Dormancy and Germination. Genes 2021, 12, 52. [Google Scholar] [CrossRef]
- Hu, Y.R.; Han, X.; Yang, M.L.; Zhang, M.H.; Pan, J.J.; Yu, D.Q. The Transcription Factor INDUCER OF CBF EXPRESSION1 Interacts with ABSCISIC ACID INSENSITIVE5 and DELLA Proteins to Fine-Tune Abscisic Acid Signaling During Seed Germination in Arabidopsis. Plant Cell 2019, 31, 1520–1538. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Zhang, Z.L.; Hanzlik, S.; Cook, E.; Chen, Q.J. Salicylic acid inhibits gibberellin-induced alpha-amylase expression and sed germination via a pathway involving an abscisic-acid-inducible WRKY gene. Plant Mol. Biol. 2007, 64, 293–303. [Google Scholar] [CrossRef] [PubMed]
- Nee, G.; Kramer, K.; Nakabayashi, K.; Yuan, B.; Xiang, Y.; Miatton, E.; Finkemeier, I.; Soppe, W.J.J. DELAY OF GERMINATION1 requires PP2C phosphatases of the ABA signaling pathway to control seed dormancy. Nat. Commun. 2017, 8, 72. [Google Scholar] [CrossRef] [PubMed]
- Carrillo-Barral, N.; Rodríguez-Gacio, M.C.; Matilla, A.J. Delay of germination-1 (DOG1): A key to understanding seed dormancy. Plants 2020, 9, 480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, X.; Welti, R.; Wang, X. Quantitative analysis of major plant hormones in crude plant extracts by high-performance liquid chromatography-mass spectrometry. Nat. Protoc. 2010, 5, 986. [Google Scholar] [CrossRef] [PubMed]
- Jeong, W.H.; Harada, K.; Yamada, T.; Abe, J.; Kitamura, K. Establishment of new method for analysis of starch contents and varietal differences in soybean seeds. Breed. Sci. 2010, 60, 160–163. [Google Scholar] [CrossRef] [Green Version]
- Miao, M.M.; Xu, X.F.; Chen, X.H.; Xue, L.B.; Cao, B.S. Cucumber carbohydrate metabolism and translocation under chilling night temperature. J. Plant Physiol. 2007, 164, 621–628. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, Y.F.; Zhang, P.F.; Bian, Y.H.; Liu, Z.Y.; Zhang, C.; Liu, X.; Wang, C.L. An integrated metabolic and transcriptomic analysis reveals the mechanism through which fruit bagging alleviates exocarp semi-russeting in pear fruit. Tree Physiol. 2021, 41, 1306–1318. [Google Scholar] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Qian, J.-Y.; Bian, Y.-H.; Liu, X.; Wang, C.-L. Transcriptome and Metabolite Conjoint Analysis Reveals the Seed Dormancy Release Process in Callery Pear. Int. J. Mol. Sci. 2022, 23, 2186. https://doi.org/10.3390/ijms23042186
Zhang J, Qian J-Y, Bian Y-H, Liu X, Wang C-L. Transcriptome and Metabolite Conjoint Analysis Reveals the Seed Dormancy Release Process in Callery Pear. International Journal of Molecular Sciences. 2022; 23(4):2186. https://doi.org/10.3390/ijms23042186
Chicago/Turabian StyleZhang, Jing, Jia-Yi Qian, Yue-Hong Bian, Xiao Liu, and Chun-Lei Wang. 2022. "Transcriptome and Metabolite Conjoint Analysis Reveals the Seed Dormancy Release Process in Callery Pear" International Journal of Molecular Sciences 23, no. 4: 2186. https://doi.org/10.3390/ijms23042186
APA StyleZhang, J., Qian, J.-Y., Bian, Y.-H., Liu, X., & Wang, C.-L. (2022). Transcriptome and Metabolite Conjoint Analysis Reveals the Seed Dormancy Release Process in Callery Pear. International Journal of Molecular Sciences, 23(4), 2186. https://doi.org/10.3390/ijms23042186





