Population Analysis of Diospyros lotus in the Northwestern Caucasus Based on Leaf Morphology and Multilocus DNA Markers
Abstract
:1. Introduction
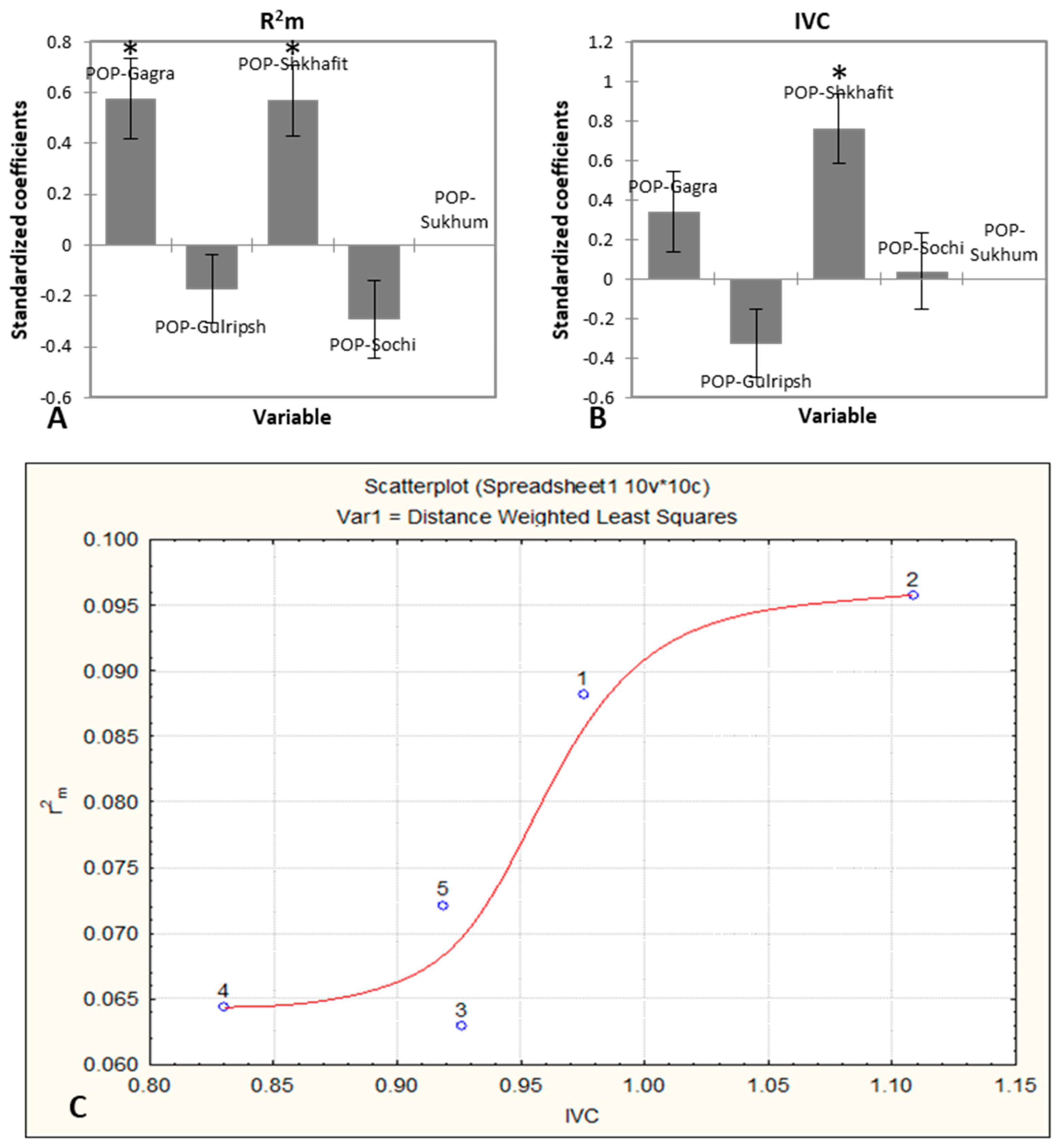
2. Results
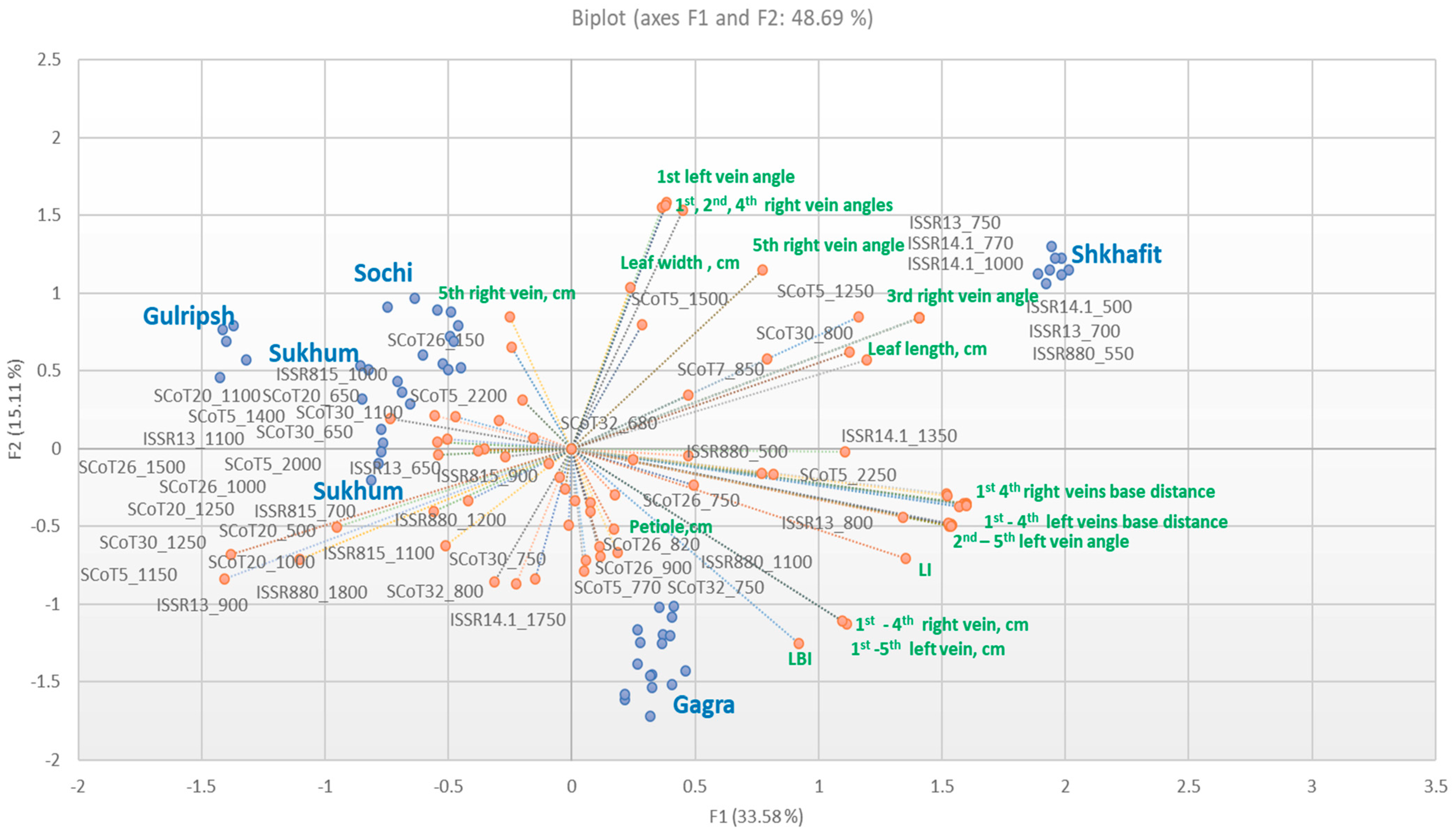
2.1. Morphological Variability, Trait Determination and Ontogenetic Strategy of D. lotus Populations
- I.
- Bio-ecological indicators—traits with a high CV and high : length of first and second veins, distance between veins.
- II.
- Biological indicators –traits with a low CV and high . These are the most reliable indicators representing the general state of the plant organism under the current environment. These indicators are the leaf length, leaf width, LI and the length of the fourth veins.
- III.
- Genotype-specific indicators –traits with a low CV and low : petiole length, LBI, length of the fifth right vein, and the angles of the second, third, fourth and fifth veins.
- IV.
- Ecological indicators—traits with a high CV and low , for which variability is weakly consistent with the plant organism: length of the fifth left vein, the angles of the first veins, and the distance between the fourth and fifth left veins.
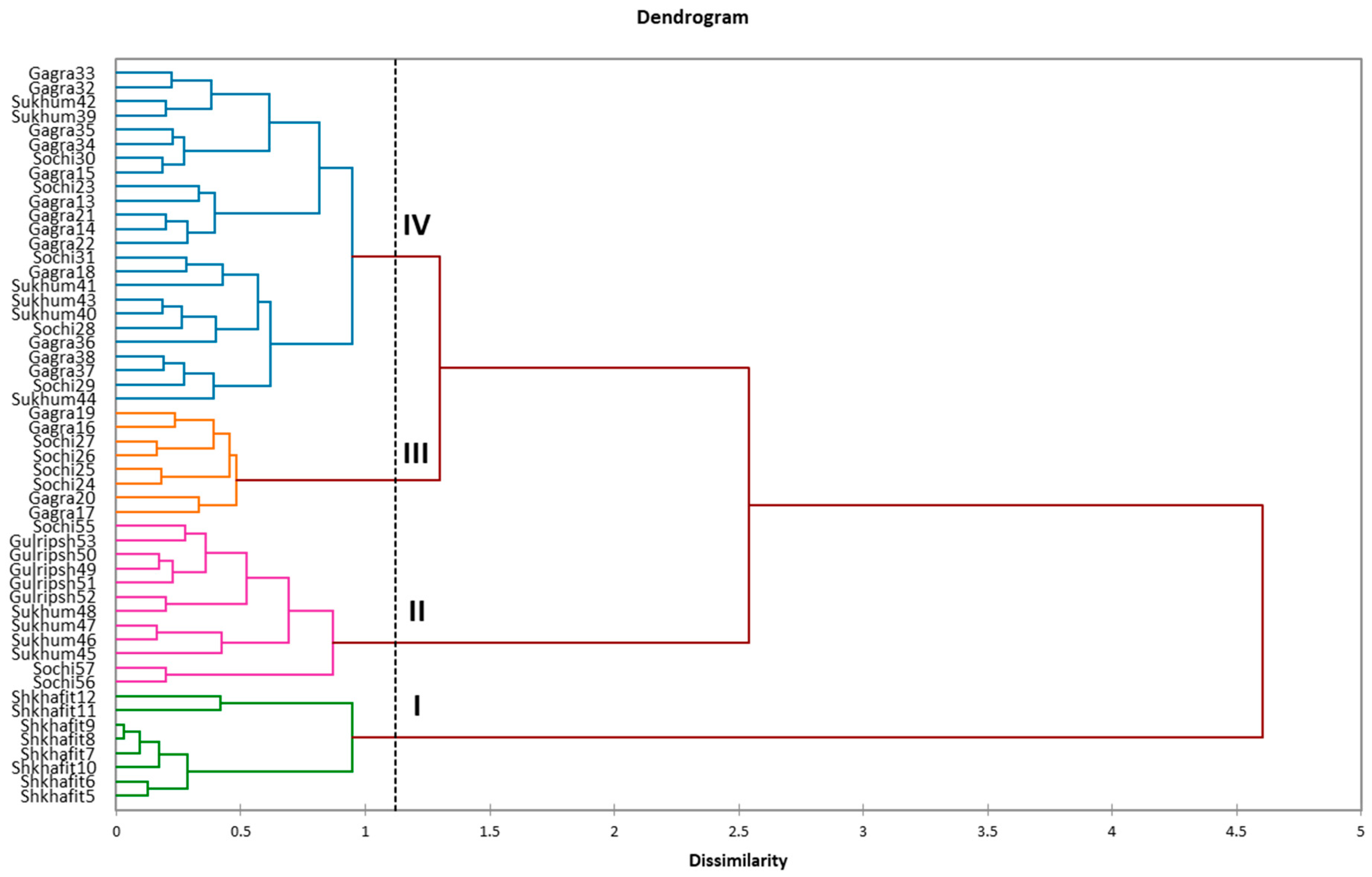
2.2. Efficiency of ISSR and SCoT Markers, Intraspecific Diversity of D. lotus and Correspondence with Morphological Traits
3. Discussion
3.1. Morphological Variability, Trait Determination and Ontogenetic Strategy of D. lotus Populations
3.2. Efficiency of ISSR and SCoT Markers, Intraspecific Diversity of D. lotus and Correspondence with Morphological Traits
4. Materials and Methods
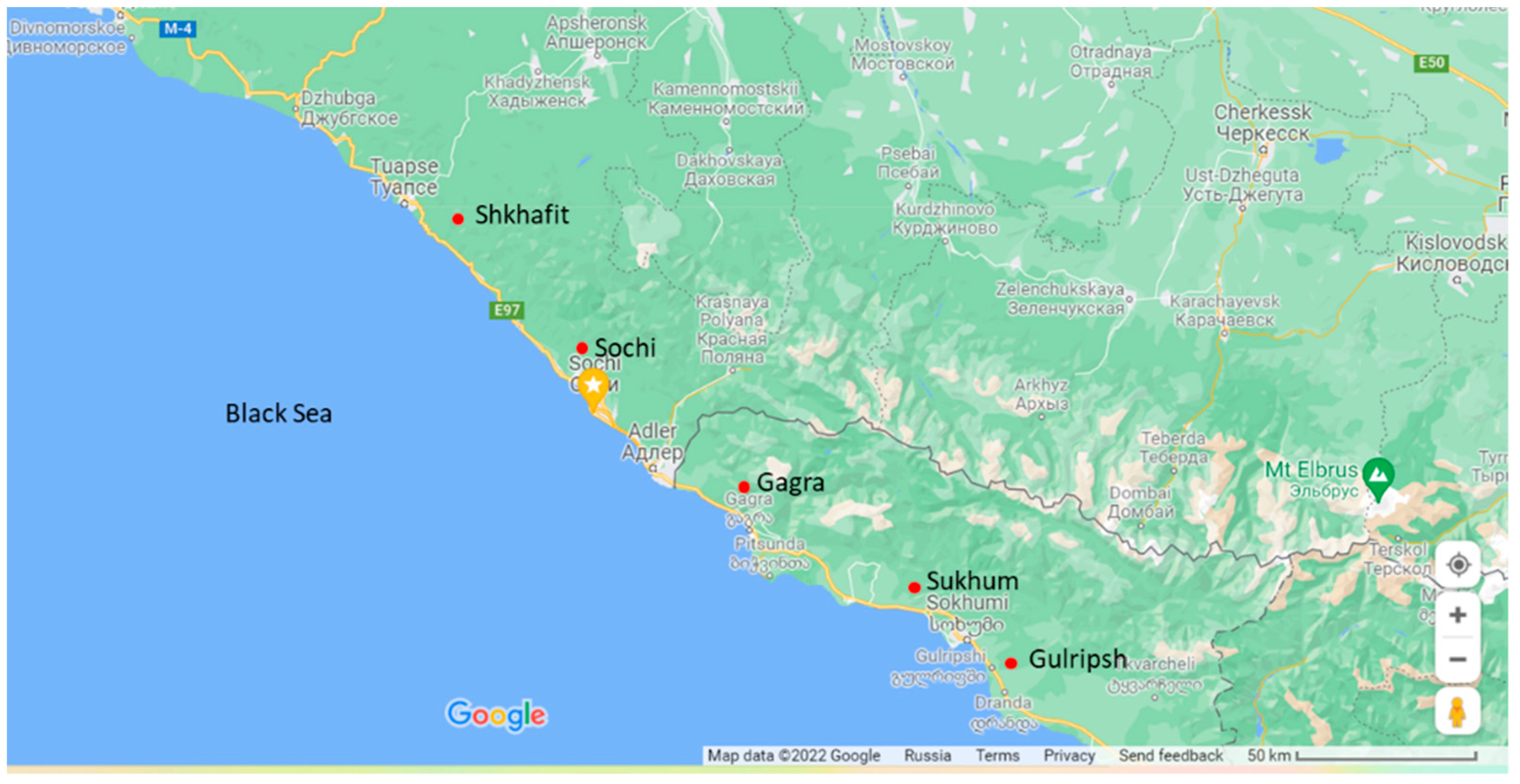
4.1. Plant Material and Morphological Evaluations
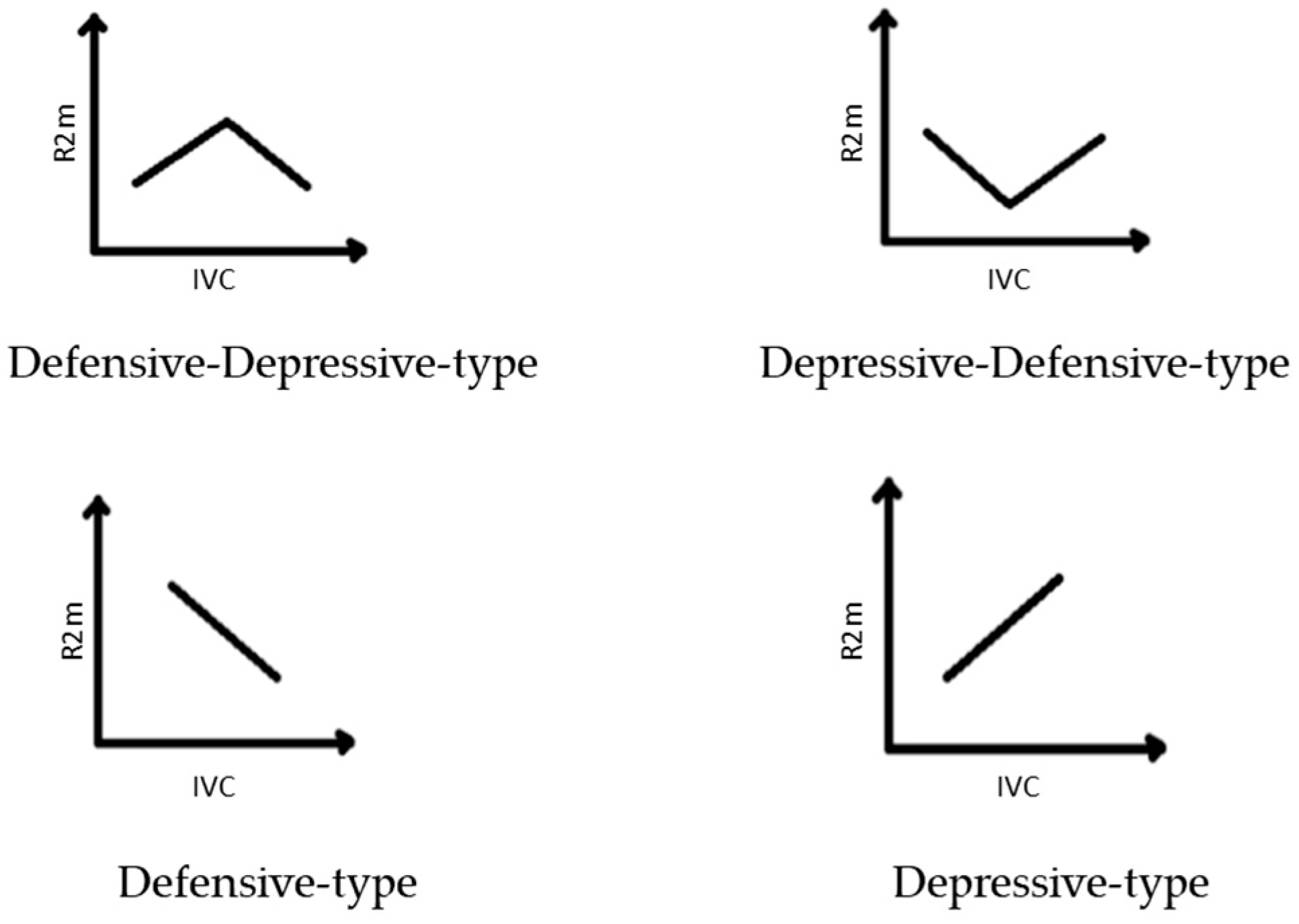
- Defensive-type strategy: the increase of stress leads to the strengthening of morphological integrity and plant development.
- Depressive-type strategy: the increase of stress leads to the weakening of morphological integrity and plant development.
- Defensive–depressive-type strategy: the increase of stress leads first to the strengthening and then to the weakening of morphological integrity.
- Depressive–defensive-type strategy: the increase of stress leads first to the weakening and then to the strengthening of morphological integrity.
4.2. DNA Extraction and Genetic Analysis
4.3. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yang, Y.; Ruan, X.; Wang, R. Indigenous Persimmon Germplasm Resources in China. Acta Hortic. 2013, 996, 89–96. [Google Scholar] [CrossRef]
- Yildirim, N.; Ercisli, S.; Agar, G.; Orhan, E.; Hizarci, Y. Genetic variation among date plum (Diospyros lotus) genotypes in Turkey. Genet. Mol. Res. 2010, 9, 981–986. [Google Scholar] [CrossRef]
- Guan, C.; Zhang, P.; Hu, C.; Chachar, S.; Riaz, A.; Wang, R.; Yang, Y. Genetic diversity, germplasm identification and population structure of Diospyros kaki Thunb. from different geographic regions in China using SSR markers. Sci. Hortic. 2019, 251, 233–240. [Google Scholar] [CrossRef]
- Omarov, M.D. Yield of different cultivars of oriental persimmon in the humid subtropics of Russia. Subtrop. Ornam. Hortic. 2018, 65, 137–141. [Google Scholar] [CrossRef]
- Omarov, M.D.; Zagirov, N.G.; Omarova, Z.M.; Avidzba, M.A. Atlas of Cultivars and Hybrids of Oriental Persimmon; Ryndin, A.V., Ed.; Russian Research Institute of Floriculture and Subtropical Crops: Sochi, Russia, 2014; p. 93. [Google Scholar]
- Gil-Muñoz, F.; Delhomme, N.; Quiñones, A.; Naval, M.d.M.; Badenes, M.L.; García-Gil, M.R. Transcriptomic Analysis Reveals Salt Tolerance Mechanisms Present in Date-Plum Persimmon Rootstock (Diospyros lotus L.). Agronomy 2020, 10, 1703. [Google Scholar] [CrossRef]
- Franks, S.J.; Weber, J.J.; Aitken, S.N. Evolutionary and plastic responses to climate change in terrestrial plant populations. Evol. Appl. 2014, 7, 123–139. [Google Scholar] [CrossRef]
- Radersma, R.; Noble, D.W.A.; Uller, T. Plasticity leaves a phenotypic signature during local adaptation. Evol. Lett. 2020, 4, 360–370. [Google Scholar] [CrossRef]
- Ramírez-Valiente, J.A.; Sánchez-Gómez, D.; Aranda, I.; Valladares, F. Phenotypic plasticity and local adaptation in leaf ecophysiological traits of 13 contrasting cork oak populations under different water availabilities. Tree Physiol. 2010, 30, 618–627. [Google Scholar] [CrossRef] [Green Version]
- Engel, K.; Tollrian, R.; Jeschke, J.M. Integrating biological invasions, climate change and phenotypic plasticity. Commun. Integ. Biol. 2011, 4, 247–250. [Google Scholar] [CrossRef] [Green Version]
- Henn, J.J.; Buzzard, B.; Enquist, B.J.; Halbritter, A.H.; Klanderud, K.; Maitner, B.S.; Michaletz, S.T.; Pötsch, C.; Seltzer, L.; Telford, R.J.; et al. Intraspecific Trait Variation and Phenotypic Plasticity Mediate Alpine Plant Species Response to Climate Change. Front. Plant Sci. 2018, 9, 1548. [Google Scholar] [CrossRef]
- Wang, M.; Zhang, J.; Guo, Z.; Guan, Y.; Qu, G.; Liu, J.; Guo, Y.; Yan, X. Morphological variation in Cynodon dactylon (L.) Pers., and its relationship with the environment along a longitudinal gradient. Hereditas 2020, 157, 4. [Google Scholar] [CrossRef] [Green Version]
- Arantes, M.K.; da Silva Filho, M.P.; Pennacchi, J.P.; Mendonca, A.M.C.; Barbosa, J.P.R.A.D. Phenotypic plasticity of leaf anatomical traits helps to explain gas-exchange response to water shortage in grasses of different photosynthetic types. Theor. Exp. Plant Physiol. 2020, 32, 341–356. [Google Scholar] [CrossRef]
- Nicotra, A.B.; Atkin, O.K.; Bonser, S.P.; Davidson, A.M.; Finnegan, E.J.; Mathesius, U.; Poot, P.; Purugganan, M.D.; Richards, C.L.; Valladares, F.; et al. Plant phenotypic plasticity in a changing climate. Trends Plant Sci. 2010, 15, 648–692. [Google Scholar] [CrossRef]
- Liu, Y.; El-Kassaby, Y.A. Phenotypic plasticity of natural Populus trichocarpa populations in response to temporally environmental change in a common garden. BMC Evol. Biol. 2019, 19, 231. [Google Scholar] [CrossRef]
- Gentili, R.; Ambrosini, R.; Augustinus, B.A.; Caronni, S.; Cardarelli, E.; Montagnani, C.; Müller-Schärer, H.; Schaffner, U.; Citterio, S. High Phenotypic Plasticity in a Prominent Plant Invader along Altitudinal and Temperature Gradients. Plants 2021, 10, 2144. [Google Scholar] [CrossRef]
- Alberto, F.J.; Aitken, S.N.; Ali, R.; Gonzalez-Martinez, S.C.; Hänninenk, H.; Kremer, A.; Lefèvre, F.; Lenormand, T.; Yeaman, S.; Whetten, R.; et al. Potential for evolutionary responses to climate change—Evidence from tree populations. Glob. Change Biol. 2013, 19, 1645–1661. [Google Scholar] [CrossRef] [Green Version]
- Rostova, N.S. Correlations: Structure and Variability; SPBGU: Saint Petersburg, Russia, 2002; p. 308. [Google Scholar]
- Gratani, L. Article Plant Phenotypic Plasticity in Response to Environmental Factors Review. Adv. Bot. 2014, 2014, 17. [Google Scholar] [CrossRef] [Green Version]
- Pérez-Ramos, I.M.; Matías, L.; Gómez-Aparicio, L.; Godoy, Ó. Functional traits and phenotypic plasticity modulate species coexistence across contrasting climatic conditions. Nat. Commun. 2019, 10, 2555. [Google Scholar] [CrossRef]
- Niinemets, U. Leaf Trait Plasticity and Evolution in Different Plant Functional Types. Annu. Plant Rev. 2020, 4, 473–522. [Google Scholar] [CrossRef]
- Flury, B.N. Common Principal Components and Related Multivariate Models; Wiley: Hoboken, NJ, USA, 1988; p. 258. [Google Scholar]
- Ishbirdin, A.R.; Ishmuratova, M.M.; Zhirnova, T.V. Life strategies of cenopopulation Cephalanthera rubra (L.) Rich. on the territory of the Bashkir State Reserve. Bull. Nizhny Novgorod Univ. N.I. Lobachevsky Ser. Biol. 2005, 1, 85–98. [Google Scholar]
- Le Corre, V.; Kremer, A. The genetic differentiation at quantitative trait loci under local adaptation. Mol. Ecol. 2012, 21, 1548–1566. [Google Scholar] [CrossRef]
- Andrews, C.A. Natural Selection, Genetic Drift, and Gene Flow Do Not Act in Isolation in Natural Populations. Nat. Educ. Knowl. 2010, 3, 5. [Google Scholar]
- Raddová, J.; Ptáčková, H.; Čechová, J.; Ondrášek, I. Genetic analysis of the genus Diospyros ssp. using RAPD and i-PBS methods. Acta Univ. Agric. Silvic. Mendel. Brun. 2012, 60, 205–216. [Google Scholar] [CrossRef] [Green Version]
- Liang, Y.; Han, W.; Sun, P.; Liang, J.; Wuyun, T.; Li, F.; Fu, J. Genetic diversity among germplasms of Diospyros kaki based on SSR markers. Sci. Hortic. 2015, 186, 180–189. [Google Scholar] [CrossRef]
- Guo, D.-L.; Luo, Z.R. Genetic relationships of the Japanese persimmon Diospyros kaki (Ebenaceae) and related species revealed by SSR analysis. Genet. Mol. Res. 2011, 10, 1060–1068. [Google Scholar] [CrossRef]
- Pinar, H.; Yildiz, E.; Kaplankiran, M.; Toplu, C.; Unlu, M.; Serce, S.; Ercisli, S. Molecular characterization of some selected persimmon genotypes and cultivars by srap and ssr markers. Genetika 2017, 49, 693–704. [Google Scholar] [CrossRef]
- Soriano, J.M.; Pecchioli, S.; Romero, C.; Vilanova, S.; Llacer, G.; Giordani, E.; Badenes, M.L. Development of microsatellite markers in polyploid persimmon (Diospyros kaki L.) from an enriched genomic library. Mol. Ecol. Notes 2006, 6, 368–370. [Google Scholar] [CrossRef]
- Naval, M.; Zuriaga, E.; Pecchioli, S.; Llácer, G.; Giordani, E.; Badenes, M.L. Analysis of genetic diversity among persimmon cultivars using microsatellite markers. Tree Genet. Genomes 2010, 6, 677–687. [Google Scholar] [CrossRef] [Green Version]
- Yonemori, K.; Honsho, C.; Kanzaki, S.; Ino, H.; Ikegami, A.; Kitajima, A.; Sugiura, A.; Parfitt, D.E. Sequence analyses of the ITS regions and the matK gene for determining phylogenetic relationships of Diospyros kaki (persimmon) with other wild Diospyros (Ebenaceae) species. Tree Genet. Genomes 2007, 4, 149–158. [Google Scholar] [CrossRef]
- Jing, Z.; Ruan, X.; Wang, R.; Yang, Y. Genetic diversity and relationships between and within persimmon (Diospyros L.) wild species and cultivated varieties by SRAP markers. Plant. Syst. Evol. 2013, 299, 1485–1492. [Google Scholar] [CrossRef]
- Guan, C.; Chachar, S.; Zhang, P.; Hu, C.; Wang, R.; Yang, Y. Inter- and Intra-specific Genetic Diversity in Diospyros Using SCoT and IRAP Markers. Hortic. Plant J. 2020, 6, 71–80. [Google Scholar] [CrossRef]
- Deng, L.; Liang, Q.; He, X.; Luo, C.; Chen, H.; Qin, Z. Investigation and Analysis of Genetic Diversity of Diospyros Germplasms Using SCoT Molecular Markers in Guangxi. PLoS ONE 2015, 10, e0136510. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Yang, T.; Jing, Z. Genetic diversity and taxonomic studies of date plum (Diospyros lotus L.) using morphological traits and SCoT markers. Biochem. Syst. Ecol. 2015, 61, 253–259. [Google Scholar] [CrossRef]
- Samarina, L.S.; Malyarovskaya, V.I.; Reim, S.; Koninskaya, N.G.; Matskiv, A.O.; Tsaturyan, G.A.; Rakhmangulov, R.S.; Shkhalakhova, R.M.; Shurkina, E.S.; Kulyan, R.V.; et al. Genetic Diversity in Diospyros Germplasm in the Western Caucasus Based on SSR and ISSR Polymorphism. Biology 2021, 10, 341. [Google Scholar] [CrossRef]
- Fu, J.; Liu, H.; Hu, J.; Liang, Y.; Liang, J.; Wuyun, T.; Tan, X. Five Complete Chloroplast Genome Sequences from Diospyros: Genome Organization and Comparative Analysis. PLoS ONE 2016, 11, e0159566. [Google Scholar] [CrossRef] [Green Version]
- Reddy, P.M.; Sarla, N.; Siddiq, E. Inter simple sequence repeat (ISSR) polymorphism and its application in plant breeding. Euphytica 2002, 128, 9–17. [Google Scholar] [CrossRef]
- Azhar, M.; Muhammad, H.A.; Siti, N.I.; Parween, K.S.A.S. Optimization of ISSR Markers for Molecular DNA Fingerprinting in Aquilaria sp. In Nuclear Technical Convention; IAEA: Bangi, Malaysia, 2013. [Google Scholar]
- Collard, B.C.Y.; Mackill, D.J. Start Codon Targeted (SCoT) Polymorphism: A simple, novel DNA marker technique for generating gene-targeted markers in plants. Plant Mol. Biol. Rep. 2009, 27, 86–93. [Google Scholar] [CrossRef]
- Etminan, A.; Pour-Aboughadareh, A.; Mohammadi, R.; Ahmadi-Rad, A.; Noori, A.; Mahdavian, Z.; Moradi, Z. Applicability of start codon targeted (SCoT) and inter-simple sequence repeat (ISSR) markers for genetic diversity analysis in durum wheat genotypes. Biotechnol. Biotechnol. Equip. 2016, 30, 1075–1081. [Google Scholar] [CrossRef] [Green Version]
- Robert, H.G.; Faik, A.A.; Millson, M.; Huang, H.S.; Chuang, L.T.; Sanz, C.; Golding, J.B. Changes in sugars, acids and fatty acids in naturally parthenocarpic date plum persimmon (Diospyros lotus L.) fruit during maturation and ripening. Eur. Food Res. Technol. 2005, 221, 113–118. [Google Scholar] [CrossRef]
- Conner, J.K.; Hartl, D.L. A Primer of Ecological Genetics; Sinauer: Sunderland, MA, USA, 2004; p. 304. [Google Scholar]
- Rubio De Casas, R.; Vargas, P.; Pe’rez-Corona, E.; Manrique, E.; Quintana, J.; Garcı’a-Verdugo, C.; Balaguer, L. Field patterns of leaf plasticity in adults of the long-lived evergreen Quercus coccifera. Ann. Bot. 2007, 100, 325–334. [Google Scholar] [CrossRef] [Green Version]
- Rehfeldt, G.E. A model of genetic variation for Pinus ponderosa in the inland northwest (USA): Applications in gene resource management. Can. J. Res. 1991, 21, 1491–1500. [Google Scholar] [CrossRef]
- Gorji, A.M.; Poczai, P.; Polgar, Z.; Taller, J. Efficiency of arbitrarily amplified dominant markers (SCoT, ISSR and RAPD) for diagnostic fingerprinting in tetraploid potato. Am. J. Potato Res. 2011, 88, 226–237. [Google Scholar] [CrossRef]
- Zeng, B.; Zhang, Y.; Huang, L.K.; Jiang, X.M.; Luo, D.; Yin, G.H. Genetic diversity of orchardgrass (Dactylis glomerata L.) germplasms with resistance to rust diseases revealed by StartCodon Targeted (SCoT) markers. Biochem. Syst. Ecol. 2014, 54, 96–102. [Google Scholar] [CrossRef] [Green Version]
- Kumar, J.; Agrawal, V. Assessment of genetic diversity, population structure and sex identification in dioecious crop, Trichosanthes dioica employing ISSR, SCoT and SRAP markers. Heliyon 2019, 5, e01346. [Google Scholar] [CrossRef] [Green Version]
- Samarina, L.S.; Matskiv, A.O.; Koninskaya, N.G.; Shkhalakhova, R.M.; Gvasaliya, M.V.; Tsaturyan, G.A.; Ryndin, A.V.; Pchikhachev, E.K.; Manakhova, K.A.; Shumeev, A.N.; et al. Genetic diversity and genome size variability in core collection of tea plant (Camellia sinensis L. Kuntze) in Russia. Front. Plant Sci. 2022, 12, 800141. [Google Scholar] [CrossRef]
- Samarina, L.S.; Malyarovskaya, V.I.; Reim, S.; Yakushina, L.G.; Koninskaya, N.G.; Klemeshova, K.V.; Shkhalakhova, R.M.; Matskiv, A.O.; Shurkina, E.S.; Gabueva, T.Y.; et al. Transferability of ISSR, SCoT and SSR Markers for Chrysanthemum × Morifolium Ramat and Genetic Relationships among Commercial Russian Cultivars. Plants 2021, 10, 1302. [Google Scholar] [CrossRef]
- Stojnić, S.; Avramidou, E.V.; Fussi, B.; Westergren, M.; Orlović, S.; Matović, B.; Trudić, B.; Kraigher, H.; Aravanopoulos, F.A.; Konnert, M. Assessment of genetic diversity and population genetic structure of norway spruce (Picea abies (L.) Karsten) at its southern Lineage in Europe. Implications for conservation of forest genetic resources. Forests 2019, 10, 258. [Google Scholar] [CrossRef] [Green Version]
- Konopiński, M.K. Shannon diversity index: A call to replace the original Shannon’s formula with unbiased estimator in the population genetics studies. PeerJ 2020, 8, e9391. [Google Scholar] [CrossRef]
- Abramoff, M.D.; Magalhães, P.; Ram, S.J. Image Processing with Image. J. Biophotonics Int. 2004, 11, 36–42. [Google Scholar]
- Nouri, A.; Golabadi, M.; Etminan, A.; Rezaei, A.; Mehrabi, A. Comparative assessment of SCoT and ISSR markers for analysis of genetic diversity and population structure in some Aegilops tauschii Coss. accessions. Plant Genet. Resour. Charact. Util. 2021, 19, 375–383. [Google Scholar] [CrossRef]
- Bogoslov, A.V.; Kashin, A.S.; Parkhomenko, A.S.; Kulikova, L.V.; Shilova, I.V.; Knjazeva, A.K. Vitality Structure of Colchicum bulbocodium subsp. versicolor (Colchicaceae, Liliopsida) Populations in the Lower Volga Region. Biol Bull. Russ. Acad. Sci. 2021, 48, 1953–1963. [Google Scholar] [CrossRef]
- Kashin, A.S.; Petrova, N.A.; Shilova, I.V. Some features of the environmental strategy of Tulipa gesneriana L. (Liliaceae, Liliopsida). Povolzhskiy Ekol. Zhurnal. 2016, 2, 209–221. [Google Scholar] [CrossRef]
- Shabanov, D.A.; Korshunov, A.V.; Kravchenko, M.A.; Meleshko, E.V.; Shabanova, A.V.; Usova, E.E. The intrapopulation developmental strategies of precocity and stuntedity: Determination by the example of anurans. J. V.N. Karazin Kharkiv Natl. Univ. 2014, 1126, 138–147. [Google Scholar]
- Zlobin, Y.; Kovalenko, I.; Klymenko, H.; Kyrylchuk, K.; Bondarieva, L.; Tykhonova, O.; Zubtsova, I. Vitality Analysis Algorithm in the Study of Plant Individuals and Populations. Open Agric. J. 2021, 15, 119–129. [Google Scholar] [CrossRef]
- Zlobin, Y.A. Principles and Methods of Studying Coenopopulations; Kazan University Press: Kazan, Russia, 1989; 147p. [Google Scholar]
- Ishbirdin, A.R.; Ishmuratova, M.M. Adaptive morphogenesis and ecological-coenotic strategies for the survival of herbaceous plants. In Methods of Population Biology: Materials of VII All-Russian; Population Workshop: Syktyvkar, Russia, 2004. [Google Scholar]
- Doyle, J.J.; Doyle, J.L. Isolation of plant DNA from fresh tissue. Focus 1991, 12, 13–15. [Google Scholar]
- Mondal, T.K. Assessment of genetic diversity of tea (Camellia sinensis (L.) O. Kuntze) by inter-simple sequence repeat polymerase chain reaction. Euphytica 2002, 128, 307–315. [Google Scholar] [CrossRef]
- Roy, S.C.; Chakraborty, B.N. Genetic diversity and relationsips among tea (Camellia sinensis) cultivars revealed by RAPD and ISSR based fingerprinting. Indian J. Biotechnol. 2009, 8, 370–376. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [Green Version]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Amiryousefi, A.; Hyvönen, J.; Poczai, P. iMEC: Online Marker Efficiency Calculator. Appl. Plant Sci. 2018, 6, e01159. [Google Scholar] [CrossRef] [PubMed]

| Marker | Primer 5′-3′ | Na | P. Bands, % | PIC | D |
| ISSR815 | (CT)8G | 10 | 50.00 | 0.35 | 0.13 |
| ISSR880 | (GGAGA)3 | 13 | 38.46 | 0.34 | 0.13 |
| ISSR13 | (AC)8C | 7 | 14.29 | 0.33 | 0.27 |
| ISSR14.1 | (CT)8TG | 9 | 44.44 | 0.34 | 0.24 |
| Mean ± SD | 9.75 ± 2.5 | 36.80 ± 15.7 | 0.34 ± 0.01 | 0.19 ± 0.07 | |
| SCoT05 | CAACAATGGCTACCACGA | 10 | 10.00 | 0.38 | 0.76 |
| SCoT07 | CAACAATGGCTACCACGG | 4 | 50.00 | 0.46 | 0.35 |
| SCoT20 | ACCATGGCTACCACCGCG | 12 | 41.67 | 0.39 | 0.63 |
| SCoT26 | ACCATGGCTACCACCGTC | 9 | 11.11 | 0.39 | 0.75 |
| SCoT30 | CCATGGCTACCACCGGCG | 11 | 27.27 | 0.43 | 0.61 |
| SCoT32 | CCATGGCTACCACCGCAC | 6 | 50.00 | 0.39 | 0.64 |
| Mean ± SD | 8.67 ± 3.08 | 31.68 ± 18.35 | 0.41 ± 0.03 | 0.62 ± 0.15 |
| POP | N | P ISSR | P SCOT | RELATIVE P ISSR | RELATIVE P SCOT |
| SHKHAFIT | 9 | 7.69% | 30.77% | 0.85% | 3.42% |
| GAGRA | 18 | 46.15% | 59.62% | 2.56% | 3.31% |
| SOCHI | 9 | 17.95% | 50.00% | 1.99% | 5.55% |
| SUKHUM | 10 | 12.82% | 53.85% | 1.28% | 5.39% |
| GULRIPSH | 6 | 2.56% | 32.69% | 0.42% | 5.45% |
| MEAN | 10.4 ± 4.5 | 17.44% ± 7.62% | 45.38% ± 5.79% | 1.42% ± 0.11% | 4.62% ± 0.15% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Samarina, L.S.; Malyarovskaya, V.I.; Rakhmangulov, R.S.; Koninskaya, N.G.; Matskiv, A.O.; Shkhalakhova, R.M.; Orlov, Y.L.; Tsaturyan, G.A.; Shurkina, E.S.; Gvasaliya, M.V.; et al. Population Analysis of Diospyros lotus in the Northwestern Caucasus Based on Leaf Morphology and Multilocus DNA Markers. Int. J. Mol. Sci. 2022, 23, 2192. https://doi.org/10.3390/ijms23042192
Samarina LS, Malyarovskaya VI, Rakhmangulov RS, Koninskaya NG, Matskiv AO, Shkhalakhova RM, Orlov YL, Tsaturyan GA, Shurkina ES, Gvasaliya MV, et al. Population Analysis of Diospyros lotus in the Northwestern Caucasus Based on Leaf Morphology and Multilocus DNA Markers. International Journal of Molecular Sciences. 2022; 23(4):2192. https://doi.org/10.3390/ijms23042192
Chicago/Turabian StyleSamarina, Lidia S., Valentina I. Malyarovskaya, Ruslan S. Rakhmangulov, Natalia G. Koninskaya, Alexandra O. Matskiv, Ruset M. Shkhalakhova, Yuriy L. Orlov, Gregory A. Tsaturyan, Ekaterina S. Shurkina, Maya V. Gvasaliya, and et al. 2022. "Population Analysis of Diospyros lotus in the Northwestern Caucasus Based on Leaf Morphology and Multilocus DNA Markers" International Journal of Molecular Sciences 23, no. 4: 2192. https://doi.org/10.3390/ijms23042192
APA StyleSamarina, L. S., Malyarovskaya, V. I., Rakhmangulov, R. S., Koninskaya, N. G., Matskiv, A. O., Shkhalakhova, R. M., Orlov, Y. L., Tsaturyan, G. A., Shurkina, E. S., Gvasaliya, M. V., Kuleshov, A. S., & Ryndin, A. V. (2022). Population Analysis of Diospyros lotus in the Northwestern Caucasus Based on Leaf Morphology and Multilocus DNA Markers. International Journal of Molecular Sciences, 23(4), 2192. https://doi.org/10.3390/ijms23042192







