Transcriptomic Analysis of the Host Response to Mild and Severe CTV Strains in Naturally Infected Citrus sinensis Orchards
Abstract
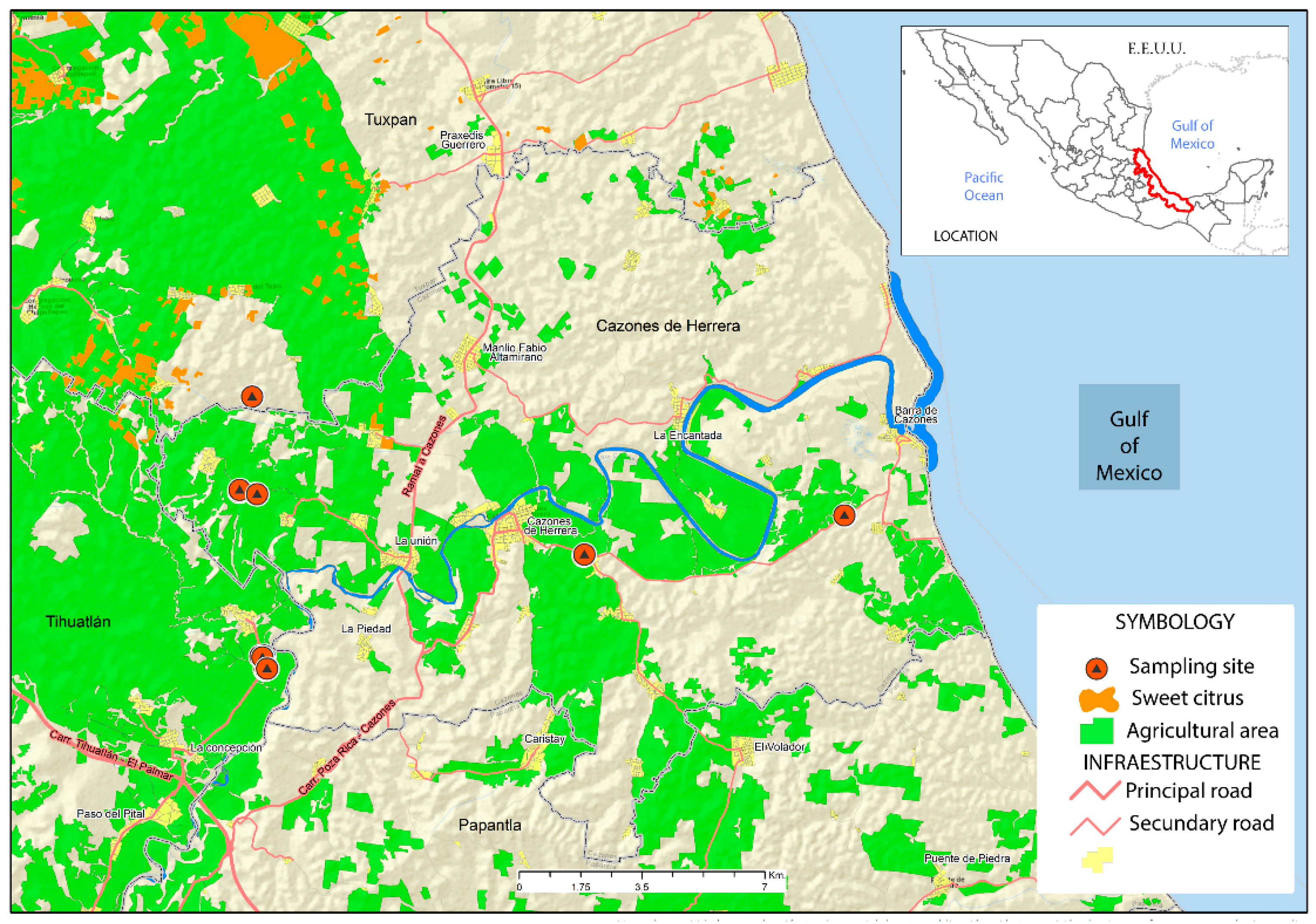
:1. Introduction
2. Results
2.1. Detection of Different CTV Isolates in C. Sinensis Showing Mild and Severe Symptoms
2.2. Phylogeny of Mild and Severe CTV Isolates
2.3. Bioinformatic Analysis of Naturally Infected Citrus sinensis Trees
2.4. Identification of Differentially Expressed Genes
2.4.1. Functional Classification by Gene Ontology (GO)
2.4.2. Gene Enrichment Analysis by PlantGSEA and Functional Classification by KEGG
2.5. Viroid-Associated Sequence Reads
2.6. Validation of Differentially Accumulated Transcripts by RT-qPCR
3. Discussion
4. Materials and Methods
4.1. Plant Material Collection
4.2. RNA Extraction
4.3. CTV Molecular Detection
4.4. Cloning, Sequencing and Phylogenetic Analysis of the CTV Isolates
4.5. Library Preparation and Sequencing for Massive RNA Sequencing (RNA-Seq)
4.5.1. Bioinformatic Analysis
4.5.2. Gene Enrichment Analysis
4.5.3. Assignment of Specific Metabolic Pathways
4.5.4. Viroid-Associated Sequence Reads
4.6. Validation of Differential Gene Expression with RT-qPCR
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAOSTAT. Available online: http://www.fao.org/faostat/en/#home (accessed on 9 October 2021).
- Dolja, V.V.; Kreuze, J.F.; Valkonen, J.P.T. Comparative and Functional Genomics of Closteroviruses. Virus Res. 2006, 117, 38–51. [Google Scholar] [CrossRef]
- ICTV. International Committee on Taxonomy of Viruses. Available online: https://talk.ictvonline.org (accessed on 6 November 2021).
- Moreno, P.; Ambrós, S.; Albiach-Martí, M.R.; Guerri, J.; Peña, L. Citrus Tristeza Virus: A Pathogen That Changed the Course of the Citrus Industry. Mol. Plant Pathol. 2008, 9, 251–268. [Google Scholar] [CrossRef]
- Secretaría de Agrícultura, Ganadería, Desarrollo Rural, Pesca y Alimentación. NOM-011-FITO-1995. Diario Oficial de la Federación, 24 September 1996.
- Secretaría de Agrícultura, Ganadería, Desarrollo Rural, Pesca y Alimentación. NOM-031-FITO-2000. Diario Oficial de la Federación, 10 August 2001.
- Secretaría de Agrícultura, Ganadería, Desarrollo Rural, Pesca y Alimentación. NOM-079-FITO-2002. Diario Oficial de la Federación, 22 May 2002.
- Dawson, W.O.; Bar-Joseph, M.; Garnsey, S.M.; Moreno, P. Citrus Tristeza Virus: Making an Ally from an Enemy. Annu. Rev. Phytopathol. 2015, 53, 137–155. [Google Scholar] [CrossRef]
- Gómez-Muñoz, N.; Velázquez, K.; Vives, M.C.; Ruiz-Ruiz, S.; Pina, J.A.; Flores, R.; Moreno, P.; Guerri, J. The Resistance of Sour Orange to Citrus Tristeza Virus Is Mediated by Both the Salicylic Acid and RNA Silencing Defence Pathways: Study of the Resistance of Sour Orange to CTV. Mol. Plant Pathol. 2017, 18, 1253–1266. [Google Scholar] [CrossRef]
- Sun, T.; Zhang, Y. Short- and Long-Distance Signaling in Plant Defense. Plant J. 2021, 105, 505–517. [Google Scholar] [CrossRef]
- Durrant, W.E.; Dong, X. Systemic Acquired Resistance. Annu. Rev. Phytopathol. 2004, 42, 185–209. [Google Scholar] [CrossRef]
- Hadidi, A.; Flores, R.; Randles, J.; Palukaitis, P. Viroids and Satellites, 1st ed.; Elsevier: Amsterdam, The Netherlands, 2017. [Google Scholar]
- Fu, S.; Shao, J.; Zhou, C.; Hartung, J.S. Transcriptome Analysis of Sweet Orange Trees Infected with ‘Candidatus Liberibacter Asiaticus’ and Two Strains of Citrus Tristeza Virus. BMC Genom. 2016, 17, 349. [Google Scholar] [CrossRef] [Green Version]
- Fu, S.; Shao, J.; Paul, C.; Zhou, C.; Hartung, J.S. Transcriptional Analysis of Sweet Orange Trees Co-Infected with ‘Candidatus Liberibacter Asiaticus’ and Mild or Severe Strains of Citrus Tristeza Virus. BMC Genom. 2017, 18, 837. [Google Scholar] [CrossRef] [Green Version]
- Mendoza, A.; Salazar, C.; Alvarado, O.; Cruz, M.A.; Barrera, H. Diferenciación molecular de razas severas y débiles de aislamientos del virus de la tristeza de los cítricos en México. Rev. Fitotec. Mex. 2003, 26, 223–230. [Google Scholar]
- Albiach-Marti, M.R.; Grosser, J.W.; Gowda, S.; Mawassi, M.; Satyanarayana, T.; Garnsey, S.M.; Dawson, W.O. Citrus Tristeza Virus Replicates and Forms Infectious Virions in Protoplasts of Resistant Citrus Relatives. Mol. Breed. 2004, 14, 117–128. [Google Scholar] [CrossRef]
- Mawassi, M.; Mietkiewska, E.; Gofman, R.; Yang, G.; Bar-Joseph, M. Unusual Sequence Relationships Between Two Isolates of Citrus Tristeza Virus. J. Gen. Virol. 1996, 77, 2359–2364. [Google Scholar] [CrossRef]
- Hagen, C.; Rojas, M.R.; Sudarshana, M.R.; Xoconostle-Cazares, B.; Natwick, E.T.; Turini, T.A.; Gilbertson, R.L. Biology and Molecular Characterization of Cucurbit Leaf Crumple Virus, an Emergent Cucurbit-Infecting Begomovirus in the Imperial Valley of California. Plant Dis. 2008, 92, 781–793. [Google Scholar] [CrossRef] [Green Version]
- Cruz-Jaramillo, J.; Ruiz-Medrano, R.; Rojas-Morales, L.; López-Buenfil, J.; Morales-Galván, O.; Chavarín-Palacio, C.; Ramírez-Pool, J.; Xoconostle-Cázares, B. Characterization of a Proposed Dichorhavirus Associated with the Citrus Leprosis Disease and Analysis of the Host Response. Viruses 2014, 6, 2602–2622. [Google Scholar] [CrossRef] [Green Version]
- Liese, A.; Romeis, T. Biochemical Regulation of in Vivo Function of Plant Calcium-Dependent Protein Kinases (CDPK). Biochim. Biophys. Acta Mol. Cell Res. 2013, 1833, 1582–1589. [Google Scholar] [CrossRef] [Green Version]
- Lincoln, J.E.; Sanchez, J.P.; Zumstein, K.; Gilchrist, D.G. Plant and Animal PR1 Family Members Inhibit Programmed Cell Death and Suppress Bacterial Pathogens in Plant Tissues. Mol. Plant Pathol. 2018, 19, 2111–2123. [Google Scholar] [CrossRef] [Green Version]
- Bent, A.F.; Kunkel, B.N.; Dahlbeck, D.; Brown, K.L.; Schmidt, R.; Giraudat, J.; Leung, J.; Staskawicz, B.J. RPS2 of Arabidopsis Thaliana: A Leucine-Rich Repeat Class of Plant Disease Resistance Genes. Science 1994, 265, 1856–1860. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG Tools for Functional Characterization of Genome and Metagenome Sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef] [Green Version]
- Montalvo-Hernandez, L.; Piedra-Ibarra, E.; Gomes-Silva, L.; Lira-Carmona, R.; Acosta-Gallegos, J.A.; Vazquez-Medrano, J.; Xoconostle-Cazares, B.; Ruiz-Medrano, R. Differential Accumulation of MRNAs in Drought-tolerant and Susceptible Common Bean Cultivars in Response to Water Deficit. New Phytopatol. 2007, 177, 102–113. [Google Scholar] [CrossRef]
- Vandelle, E.; Vannozzi, A.; Wong, D.; Danzi, D.; Digby, A.-M.; Dal Santo, S.; Astegno, A. Identification, Characterization, and Expression Analysis of Calmodulin and Calmodulin-like Genes in Grapevine (Vitis Vinifera) Reveal Likely Roles in Stress Responses. Plant Physiol. Biochem. 2018, 129, 221–237. [Google Scholar] [CrossRef]
- Wang, B.; Shi, G.-P.; Yao, P.M.; Li, Z.; Chapman, H.A.; Brömme, D. Human Cathepsin F. J. Biol. Chem. 1998, 273, 32000–32008. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.-C.; Guo, L.; Wang, X. Nuclear Moonlighting of Cytosolic Glyceraldehyde-3-Phosphate Dehydrogenase Regulates Arabidopsis Response to Heat Stress. Nat. Commun. 2020, 11, 3439. [Google Scholar] [CrossRef]
- Fedoreyeva, L.I.; Vanyushin, B.F.; Baranova, E.N.; Fedoreyeva, L.I.; Vanyushin, B.F.; Baranova, E.N. Peptide AEDL Alters Chromatin Conformation via Histone Binding. AIMS Biophys. 2020, 7, 1–16. [Google Scholar] [CrossRef]
- Ashapkin, V.V.; Kutueva, L.I.; Aleksandrushkina, N.I.; Vanyushin, B.F. Epigenetic Mechanisms of Plant Adaptation to Biotic and Abiotic Stresses. Int. J. Mol. Sci. 2020, 21, 7457. [Google Scholar] [CrossRef]
- Ramirez-Prado, J.S.; Piquerez, S.J.M.; Bendahmane, A.; Hirt, H.; Raynaud, C.; Benhamed, M. Modify the Histone to Win the Battle: Chromatin Dynamics in Plant–Pathogen Interactions. Front. Plant Sci. 2018, 9, 355. [Google Scholar] [CrossRef]
- Rivas-Valencia, P.; Domínguez-Monge, S.; Santillán-Mendoza, R.; Loeza-Kuk, E.; Pérez-Hernández, O.; Rodríguez-Quibrera, C.G.; Lomas-Barrié, C. Severe "Citrus Tristeza Virus" Isolates from Eastern Mexico Are Related to the T36 Genotype Group. AJPS 2020, 11, 1521–1532. [Google Scholar] [CrossRef]
- Chen, A.Y.S.; Peng, J.H.C.; Polek, M.; Tian, T.; Ludman, M.; Fátyol, K.; Ng, J.C.K. Comparative Analysis Identifies Amino Acids Critical for Citrus Tristeza Virus (T36CA) Encoded Proteins Involved in Suppression of RNA Silencing and Differential Systemic Infection in Two Plant Species. Mol. Plant Pathol. 2021, 22, 64–76. [Google Scholar] [CrossRef]
- Ruiz-Medrano, R.; Moya, J.H.; Xoconostle-Cázares, B.; Lucas, W.J. Influence of Cucumber Mosaic Virus Infection on the MRNA Population Present in the Phloem Translocation Stream of Pumpkin Plants. Funct. Plant Biol. 2007, 34, 292. [Google Scholar] [CrossRef]
- Chen, D.; Hao, F.; Mu, H.; Ahsan, N.; Thelen, J.J.; Stacey, G. S-Acylation of P2K1 Mediates Extracellular ATP-Induced Immune Signaling in Arabidopsis. Nat. Commun. 2021, 12, 2750. [Google Scholar] [CrossRef]
- Molina, A.; Miedes, E.; Bacete, L.; Rodríguez, T.; Mélida, H.; Denancé, N.; Sánchez-Vallet, A.; Rivière, M.-P.; López, G.; Freydier, A.; et al. Arabidopsis Cell Wall Composition Determines Disease Resistance Specificity and Fitness. Proc. Natl. Acad. Sci. USA 2021, 118, e2010243118. [Google Scholar] [CrossRef]
- Logemann, J.; Schell, J.; Willmitzer, L. Improved Method for the Isolation of RNA from Plant Tissues. Anal. Biochem. 1987, 163, 16–20. [Google Scholar] [CrossRef]
- Stucky, B.J. SeqTrace: A Graphical Tool for Rapidly Processing DNA Sequencing Chromatograms. J. Biomol. Tech. 2012, 23, 90–93. [Google Scholar] [CrossRef] [Green Version]
- Harper, S.J. Citrus Tristeza Virus: Evolution of Complex and Varied Genotypic Groups. Front. Microbiol. 2013, 4, 93. [Google Scholar] [CrossRef] [Green Version]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI Search and Sequence Analysis Tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-Length Transcriptome Assembly from RNA-Seq Data without a Reference Genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [Green Version]
- Schmieder, R.; Edwards, R. Fast Identification and Removal of Sequence Contamination from Genomic and Metagenomic Datasets. PLoS ONE 2011, 6, e17288. [Google Scholar] [CrossRef] [Green Version]
- Evans, T.; Loose, M. AlignWise: A Tool for Identifying Protein-Coding Sequence and Correcting Frame-Shifts. BMC Bioinform. 2015, 16, 376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alva, V.; Nam, S.-Z.; Söding, J.; Lupas, A.N. The MPI Bioinformatics Toolkit as an Integrative Platform for Advanced Protein Sequence and Structure Analysis. Nucleic Acids Res. 2016, 44, W410–W415. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate Transcript Quantification from RNA-Seq Data with or without a Reference Genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [Green Version]
- Anders, S.; Huber, W. Differential Expression Analysis for Sequence Count Data. Nat. Preced. 2010, 12, 1. [Google Scholar]
- Bardou, P.; Mariette, J.; Escudié, F.; Djemiel, C.; Klopp, C. Jvenn: An Interactive Venn Diagram Viewer. BMC Bioinform. 2014, 15, 293. [Google Scholar] [CrossRef] [Green Version]
- Yi, X.; Du, Z.; Su, Z. PlantGSEA: A Gene Set Enrichment Analysis Toolkit for Plant Community. Nucleic Acids Res. 2013, 41, W98–W103. [Google Scholar] [CrossRef] [Green Version]
- Ogata, H.; Goto, S.; Sato, K.; Fujibuchi, W.; Bono, H.; Kanehisa, M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999, 27, 29–34. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Mafra, V.; Kubo, K.S.; Alves-Ferreira, M.; Ribeiro-Alves, M.; Stuart, R.M.; Boava, L.P.; Rodrigues, C.M.; Machado, M.A. Reference Genes for Accurate Transcript Normalization in Citrus Genotypes under Different Experimental Conditions. PLoS ONE 2012, 7, e31263. [Google Scholar] [CrossRef]







| Condition | TPM * | FPKM ** |
|---|---|---|
| Citrus exocortis viroid | ||
| Asymptomatic | 1,000,000 | 4,528,301.89 |
| CTV severe isolate | 1,000,000 | 4,244,325.52 |
| CTV mild isolate | 0 | 0 |
| Hop stunt viroid | ||
| Asymptomatic | 1,000,000 | 5,813,953.49 |
| CTV severe isolate | 1,000,000 | 5,639,097.74 |
| CTV mild isolate | 0 | 0 |
| Gen ID | Gen Name | Description 1 | log2 Fold Change |
|---|---|---|---|
| Downregulated genes, mild CTV strain | |||
| XP_006470053.1 | CHC1 | CHC1 is predicted to encode a protein that belongs to the chromodomain remodeling complex. | −8.1749 |
| XP_006475507.1 | ABI2 | Encodes a protein phosphatase 2C and is involved in ABA signal transduction. | −9.6421 |
| XP_024949213.1 | Loricrin-like protein | Loricrin-like protein; resistance to Phytophthora. | −7.3923 |
| Upregulated genes, mild CTV strain | |||
| XP_006490176.1 | PR-6 | PR (pathogenesis-related) peptide that belongs to the PR-6 proteinase inhibitor family. | 5.0405 |
| XP_006470311.1 | PDP6 | Histone H3-K27 trimethylation, regulation of transcription, DNA-templated. | 7.9542 |
| XP_024953872.1 | DRP | Disease resistance protein (TIR-NBS-LRR class). | 5.4683 |
| Downregulated genes, severe CTV strain | |||
| XP_006485913.1 | STIPL1 | Homologue of spliceosome disassembly factor NTR1. Required for correct expression and splicing of a regulator of seed dormancy. | −7.4838 |
| XP_006483041.1 | SAG29 | Encodes a member of the SWEET sucrose efflux transporter family proteins. | −8.6582 |
| XP_006468074.1 | BAG2 | Plant BAG proteins are multi-functional. They regulate apoptotic-like processes, pathogen attack to abiotic stress and plant development. | −7.2095 |
| Upregulated genes, severe CTV strain | |||
| XP_006471773.1 | EDR1 | Enhanced disease resistance 1 (EDR1) confers resistance to powdery mildew disease caused by the fungus Erysiphe cichoracearum. | 7.1396 |
| XP_006479423.1 | RIN13 | Encodes RPM1 Interacting Protein 13 (RIN13), a resistance protein interactor shown to positively enhance resistance function of RPM1. | 7.2574 |
| XP_024949989.1 | SYN4 | Encodes a SCC1/REC8 ortholog that may be involved in mitosis and may represent a mitotic cohesin. Plays a role in somatic DNA double-strand break damage repair. | 7.0768 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramírez-Pool, J.A.; Xoconostle-Cázares, B.; Calderón-Pérez, B.; Ibarra-Laclette, E.; Villafán, E.; Lira-Carmona, R.; Ruiz-Medrano, R. Transcriptomic Analysis of the Host Response to Mild and Severe CTV Strains in Naturally Infected Citrus sinensis Orchards. Int. J. Mol. Sci. 2022, 23, 2435. https://doi.org/10.3390/ijms23052435
Ramírez-Pool JA, Xoconostle-Cázares B, Calderón-Pérez B, Ibarra-Laclette E, Villafán E, Lira-Carmona R, Ruiz-Medrano R. Transcriptomic Analysis of the Host Response to Mild and Severe CTV Strains in Naturally Infected Citrus sinensis Orchards. International Journal of Molecular Sciences. 2022; 23(5):2435. https://doi.org/10.3390/ijms23052435
Chicago/Turabian StyleRamírez-Pool, José Abrahán, Beatriz Xoconostle-Cázares, Berenice Calderón-Pérez, Enrique Ibarra-Laclette, Emanuel Villafán, Rosalía Lira-Carmona, and Roberto Ruiz-Medrano. 2022. "Transcriptomic Analysis of the Host Response to Mild and Severe CTV Strains in Naturally Infected Citrus sinensis Orchards" International Journal of Molecular Sciences 23, no. 5: 2435. https://doi.org/10.3390/ijms23052435
APA StyleRamírez-Pool, J. A., Xoconostle-Cázares, B., Calderón-Pérez, B., Ibarra-Laclette, E., Villafán, E., Lira-Carmona, R., & Ruiz-Medrano, R. (2022). Transcriptomic Analysis of the Host Response to Mild and Severe CTV Strains in Naturally Infected Citrus sinensis Orchards. International Journal of Molecular Sciences, 23(5), 2435. https://doi.org/10.3390/ijms23052435






