Dysregulation of Human Somatic piRNA Expression in Parkinson’s Disease Subtypes and Stages
Abstract
1. Introduction
2. Materials & Methods
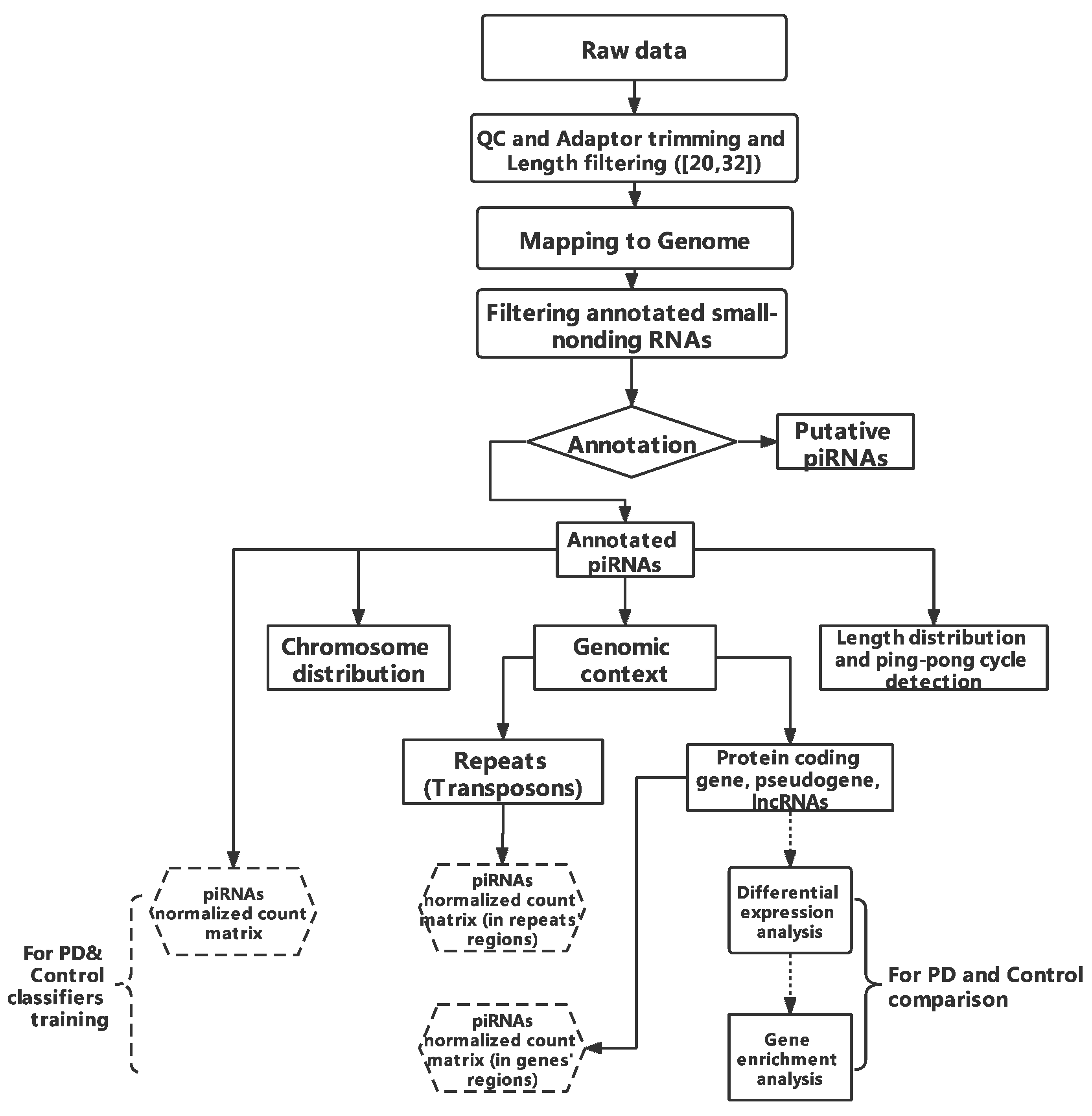
2.1. Workflow
2.2. Collection of High-Throughput Small RNA Sequencing Datasets from GEO
2.3. Pre-Processing and piRNAs Annotation
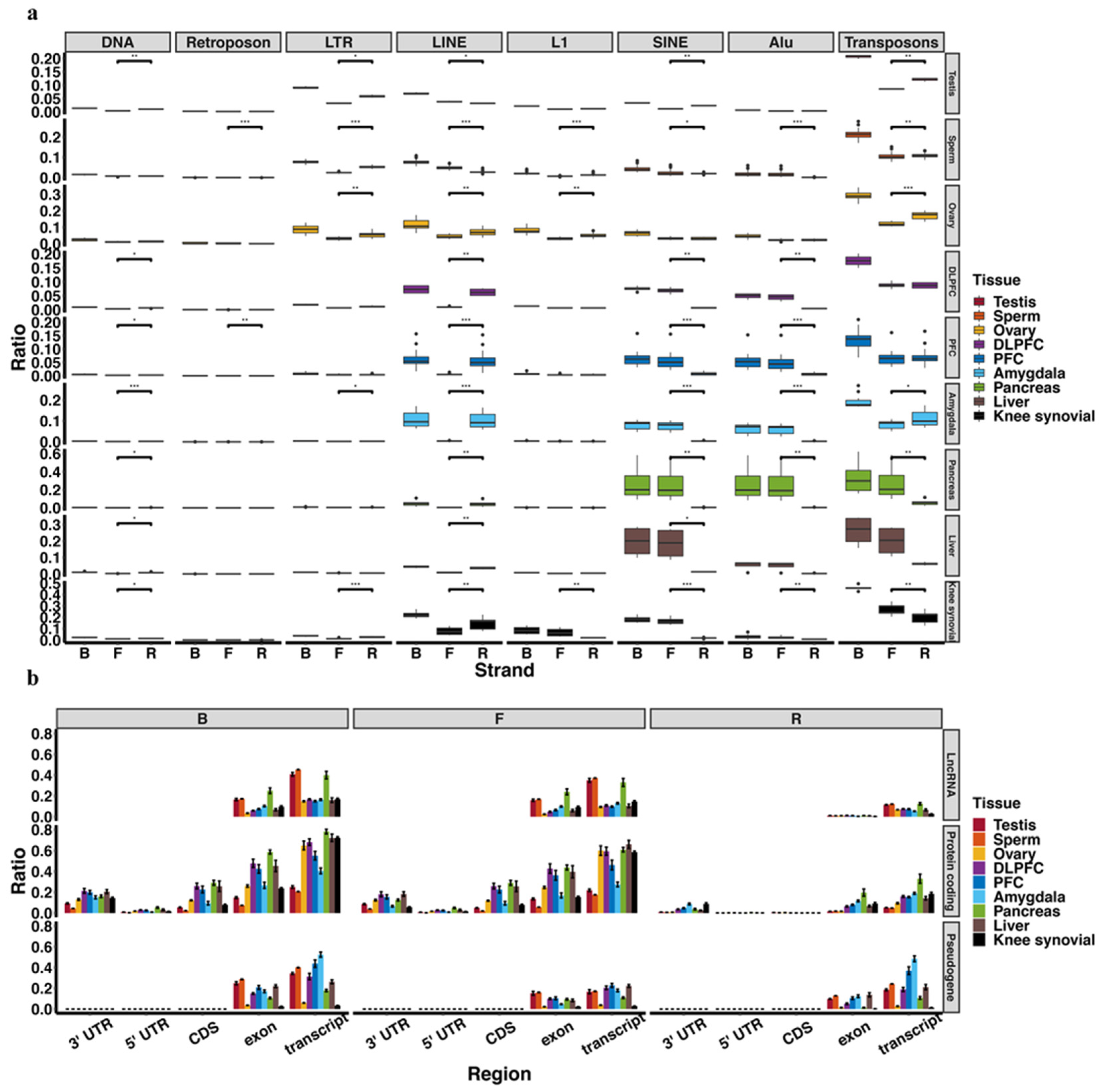
2.4. Transposons and Transcripts Annotation
2.5. Differential Analysis and Enrichment Analysis
2.6. Classifier Construction
3. Results
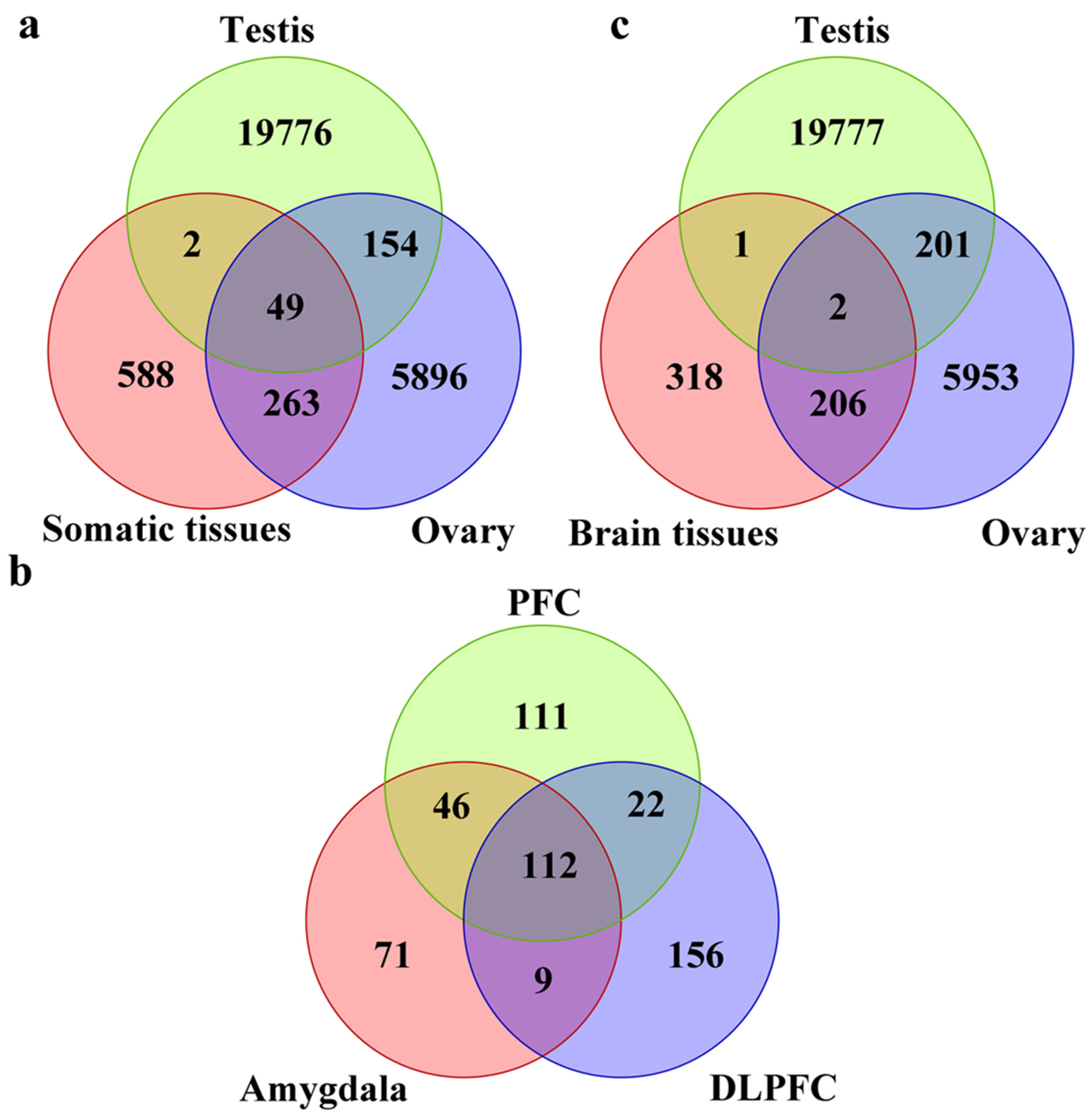
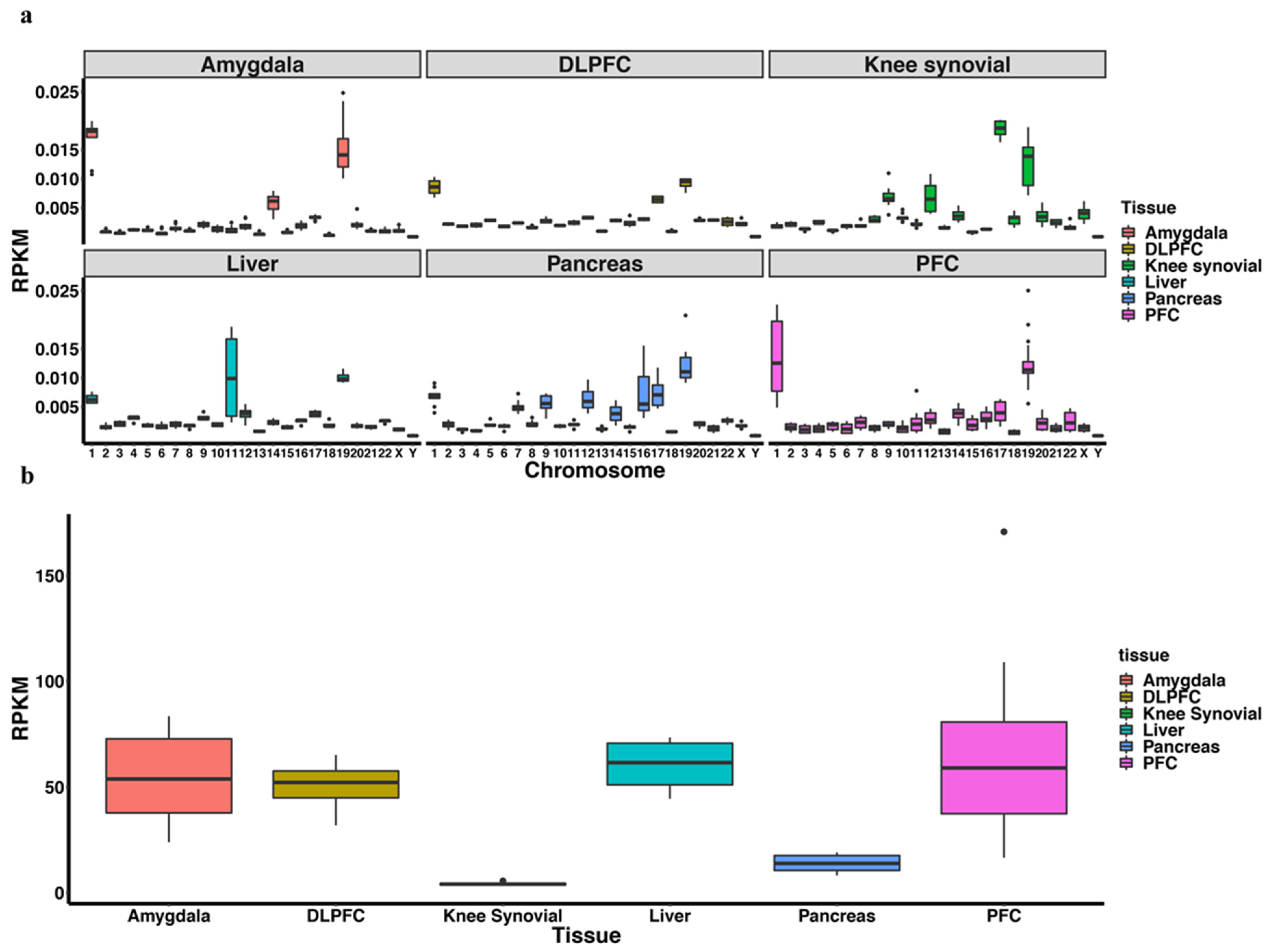
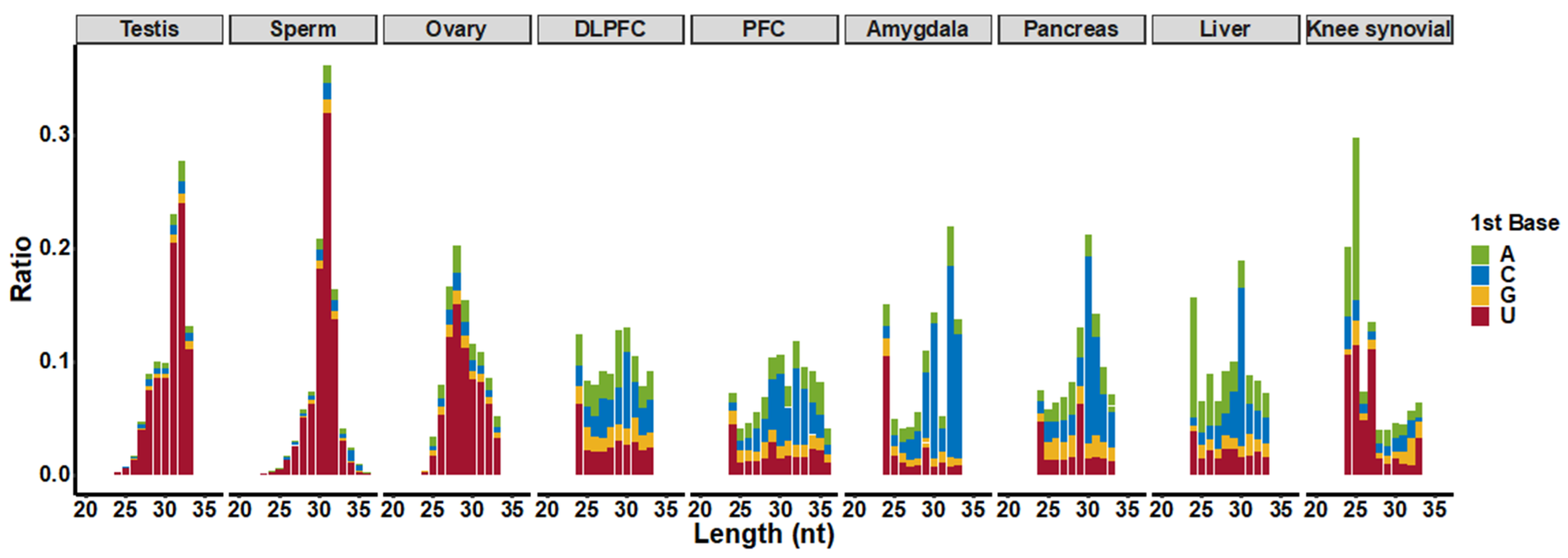
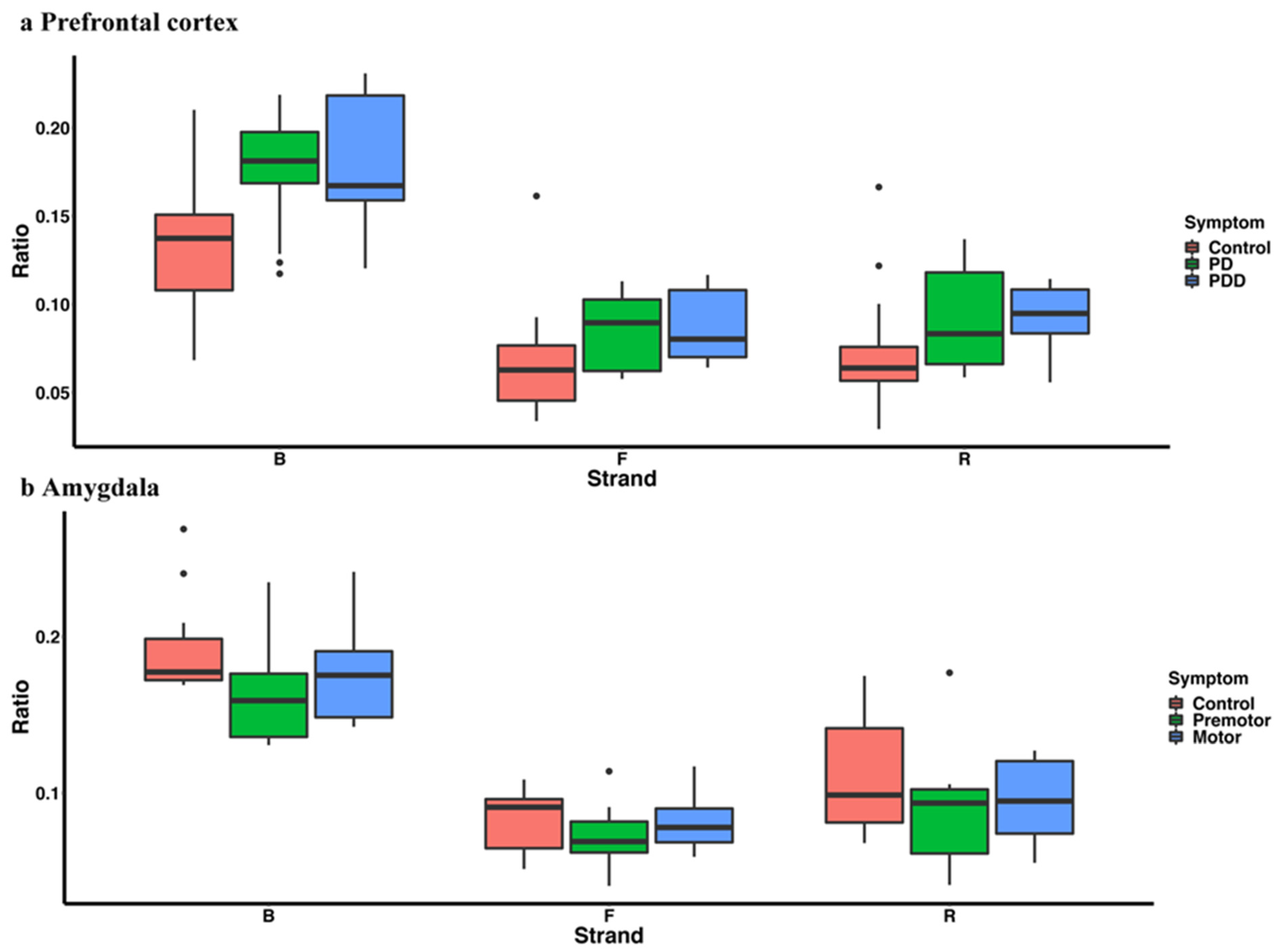
3.1. piRNA Expression in Somatic Cells and Comparison to Germline Cells
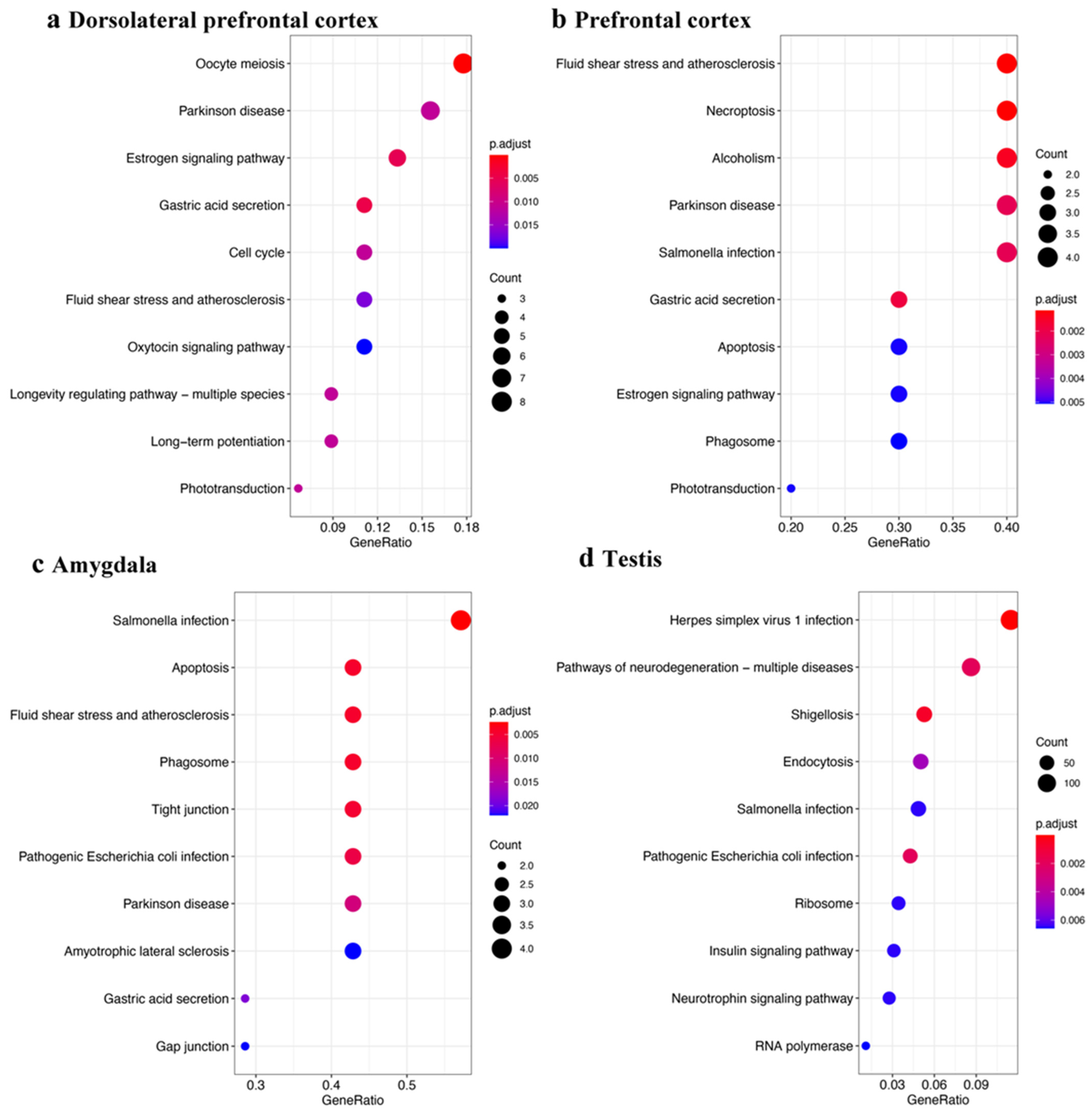
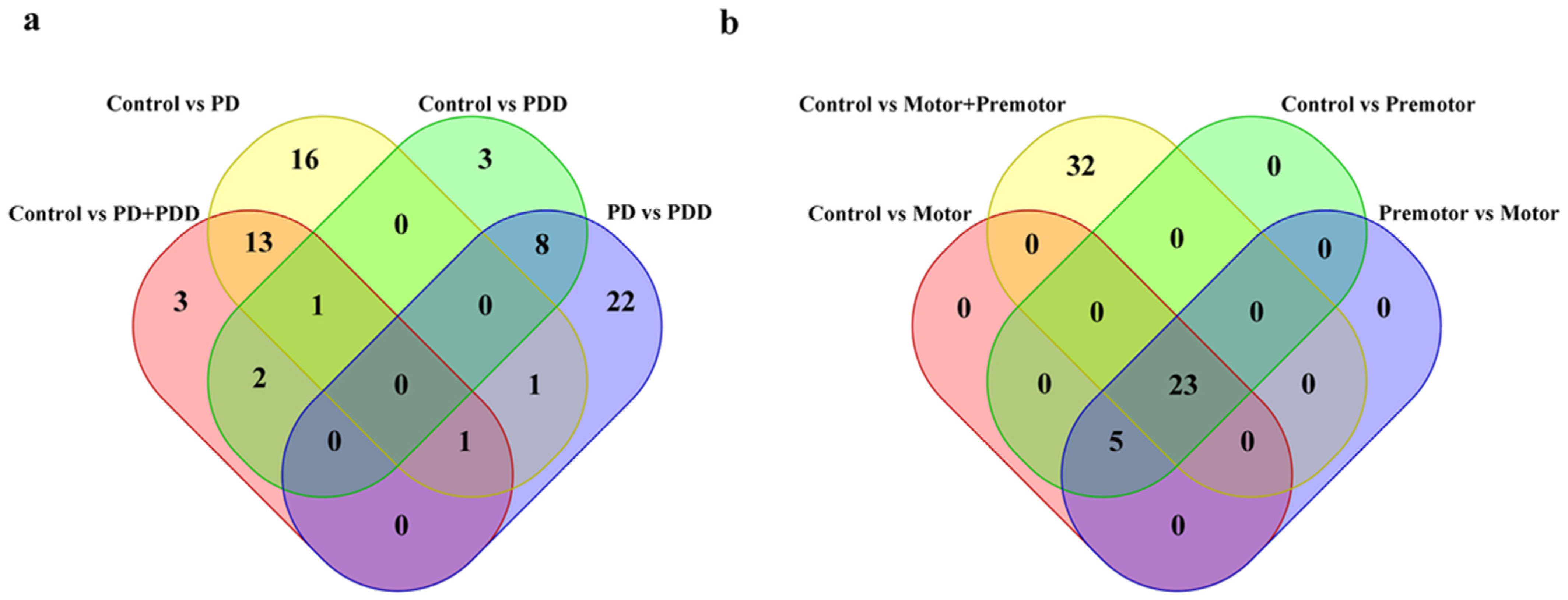
3.2. piRNAs in Parkinson’s Disease
3.3. sPLS-DA of piRNAs Expression Level in Control and PD
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ALS | amyotrophic lateral sclerosis |
| AUC | area under curve |
| DLPFC | dorsolateral prefrontal cortex |
| GEO | Gene Expression Omnibus |
| GO | gene ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| LEVs | large extracellular vesicles |
| LINE | long interspersed nuclear element |
| lncRNA | long non-coding RNA |
| LTR | Long terminal repeat |
| miRNA | microRNA |
| miscRNA | miscellaneous RNA |
| ncRNA | non-coding RNA |
| NGS | next generation sequencing |
| PD | Parkinson’s disease |
| PDD | Parkinson’s disease dementia |
| PFC | prefrontal cortex |
| piRNA | piwi-interacting RNAs |
| PIWI | P-element induced wimpy testis in Drosophila |
| ROC | receiver operating characteristic |
| rRNA | ribosomal RNA |
| scaRNA | small cajal body-specific RNA |
| scRNA | small conditional RNA |
| SEVs | small extracellular vesicles |
| SINE | short interspersed nuclear element |
| snoRNA | small nucleolar RNA |
| snRNA | small nuclear RNA |
| sPLS-DA | sparse partial least square discriminant analysis |
| sRNA | bacterial small RNA |
| tRNA | transfer RNA |
| UTR | untranslated region |
References
- Weick, E.M.; Miska, E.A. piRNAs: From biogenesis to function. Development 2014, 141, 3458–3471. [Google Scholar] [CrossRef] [PubMed]
- Hirakata, S.; Siomi, M.C. piRNA biogenesis in the germline: From transcription of piRNA genomic sources to piRNA maturation. Biochim. Biophys. Acta 2016, 1859, 82–92. [Google Scholar] [CrossRef] [PubMed]
- Czech, B.; Munafò, M.; Ciabrelli, F.; Eastwood, E.L.; Fabry, M.H.; Kneuss, E.; Hannon, G.J. piRNA-Guided Genome Defense: From Biogenesis to Silencing. Annu. Rev. Genet. 2018, 52, 131–157. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.H.; Lee, B.; Li, X.Z. The birth of piRNAs: How mammalian piRNAs are produced, originated, and evolved. Mamm. Genome 2021. [Google Scholar] [CrossRef]
- Senti, K.A.; Brennecke, J. The piRNA pathway: A fly’s perspective on the guardian of the genome. Trends Genet. 2010, 26, 499–509. [Google Scholar] [CrossRef]
- Muerdter, F.; Guzzardo, P.M.; Gillis, J.; Luo, Y.; Yu, Y.; Chen, C.; Fekete, R.; Hannon, G.J. A genome-wide RNAi screen draws a genetic framework for transposon control and primary piRNA biogenesis in Drosophila. Mol. Cell 2013, 50, 736–748. [Google Scholar] [CrossRef]
- McClintock, B. The origin and behavior of mutable loci in maize. Proc. Natl. Acad. Sci. USA 1950, 36, 344–355. [Google Scholar] [CrossRef]
- O’Donnell, K.A.; Boeke, J.D. Mighty Piwis defend the germline against genome intruders. Cell 2007, 129, 37–44. [Google Scholar] [CrossRef]
- Muñoz-Lopez, M.; Vilar-Astasio, R.; Tristan-Ramos, P.; Lopez-Ruiz, C.; Garcia-Pérez, J.L. Study of Transposable Elements and Their Genomic Impact. Methods Mol. Biol. 2016, 1400, 1–19. [Google Scholar]
- Girard, A.; Sachidanandam, R.; Hannon, G.J.; Carmell, M.A. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 2006, 442, 199–202. [Google Scholar] [CrossRef]
- Ozata, D.M.; Gainetdinov, I.; Zoch, A.; O’Carroll, D.; Zamore, P.D. PIWI-interacting RNAs: Small RNAs with big functions. Nat. Rev. Genet. 2019, 20, 89–108. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, Y.W.; Siomi, M.C.; Siomi, H. PIWI-interacting RNA: Its biogenesis and functions. Annu. Rev. Biochem. 2015, 84, 405–433. [Google Scholar] [CrossRef]
- Goh, W.S.S.; Falciatori, I.; Tam, O.H.; Burgess, R.; Meikar, O.; Kotaja, N.; Hammell, M.; Hannon, G.J. piRNA-directed cleavage of meiotic transcripts regulates spermatogenesis. Genes Dev. 2015, 29, 1032–1044. [Google Scholar] [CrossRef]
- Zhang, P.; Kang, J.-Y.; Gou, L.-T.; Wang, J.; Xue, Y.; Skogerboe, G.; Dai, P.; Huang, D.-W.; Chen, R.; Fu, X.-D. MIWI and piRNA-mediated cleavage of messenger RNAs in mouse testes. Cell Res. 2015, 25, 193–207. [Google Scholar] [CrossRef]
- Dai, P.; Wang, X.; Gou, L.-T.; Li, Z.-T.; Wen, Z.; Chen, Z.-G.; Hua, M.-M.; Zhong, A.; Wang, L.; Su, H. A translation-activating function of MIWI/piRNA during mouse spermiogenesis. Cell 2019, 179, 1566–1581.e16. [Google Scholar] [CrossRef] [PubMed]
- Zheng, K.; Wang, P.J. Blockade of pachytene piRNA biogenesis reveals a novel requirement for maintaining post-meiotic germline genome integrity. PLoS Genet. 2012, 8, e1003038. [Google Scholar] [CrossRef] [PubMed]
- Özata, D.M.; Yu, T.; Mou, H.; Gainetdinov, I.; Colpan, C.; Cecchini, K.; Kaymaz, Y.; Wu, P.H.; Fan, K.; Kucukural, A.; et al. Evolutionarily conserved pachytene piRNA loci are highly divergent among modern humans. Nat. Ecol. Evol. 2020, 4, 156–168. [Google Scholar] [CrossRef]
- Gainetdinov, I.; Colpan, C.; Arif, A.; Cecchini, K.; Zamore, P.D. A single mechanism of biogenesis, initiated and directed by PIWI proteins, explains piRNA production in most animals. Mol. Cell 2018, 71, 775–790.e5. [Google Scholar] [CrossRef]
- Robine, N.; Lau, N.C.; Balla, S.; Jin, Z.; Okamura, K.; Kuramochi-Miyagawa, S.; Blower, M.D.; Lai, E.C. A broadly conserved pathway generates 3′ UTR-directed primary piRNAs. Curr. Biol. 2009, 19, 2066–2076. [Google Scholar] [CrossRef]
- Aravin, A.; Gaidatzis, D.; Pfeffer, S.; Lagos-Quintana, M.; Landgraf, P.; Iovino, N.; Morris, P.; Brownstein, M.J.; Kuramochi-Miyagawa, S.; Nakano, T. A novel class of small RNAs bind to MILI protein in mouse testes. Nature 2006, 442, 203–207. [Google Scholar] [CrossRef]
- Gou, L.-T.; Dai, P.; Yang, J.-H.; Xue, Y.; Hu, Y.-P.; Zhou, Y.; Kang, J.-Y.; Wang, X.; Li, H.; Hua, M.-M. Pachytene piRNAs instruct massive mRNA elimination during late spermiogenesis. Cell Res. 2014, 24, 680–700. [Google Scholar] [CrossRef] [PubMed]
- Saito, K.; Inagaki, S.; Mituyama, T.; Kawamura, Y.; Ono, Y.; Sakota, E.; Kotani, H.; Asai, K.; Siomi, H.; Siomi, M.C. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature 2009, 461, 1296–1299. [Google Scholar] [CrossRef] [PubMed]
- Klattenhoff, C.; Xi, H.; Li, C.; Lee, S.; Xu, J.; Khurana, J.S.; Zhang, F.; Schultz, N.; Koppetsch, B.S.; Nowosielska, A.; et al. The Drosophila HP1 homolog Rhino is required for transposon silencing and piRNA production by dual-strand clusters. Cell 2009, 138, 1137–1149. [Google Scholar] [CrossRef] [PubMed]
- Malone, C.D.; Brennecke, J.; Dus, M.; Stark, A.; McCombie, W.R.; Sachidanandam, R.; Hannon, G.J. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 2009, 137, 522–535. [Google Scholar] [CrossRef]
- Théron, E.; Dennis, C.; Brasset, E.; Vaury, C. Distinct features of the piRNA pathway in somatic and germ cells: From piRNA cluster transcription to piRNA processing and amplification. Mob. DNA 2014, 5, 28. [Google Scholar] [CrossRef]
- Ross, R.J.; Weiner, M.M.; Lin, H. PIWI proteins and PIWI-interacting RNAs in the soma. Nature 2014, 505, 353–359. [Google Scholar] [CrossRef]
- Rinkevich, Y.; Rosner, A.; Rabinowitz, C.; Lapidot, Z.; Moiseeva, E.; Rinkevich, B. Piwi positive cells that line the vasculature epithelium, underlie whole body regeneration in a basal chordate. Dev. Biol. 2010, 345, 94–104. [Google Scholar] [CrossRef]
- Martinez, V.D.; Vucic, E.A.; Thu, K.L.; Hubaux, R.; Enfield, K.S.; Pikor, L.A.; Becker-Santos, D.D.; Brown, C.J.; Lam, S.; Lam, W.L. Unique somatic and malignant expression patterns implicate PIWI-interacting RNAs in cancer-type specific biology. Sci. Rep. 2015, 5, 10423. [Google Scholar] [CrossRef]
- Halbach, R.; Miesen, P.; Joosten, J.; Taşköprü, E.; Rondeel, I.; Pennings, B.; Vogels, C.B.; Merkling, S.H.; Koenraadt, C.J.; Lambrechts, L. A satellite repeat-derived piRNA controls embryonic development of Aedes. Nature 2020, 580, 274–277. [Google Scholar] [CrossRef]
- Kim, K.W. PIWI Proteins and piRNAs in the Nervous System. Mol. Cells 2019, 42, 828. [Google Scholar]
- Perrat, P.N.; DasGupta, S.; Wang, J.; Theurkauf, W.; Weng, Z.; Rosbash, M.; Waddell, S. Transposition-driven genomic heterogeneity in the Drosophila brain. Science 2013, 340, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Faulkner, G.J.; Garcia-Perez, J.L. L1 mosaicism in mammals: Extent, effects, and evolution. Trends Genet. 2017, 33, 802–816. [Google Scholar] [CrossRef]
- Larsen, P.A.; Hunnicutt, K.E.; Larsen, R.J.; Yoder, A.D.; Saunders, A.M. Warning SINEs: Alu elements, evolution of the human brain, and the spectrum of neurological disease. Chromosome Res. 2018, 26, 93–111. [Google Scholar] [CrossRef] [PubMed]
- Olivieri, D.; Sykora, M.M.; Sachidanandam, R.; Mechtler, K.; Brennecke, J. An in vivo RNAi assay identifies major genetic and cellular requirements for primary piRNA biogenesis in Drosophila. EMBO J. 2010, 29, 3301–3317. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.C.; Lin, H. Beyond transposons: The epigenetic and somatic functions of the Piwi-piRNA mechanism. Curr. Opin. Cell Biol. 2013, 25, 190–194. [Google Scholar] [CrossRef] [PubMed]
- Ishizu, H.; Iwasaki, Y.W.; Hirakata, S.; Ozaki, H.; Iwasaki, W.; Siomi, H.; Siomi, M.C. Somatic Primary piRNA Biogenesis Driven by cis-Acting RNA Elements and trans-Acting Yb. Cell Rep. 2015, 12, 429–440. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.W.; Tang, N.H.; Andrusiak, M.G.; Wu, Z.; Chisholm, A.D.; Jin, Y. A Neuronal piRNA Pathway Inhibits Axon Regeneration in C. elegans. Neuron 2018, 97, 511–519.e6. [Google Scholar] [CrossRef]
- Lee, D.; Yang, H.; Kim, J.; Brady, S.; Zdraljevic, S.; Zamanian, M.; Kim, H.; Paik, Y.K.; Kruglyak, L.; Andersen, E.C.; et al. The genetic basis of natural variation in a phoretic behavior. Nat. Commun. 2017, 8, 273. [Google Scholar] [CrossRef]
- Rajasethupathy, P.; Antonov, I.; Sheridan, R.; Frey, S.; Sander, C.; Tuschl, T.; Kandel, E.R. A role for neuronal piRNAs in the epigenetic control of memory-related synaptic plasticity. Cell 2012, 149, 693–707. [Google Scholar] [CrossRef]
- Leighton, L.J.; Wei, W.; Marshall, P.R.; Ratnu, V.S.; Li, X.; Zajaczkowski, E.L.; Spadaro, P.A.; Khandelwal, N.; Kumar, A.; Bredy, T.W. Disrupting the hippocampal Piwi pathway enhances contextual fear memory in mice. Neurobiol. Learn. Mem 2019, 161, 202–209. [Google Scholar] [CrossRef]
- Nandi, S.; Chandramohan, D.; Fioriti, L.; Melnick, A.M.; Hébert, J.M.; Mason, C.E.; Rajasethupathy, P.; Kandel, E.R. Roles for small noncoding RNAs in silencing of retrotransposons in the mammalian brain. Proc. Natl. Acad. Sci. USA 2016, 113, 12697–12702. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.J.; Banerjee, S.; Zhou, H.; Jammalamadaka, A.; Arcila, M.; Manjunath, B.; Kosik, K.S. Identification of piRNAs in the central nervous system. RNA 2011, 17, 1090–1099. [Google Scholar] [CrossRef] [PubMed]
- Kalia, L.V.; Lang, A.E. Parkinson’s disease. Lancet 2015, 386, 896–912. [Google Scholar] [CrossRef]
- Bloem, B.R.; Okun, M.S.; Klein, C. Parkinson’s disease. Lancet 2021, 397, 2284–2303. [Google Scholar] [CrossRef]
- Bernheimer, H.; Birkmayer, W.; Hornykiewicz, O.; Jellinger, K.; Seitelberger, F. Brain dopamine and the syndromes of Parkinson and Huntington. Clinical, morphological and neurochemical correlations. J. Neurol. Sci. 1973, 20, 415–455. [Google Scholar] [CrossRef]
- Belvisi, D.; Pellicciari, R.; Fabbrini, A.; Costanzo, M.; Pietracupa, S.; De Lucia, M.; Modugno, N.; Magrinelli, F.; Dallocchio, C.; Ercoli, T.; et al. Risk factors of Parkinson disease: Simultaneous assessment, interactions, and etiologic subtypes. Neurology 2020, 95, e2500–e2508. [Google Scholar] [CrossRef]
- Moustafa, A.A.; Chakravarthy, S.; Phillips, J.R.; Gupta, A.; Keri, S.; Polner, B.; Frank, M.J.; Jahanshahi, M. Motor symptoms in Parkinson’s disease: A unified framework. Neurosci. Biobehav. Rev. 2016, 68, 727–740. [Google Scholar] [CrossRef]
- Schapira, A.H.V.; Chaudhuri, K.R.; Jenner, P. Non-motor features of Parkinson disease. Nat. Rev. Neurosci. 2017, 18, 435–450. [Google Scholar] [CrossRef]
- Solla, P.; Masala, C.; Pinna, I.; Ercoli, T.; Loy, F.; Orofino, G.; Fadda, L.; Defazio, G. Frequency and Determinants of Olfactory Hallucinations in Parkinson’s Disease Patients. Brain Sci. 2021, 11, 841. [Google Scholar] [CrossRef]
- Dickson, D.W. Parkinson’s disease and parkinsonism: Neuropathology. Cold Spring Harb. Perspect. Med. 2012, 2, a009258. [Google Scholar] [CrossRef]
- Braak, H.; Ghebremedhin, E.; Rüb, U.; Bratzke, H.; Del Tredici, K. Stages in the development of Parkinson’s disease-related pathology. Cell Tissue Res. 2004, 318, 121–134. [Google Scholar] [CrossRef]
- Wakabayashi, K.; Tanji, K.; Odagiri, S.; Miki, Y.; Mori, F.; Takahashi, H. The Lewy body in Parkinson’s disease and related neurodegenerative disorders. Mol. Neurobiol. 2013, 47, 495–508. [Google Scholar] [CrossRef] [PubMed]
- Gibb, W.R.; Lees, A.J. The relevance of the Lewy body to the pathogenesis of idiopathic Parkinson’s disease. J. Neurol. Neurosurg. Psychiatry 1988, 51, 745–752. [Google Scholar] [CrossRef] [PubMed]
- Trojanowski, J.Q.; Lee, V.M. Aggregation of neurofilament and alpha-synuclein proteins in Lewy bodies: Implications for the pathogenesis of Parkinson disease and Lewy body dementia. Arch. Neurol. 1998, 55, 151–152. [Google Scholar] [CrossRef]
- Spillantini, M.G.; Crowther, R.A.; Jakes, R.; Hasegawa, M.; Goedert, M. alpha-Synuclein in filamentous inclusions of Lewy bodies from Parkinson’s disease and dementia with lewy bodies. Proc. Natl. Acad. Sci. USA 1998, 95, 6469–6473. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.; Fiesel, F.C.; Truban, D.; Castanedes Casey, M.; Lin, W.L.; Soto, A.I.; Tacik, P.; Rousseau, L.G.; Diehl, N.N.; Heckman, M.G.; et al. Age- and disease-dependent increase of the mitophagy marker phospho-ubiquitin in normal aging and Lewy body disease. Autophagy 2018, 14, 1404–1418. [Google Scholar] [CrossRef]
- Minami, A.; Nakanishi, A.; Matsuda, S.; Kitagishi, Y.; Ogura, Y. Function of α-synuclein and PINK1 in Lewy body dementia (Review). Int. J. Mol. Med. 2015, 35, 3–9. [Google Scholar] [CrossRef]
- Beyer, K.; Domingo-Sàbat, M.; Ariza, A. Molecular pathology of Lewy body diseases. Int. J. Mol. Sci. 2009, 10, 724–745. [Google Scholar] [CrossRef]
- King, E.; Thomas, A. Systemic Inflammation in Lewy Body Diseases: A Systematic Review. Alzheimer Dis. Assoc. Disord. 2017, 31, 346–356. [Google Scholar] [CrossRef]
- Lazdon, E.; Stolero, N.; Frenkel, D. Microglia and Parkinson’s disease: Footprints to pathology. J. Neural. Transm. 2020, 127, 149–158. [Google Scholar] [CrossRef]
- Schulze, M.; Sommer, A.; Plötz, S.; Farrell, M.; Winner, B.; Grosch, J.; Winkler, J.; Riemenschneider, M.J. Sporadic Parkinson’s disease derived neuronal cells show disease-specific mRNA and small RNA signatures with abundant deregulation of piRNAs. Acta Neuropathol. Commun. 2018, 6, 58. [Google Scholar] [CrossRef] [PubMed]
- Wakisaka, K.T.; Tanaka, R.; Hirashima, T.; Muraoka, Y.; Azuma, Y.; Yoshida, H.; Tokuda, T.; Asada, S.; Suda, K.; Ichiyanagi, K. Novel roles of Drosophila FUS and Aub responsible for piRNA biogenesis in neuronal disorders. Brain Res. 2019, 1708, 207–219. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Samimi, H.; Gamez, M.; Zare, H.; Frost, B. Pathogenic tau-induced piRNA depletion promotes neuronal death through transposable element dysregulation in neurodegenerative tauopathies. Nat. Neurosci. 2018, 21, 1038–1048. [Google Scholar] [CrossRef]
- Huang, X.; Wong, G. An old weapon with a new function: PIWI-interacting RNAs in neurodegenerative diseases. Transl. Neurodegener. 2021, 10, 9. [Google Scholar] [CrossRef]
- Tosar, J.P.; Rovira, C.; Cayota, A. Non-coding RNA fragments account for the majority of annotated piRNAs expressed in somatic non-gonadal tissues. Commun. Biol. 2018, 1, 2. [Google Scholar] [CrossRef] [PubMed]
- Freedman, J.E.; Gerstein, M.; Mick, E.; Rozowsky, J.; Levy, D.; Kitchen, R.; Das, S.; Shah, R.; Danielson, K.; Beaulieu, L. Diverse human extracellular RNAs are widely detected in human plasma. Nat. Commun. 2016, 7, 11106. [Google Scholar] [CrossRef]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets—Update. Nucleic Acids Res. 2013, 4, D991–D995. [Google Scholar] [CrossRef]
- Morin, R.D.; O’Connor, M.D.; Griffith, M.; Kuchenbauer, F.; Delaney, A.; Prabhu, A.-L.; Zhao, Y.; McDonald, H.; Zeng, T.; Hirst, M. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008, 18, 610–621. [Google Scholar] [CrossRef]
- Ha, H.; Song, J.; Wang, S.; Kapusta, A.; Feschotte, C.; Chen, K.C.; Xing, J. A comprehensive analysis of piRNAs from adult human testis and their relationship with genes and mobile elements. BMC Genom. 2014, 15, 545. [Google Scholar] [CrossRef]
- Xu, H.; Wang, X.; Wang, Z.; Li, J.; Xu, Z.; Miao, M.; Chen, G.; Lei, X.; Wu, J.; Shi, H. MicroRNA expression profile analysis in sperm reveals hsa-mir-191 as an auspicious omen of in vitro fertilization. BMC Genom. 2020, 21, 165. [Google Scholar] [CrossRef]
- Roovers, E.F.; Rosenkranz, D.; Mahdipour, M.; Han, C.-T.; He, N.; de Sousa Lopes, S.M.C.; van der Westerlaken, L.A.; Zischler, H.; Butter, F.; Roelen, B.A. Piwi proteins and piRNAs in mammalian oocytes and early embryos. Cell Rep. 2015, 10, 2069–2082. [Google Scholar] [CrossRef]
- Ryvkin, P.; Leung, Y.Y.; Silverman, I.M.; Childress, M.; Valladares, O.; Dragomir, I.; Gregory, B.D.; Wang, L.-S. HAMR: High-throughput annotation of modified ribonucleotides. RNA 2013, 19, 1684–1692. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Wu, Y.J.; Liang, X.; Ji, M.; Ying, H.M.; Wang, X.Y.; Sun, X.; Shao, C.H.; Zhan, L.X.; Zhang, Y. Network-based integration of mRNA and miRNA profiles reveals new target genes involved in pancreatic cancer. Mol. Carcinog. 2019, 58, 206–218. [Google Scholar] [CrossRef] [PubMed]
- Bratus-Neuenschwander, A.; Castro-Giner, F.; Frank-Bertoncelj, M.; Aluri, S.; Fucentese, S.F.; Schlapbach, R.; Sprott, H. Pain-associated transcriptome changes in synovium of knee osteoarthritis patients. Genes 2018, 9, 338. [Google Scholar] [CrossRef] [PubMed]
- Yamane, D.; Selitsky, S.R.; Shimakami, T.; Li, Y.; Zhou, M.; Honda, M.; Sethupathy, P.; Lemon, S.M. Differential hepatitis C virus RNA target site selection and host factor activities of naturally occurring miR-122 3’ variants. Nucleic Acids Res. 2017, 45, 4743–4755. [Google Scholar] [CrossRef][Green Version]
- Hoss, A.G.; Labadorf, A.; Beach, T.G.; Latourelle, J.C.; Myers, R.H. microRNA profiles in Parkinson’s disease prefrontal cortex. Front. Aging Neurosci. 2016, 8, 36. [Google Scholar] [CrossRef]
- Hoss, A.G.; Labadorf, A.; Latourelle, J.C.; Kartha, V.K.; Hadzi, T.C.; Gusella, J.F.; MacDonald, M.E.; Chen, J.-F.; Akbarian, S.; Weng, Z. miR-10b-5p expression in Huntington’s disease brain relates to age of onset and the extent of striatal involvement. BMC Med. Genom. 2015, 8, 10. [Google Scholar] [CrossRef]
- Wake, C.; Labadorf, A.; Dumitriu, A.; Hoss, A.G.; Bregu, J.; Albrecht, K.H.; DeStefano, A.L.; Myers, R.H. Novel microRNA discovery using small RNA sequencing in post-mortem human brain. BMC Genom. 2016, 17, 776. [Google Scholar] [CrossRef]
- Pantano, L.; Friedlaender, M.R.; Escaramis, G.; Lizano, E.; Pallares-Albanell, J.; Ferrer, I.; Estivill, X.; Martí, E. Specific small-RNA signatures in the amygdala at premotor and motor stages of Parkinson’s disease revealed by deep sequencing analysis. Bioinformatics 2016, 32, 673–681. [Google Scholar] [CrossRef]
- Pantano, L.; Pantano, F.; Marti, E.; Sui, S.H. Visualization of the small RNA transcriptome using seqclusterViz. F1000Research 2019, 8, 232. [Google Scholar]
- Sproviero, D.; Gagliardi, S.; Zucca, S.; Arigoni, M.; Giannini, M.; Garofalo, M.; Olivero, M.; Dell’Orco, M.; Pansarasa, O.; Bernuzzi, S. Different miRNA Profiles in Plasma Derived Small and Large Extracellular Vesicles from Patients with Neurodegenerative Diseases. Int. J. Mol. Sci. 2021, 22, 2737. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Yates, A.D.; Achuthan, P.; Akanni, W.; Allen, J.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Azov, A.G.; Bennett, R. Ensembl 2020. Nucleic Acids Res. 2020, 48, D682–D688. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B. Aligning short sequencing reads with Bowtie. Curr. Protoc. Bioinform. 2010, 32, 11.7.1–11.7.14. [Google Scholar] [CrossRef]
- Lee, C.M.; Barber, G.P.; Casper, J.; Clawson, H.; Diekhans, M.; Gonzalez, J.N.; Hinrichs, A.S.; Lee, B.T.; Nassar, L.R.; Powell, C.C. UCSC Genome Browser enters 20th year. Nucleic Acids Res. 2020, 48, D756–D761. [Google Scholar] [CrossRef]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016, 44, D733–D745. [Google Scholar] [CrossRef]
- Frankish, A.; Diekhans, M.; Ferreira, A.-M.; Johnson, R.; Jungreis, I.; Loveland, J.; Mudge, J.M.; Sisu, C.; Wright, J.; Armstrong, J. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019, 47, D766–D773. [Google Scholar] [CrossRef]
- Leung, Y.Y.; Kuksa, P.P.; Amlie-Wolf, A.; Valladares, O.; Ungar, L.H.; Kannan, S.; Gregory, B.D.; Wang, L.-S. DASHR: Database of small human noncoding RNAs. Nucleic Acids Res. 2016, 44, D216–D222. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, P.; Lu, Y.; Li, Y.; Zheng, Y.; Kan, Y.; Chen, R.; He, S. piRBase: A comprehensive database of piRNA sequences. Nucleic Acids Res. 2019, 47, D175–D180. [Google Scholar] [CrossRef]
- Fernandes, J.D.; Zamudio-Hurtado, A.; Clawson, H.; Kent, W.J.; Haussler, D.; Salama, S.R.; Haeussler, M. The UCSC repeat browser allows discovery and visualization of evolutionary conflict across repeat families. Mob. DNA 2020, 11, 13. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Stein, C.K.; Qu, P.; Epstein, J.; Buros, A.; Rosenthal, A.; Crowley, J.; Morgan, G.; Barlogie, B. Removing batch effects from purified plasma cell gene expression microarrays with modified ComBat. BMC Bioinform. 2015, 16, 63. [Google Scholar] [CrossRef] [PubMed]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. g:Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef] [PubMed]
- Ståhle, L.; Wold, S. Partial least squares analysis with cross-validation for the two-class problem: A Monte Carlo study. J. Chemom. 1987, 1, 185–196. [Google Scholar] [CrossRef]
- Barker, M.; Rayens, W. Partial least squares for discrimination. J. Chemom. J. Chemom. Soc. 2003, 17, 166–173. [Google Scholar] [CrossRef]
- Lê Cao, K.-A.; Boitard, S.; Besse, P. Sparse PLS discriminant analysis: Biologically relevant feature selection and graphical displays for multiclass problems. BMC Bioinform. 2011, 12, 253. [Google Scholar] [CrossRef]
- Stone, M. Cross-validatory choice and assessment of statistical predictions. J. R. Stat. Soc. Ser. B 1974, 36, 111–133. [Google Scholar] [CrossRef]
- Allen, D.M. The relationship between variable selection and data agumentation and a method for prediction. Technometrics 1974, 16, 125–127. [Google Scholar] [CrossRef]
- Hanley, J.A.; McNeil, B.J. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology 1982, 143, 29–36. [Google Scholar] [CrossRef]
- Rohart, F.; Gautier, B.; Singh, A.; Lê Cao, K.-A. mixOmics: An R package for ‘omics feature selection and multiple data integration. PLoS Comput. Biol. 2017, 13, e1005752. [Google Scholar] [CrossRef] [PubMed]
- Yan, Z.; Hu, H.Y.; Jiang, X.; Maierhofer, V.; Neb, E.; He, L.; Hu, Y.; Hu, H.; Li, N.; Chen, W. Widespread expression of piRNA-like molecules in somatic tissues. Nucleic Acids Res. 2011, 39, 6596–6607. [Google Scholar] [CrossRef] [PubMed]
- Carmell, M.A.; Girard, A.; van de Kant, H.J.; Bourc’his, D.; Bestor, T.H.; de Rooij, D.G.; Hannon, G.J. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev. Cell 2007, 12, 503–514. [Google Scholar] [CrossRef]
- Perera, B.P.; Tsai, Z.T.-Y.; Colwell, M.L.; Jones, T.R.; Goodrich, J.M.; Wang, K.; Sartor, M.A.; Faulk, C.; Dolinoy, D.C. Somatic expression of piRNA and associated machinery in the mouse identifies short, tissue-specific piRNA. Epigenetics 2019, 14, 504–521. [Google Scholar] [CrossRef]
- Faulkner, G.J.; Kimura, Y.; Daub, C.O.; Wani, S.; Plessy, C.; Irvine, K.M.; Schroder, K.; Cloonan, N.; Steptoe, A.L.; Lassmann, T. The regulated retrotransposon transcriptome of mammalian cells. Nat. Genet. 2009, 41, 563. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, T.; Cheng, E.-c.; Zhong, M.; Lin, H. Retrotransposons and pseudogenes regulate mRNAs and lncRNAs via the piRNA pathway in the germline. Genome Res. 2015, 25, 368–380. [Google Scholar] [CrossRef]
- Wang, C.; Lin, H. Roles of piRNAs in transposon and pseudogene regulation of germline mRNAs and lncRNAs. Genome Biol. 2021, 22, 27. [Google Scholar] [CrossRef]
- Uhrig, S.; Klein, H. PingPongPro: A tool for the detection of piRNA-mediated transposon-silencing in small RNA-Seq data. Bioinformatics 2019, 35, 335–336. [Google Scholar] [CrossRef]
- Zhang, D.; Tu, S.; Stubna, M.; Wu, W.-S.; Huang, W.-C.; Weng, Z.; Lee, H.-C. The piRNA targeting rules and the resistance to piRNA silencing in endogenous genes. Science 2018, 359, 587–592. [Google Scholar] [CrossRef]
- Chalorak, P.; Dharmasaroja, P.; Meemon, K. Downregulation of eEF1A/EFT3-4 Enhances Dopaminergic Neurodegeneration After 6-OHDA Exposure in C. elegans Model. Front. Neurosci. 2020, 14, 303. [Google Scholar] [CrossRef]
- Dumitriu, A.; Golji, J.; Labadorf, A.T.; Gao, B.; Beach, T.G.; Myers, R.H.; Longo, K.A.; Latourelle, J.C. Integrative analyses of proteomics and RNA transcriptomics implicate mitochondrial processes, protein folding pathways and GWAS loci in Parkinson disease. BMC Med Genom. 2015, 9, 5. [Google Scholar] [CrossRef] [PubMed]
- Shen, E.-Z.; Chen, H.; Ozturk, A.R.; Tu, S.; Shirayama, M.; Tang, W.; Ding, Y.-H.; Dai, S.-Y.; Weng, Z.; Mello, C.C. Identification of piRNA binding sites reveals the argonaute regulatory landscape of the C. elegans germline. Cell 2018, 172, 937–951.e18. [Google Scholar] [CrossRef]
- Chavda, V.; Madhwani, K.; Chaurasia, B. PiWi RNA in Neurodevelopment and Neurodegenerative disorders. Curr. Mol. Pharmacol. 2021, 14, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Wakisaka, K.T.; Imai, Y. The dawn of pirna research in various neuronal disorders. Front. Biosci. (Landmark Ed.) 2019, 24, 1440–1451. [Google Scholar] [CrossRef] [PubMed]
- Hyun, S. Small RNA Pathways That Protect the Somatic Genome. Int. J. Mol. Sci. 2017, 18, 912. [Google Scholar] [CrossRef]
- Reik, W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature 2007, 447, 425–432. [Google Scholar] [CrossRef]
- Barckmann, B.; Barckmann, B.; Pierson, S.; Dufourt, J.; Papin, C.; Armenise, C.; Port, F.; Grentzinger, T.; Chambeyron, S.; Baronian, G.; et al. Aubergine iCLIP reveals piRNA-dependent decay of mRNAs involved in germ cell development in the early embryo. Cell Rep. 2015, 12, 1205–1216. [Google Scholar] [CrossRef]
- Rojas-Ríos, P.; Chartier, A.; Pierson, S.; Simonelig, M. Aubergine and piRNAs promote germline stem cell self-renewal by repressing the proto-oncogene Cbl. EMBO J. 2017, 36, 3194–3211. [Google Scholar] [CrossRef]
- Tang, W.; Seth, M.; Tu, S.; Shen, E.-Z.; Li, Q.; Shirayama, M.; Weng, Z.; Mello, C.C. A sex chromosome piRNA promotes robust dosage compensation and sex determination in C. elegans. Dev. Cell 2018, 44, 762–770.e3. [Google Scholar] [CrossRef]
- Balaratnam, S.; West, N.; Basu, S. A piRNA utilizes HILI and HIWI2 mediated pathway to down-regulate ferritin heavy chain 1 mRNA in human somatic cells. Nucleic Acids Res. 2018, 46, 10635–10648. [Google Scholar] [CrossRef]
- Wu, P.-H.; Fu, Y.; Cecchini, K.; Özata, D.M.; Arif, A.; Yu, T.; Colpan, C.; Gainetdinov, I.; Weng, Z.; Zamore, P.D. The evolutionarily conserved piRNA-producing locus pi6 is required for male mouse fertility. Nat. Genet. 2020, 52, 728–739. [Google Scholar] [CrossRef] [PubMed]
- Li, X.Z.; Roy, C.K.; Dong, X.; Bolcun-Filas, E.; Wang, J.; Han, B.W.; Xu, J.; Moore, M.J.; Schimenti, J.C.; Weng, Z. An ancient transcription factor initiates the burst of piRNA production during early meiosis in mouse testes. Mol. Cell 2013, 50, 67–81. [Google Scholar] [CrossRef] [PubMed]
- Fu, A.; Jacobs, D.I.; Zhu, Y. Epigenome-wide analysis of piRNAs in gene-specific DNA methylation. RNA Biol. 2014, 11, 1301–1312. [Google Scholar] [CrossRef] [PubMed]
- Dufourt, J.; Bontonou, G.; Chartier, A.; Jahan, C.; Meunier, A.-C.; Pierson, S.; Harrison, P.F.; Papin, C.; Beilharz, T.H.; Simonelig, M. piRNAs and Aubergine cooperate with Wispy poly (A) polymerase to stabilize mRNAs in the germ plasm. Nat. Commun. 2017, 8, 1305. [Google Scholar] [CrossRef] [PubMed]
- Vourekas, A.; Alexiou, P.; Vrettos, N.; Maragkakis, M.; Mourelatos, Z. Sequence-dependent but not sequence-specific piRNA adhesion traps mRNAs to the germ plasm. Nature 2016, 531, 390–394. [Google Scholar] [CrossRef] [PubMed]
- Klein, J.D.; Qu, C.; Yang, X.; Fan, Y.; Tang, C.; Peng, J.C. c-Fos repression by Piwi regulates Drosophila ovarian germline formation and tissue morphogenesis. PLoS Genet. 2016, 12, e1006281. [Google Scholar] [CrossRef]
- Braak, H.; Braak, E. Pathoanatomy of Parkinson’s disease. J. Neurol. 2000, 247, II3–II10. [Google Scholar] [CrossRef]
- Cools, R.; Clark, L.; Owen, A.M.; Robbins, T.W. Defining the neural mechanisms of probabilistic reversal learning using event-related functional magnetic resonance imaging. J. Neurosci. 2002, 22, 4563–4567. [Google Scholar] [CrossRef]
- Vriend, C.; Boedhoe, P.S.; Rutten, S.; Berendse, H.W.; van der Werf, Y.D.; van den Heuvel, O.A. A smaller amygdala is associated with anxiety in Parkinson’s disease: A combined FreeSurfer—VBM study. J. Neurol. Neurosurg. Psychiatry 2016, 87, 493–500. [Google Scholar] [CrossRef]
- Carey, G.; Görmezoğlu, M.; de Jong, J.J.A.; Hofman, P.A.M.; Backes, W.H.; Dujardin, K.; Leentjens, A.F.G. Neuroimaging of Anxiety in Parkinson’s Disease: A Systematic Review. Mov. Disord. 2021, 36, 327–339. [Google Scholar] [CrossRef]
- Izadpanah, M.; Seddigh, A.; Ebrahimi Barough, S.; Fazeli, S.A.S.; Ai, J. Potential of Extracellular Vesicles in Neurodegenerative Diseases: Diagnostic and Therapeutic Indications. J. Mol. Neurosci. 2018, 66, 172–179. [Google Scholar] [CrossRef] [PubMed]
- Gámez-Valero, A.; Beyer, K.; Borràs, F.E. Extracellular vesicles, new actors in the search for biomarkers of dementias. Neurobiol. Aging 2019, 74, 15–20. [Google Scholar] [CrossRef]
- Lamontagne-Proulx, J.; St-Amour, I.; Labib, R.; Pilon, J.; Denis, H.L.; Cloutier, N.; Roux-Dalvai, F.; Vincent, A.T.; Mason, S.L.; Williams-Gray, C.; et al. Portrait of blood-derived extracellular vesicles in patients with Parkinson’s disease. Neurobiol. Dis. 2019, 124, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Ohmichi, T.; Mitsuhashi, M.; Tatebe, H.; Kasai, T.; Ali El-Agnaf, O.M.; Tokuda, T. Quantification of brain-derived extracellular vesicles in plasma as a biomarker to diagnose Parkinson’s and related diseases. Parkinsonism Relat. Disord. 2019, 61, 82–87. [Google Scholar] [CrossRef] [PubMed]
- Leggio, L.; Paternò, G.; Vivarelli, S.; Falzone, G.G.; Giachino, C.; Marchetti, B.; Iraci, N. Extracellular Vesicles as Novel Diagnostic and Prognostic Biomarkers for Parkinson’s Disease. Aging Dis. 2021, 12, 1494–1515. [Google Scholar] [CrossRef]
- He, S.; Huang, L.; Shao, C.; Nie, T.; Xia, L.; Cui, B.; Lu, F.; Zhu, L.; Chen, B.; Yang, Q. Several miRNAs derived from serum extracellular vesicles are potential biomarkers for early diagnosis and progression of Parkinson’s disease. Transl. Neurodegener. 2021, 10, 25. [Google Scholar] [CrossRef]
- Lööv, C.; Scherzer, C.R.; Hyman, B.T.; Breakefield, X.O.; Ingelsson, M. α-Synuclein in Extracellular Vesicles: Functional Implications and Diagnostic Opportunities. Cell Mol. Neurobiol. 2016, 36, 437–448. [Google Scholar] [CrossRef]
- Kitamura, Y.; Kojima, M.; Kurosawa, T.; Sasaki, R.; Ichihara, S.; Hiraku, Y.; Tomimoto, H.; Murata, M.; Oikawa, S. Proteomic Profiling of Exosomal Proteins for Blood-based Biomarkers in Parkinson’s Disease. Neuroscience 2018, 392, 121–128. [Google Scholar] [CrossRef]
- Li, M.J.; Wang, P.; Liu, X.; Lim, E.L.; Wang, Z.; Yeager, M.; Wong, M.P.; Sham, P.C.; Chanock, S.J.; Wang, J. GWASdb: A database for human genetic variants identified by genome-wide association studies. Nucleic Acids Res. 2012, 40, D1047–D1054. [Google Scholar] [CrossRef]
- Peng, Q.; Long, C.L.; Malhotra, S.; Humphrey, M.B. A physical interaction between the adaptor proteins DOK3 and DAP12 is required to inhibit lipopolysaccharide signaling in macrophages. Sci. Signal 2013, 6, ra72. [Google Scholar] [CrossRef]
- Qin, L.; Wu, X.; Block, M.L.; Liu, Y. Systemic LPS causes chronic neuroinflammation and progressive neurodegeneration. Glia 2007, 55, 453–462. [Google Scholar] [CrossRef]
- Kim, S.R.; Kareva, T.; Yarygina, O.; Kholodilov, N.; Burke, R.E. AAV transduction of dopamine neurons with constitutively active Rheb protects from neurodegeneration and mediates axon regrowth. Mol. Ther. 2012, 20, 275–286. [Google Scholar] [CrossRef]
- Vanni, S.; Zattoni, M.; Moda, F.; Giaccone, G.; Tagliavini, F.; Haïk, S.; Deslys, J.-P.; Zanusso, G.; Ironside, J.W.; Carmona, M. Hemoglobin mRNA changes in the frontal cortex of patients with neurodegenerative diseases. Front. Neurosci. 2018, 12, 8. [Google Scholar] [CrossRef]
- Shen, L.; Wang, C.; Chen, L.; Wong, G. Dysregulation of MicroRNAs and PIWI-Interacting RNAs in a Caenorhabditis elegans Parkinson’s Disease Model Overexpressing Human α-Synuclein and Influence of tdp-1. Front. Neurosci. 2021, 15, 600462. [Google Scholar] [CrossRef]
- Stephenson, J.; Nutma, E.; van der Valk, P.; Amor, S. Inflammation in CNS neurodegenerative diseases. Immunology 2018, 154, 204–219. [Google Scholar] [CrossRef]
- Guo, C.; Jeong, H.H.; Hsieh, Y.C.; Klein, H.U.; Bennett, D.A.; De Jager, P.L.; Liu, Z.; Shulman, J.M. Tau Activates Transposable Elements in Alzheimer’s Disease. Cell Rep. 2018, 23, 2874–2880. [Google Scholar] [CrossRef]
- Ravel-Godreuil, C.; Znaidi, R.; Bonnifet, T.; Joshi, R.L.; Fuchs, J. Transposable elements as new players in neurodegenerative diseases. FEBS Lett. 2021, 595, 2733–2755. [Google Scholar] [CrossRef]
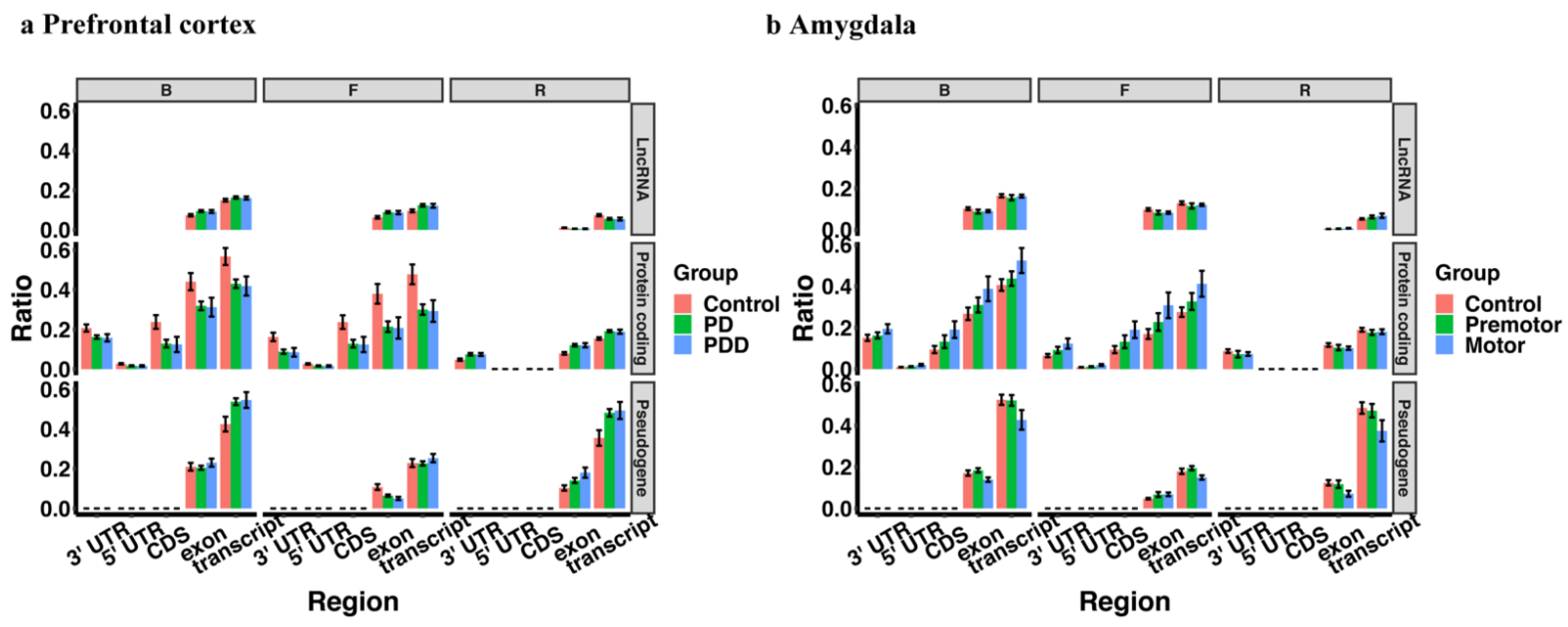
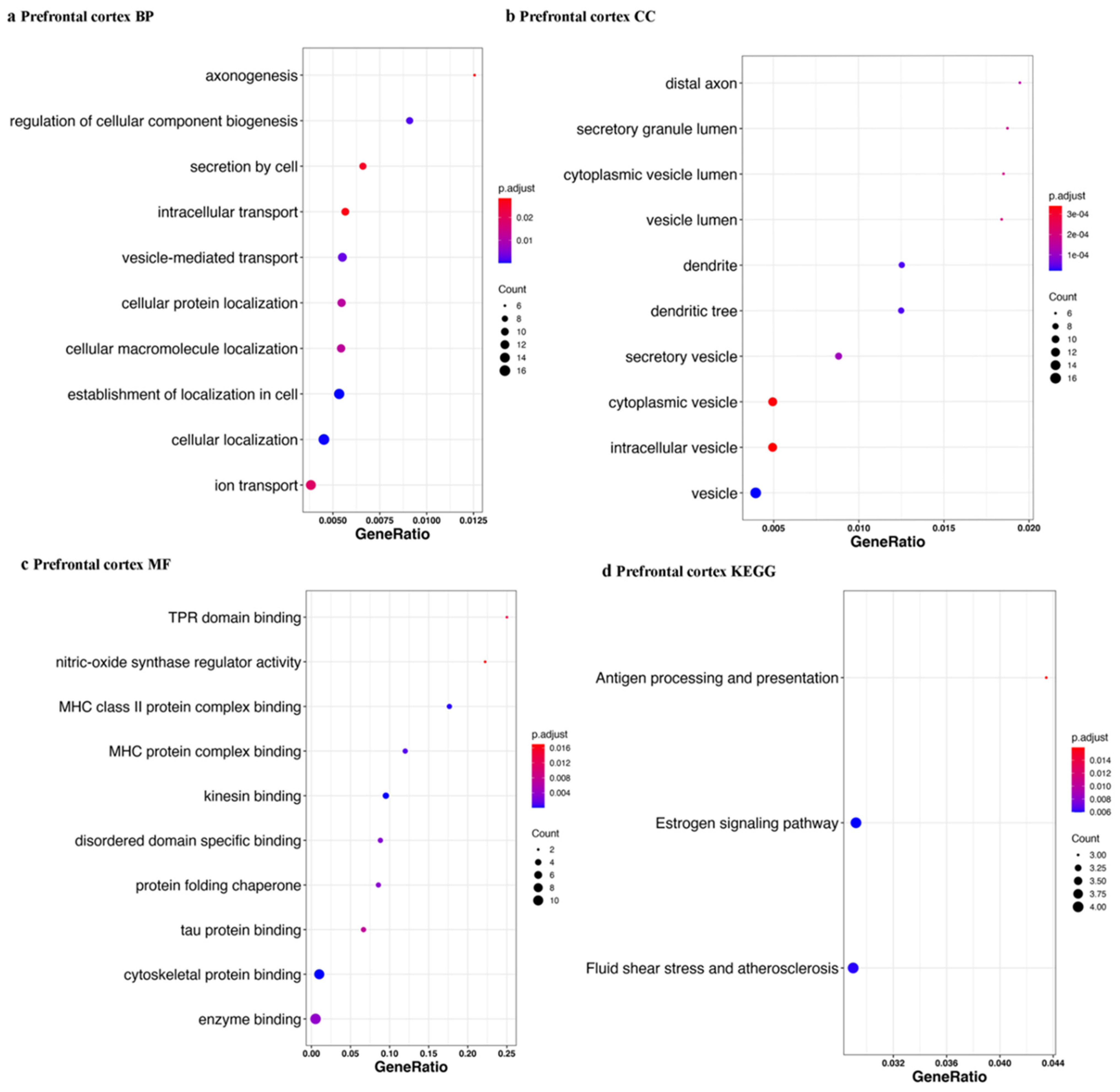
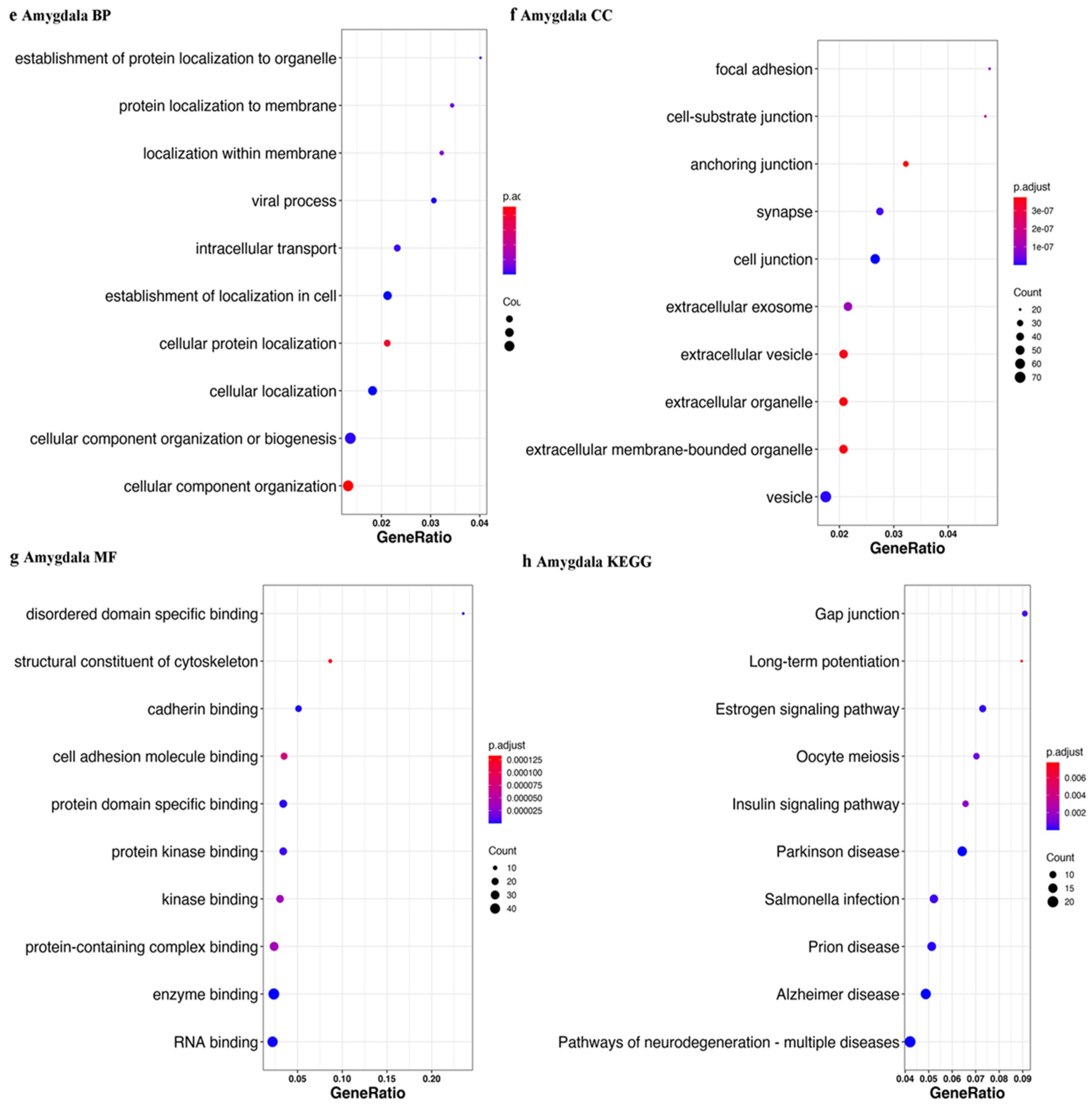
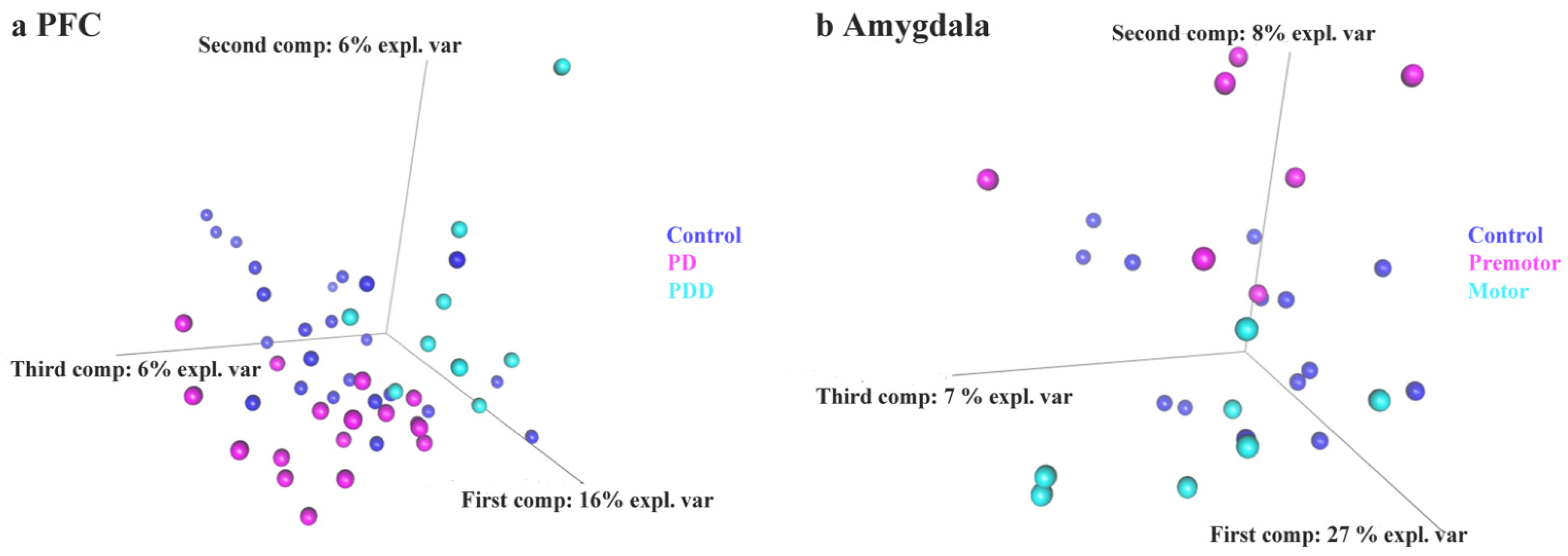
| Tissue | Accession | Sample Size | Library Size | References |
|---|---|---|---|---|
| Testis | PRJNA196749 | 3 | 6.84–25.30 million | [69] |
| Sperm | PRJNA564759 | 102 | 0.36–1.63 million | [70] |
| Ovary | PRJNA272542 | 12 Note: 4 adult ovaries; 4 ovaries from 1st trimester embryos; 4 ovaries from 2st trimester embryos | 13.3–15.8 million | [71] |
| Dorsolateral Prefrontal Cortex (DLPFC) | PRJNA185476 | 4 | 6–12.25 million | [72] |
| Pancreas | PRJNA490335 | 3 | 12.2–12.5 million | [73] |
| Knee synovial tissues | PRJNA389258 | 10 | 8.39–17.21 million | [74] |
| Liver | PRJNA246372 | 4 | 5.83–40.70 million | [75] |
| Prefrontal cortex (PFC) | PRJNA295431 PRJNA272617 | 26 (Parkinson’s disease) Note: including 17 Parkinson’s disease (PD); 9 Parkinson’s disease with dementia (PDD) 25 (Control) | 6.09–33.40 million | [76,77,78] |
| Amygdala | PRJNA381204 | 14 (Parkinson’s disease) Note: including 7 premotor stage; 7 motor stage 14 (Control) | 13.5–17.7 million | [79,80] |
| Blood (extracellular vesicles) | PRJNA655240 | 9 (Parkinson’s disease) Note: in each sample, both large (LEV) and small extracelluar vesciles (SEVs) were tested 6 (Control) Note: in each sample, both large (LEV) and small extracelluar vesciles (SEVs) were tested | 2.37–30.7 million | [81] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, T.; Wong, G. Dysregulation of Human Somatic piRNA Expression in Parkinson’s Disease Subtypes and Stages. Int. J. Mol. Sci. 2022, 23, 2469. https://doi.org/10.3390/ijms23052469
Zhang T, Wong G. Dysregulation of Human Somatic piRNA Expression in Parkinson’s Disease Subtypes and Stages. International Journal of Molecular Sciences. 2022; 23(5):2469. https://doi.org/10.3390/ijms23052469
Chicago/Turabian StyleZhang, Tianjiao, and Garry Wong. 2022. "Dysregulation of Human Somatic piRNA Expression in Parkinson’s Disease Subtypes and Stages" International Journal of Molecular Sciences 23, no. 5: 2469. https://doi.org/10.3390/ijms23052469
APA StyleZhang, T., & Wong, G. (2022). Dysregulation of Human Somatic piRNA Expression in Parkinson’s Disease Subtypes and Stages. International Journal of Molecular Sciences, 23(5), 2469. https://doi.org/10.3390/ijms23052469





