Structural and Kinetic Views of Molecular Chaperones in Multidomain Protein Folding
Abstract
1. Introduction
2. Folding Process of Multidomain Proteins
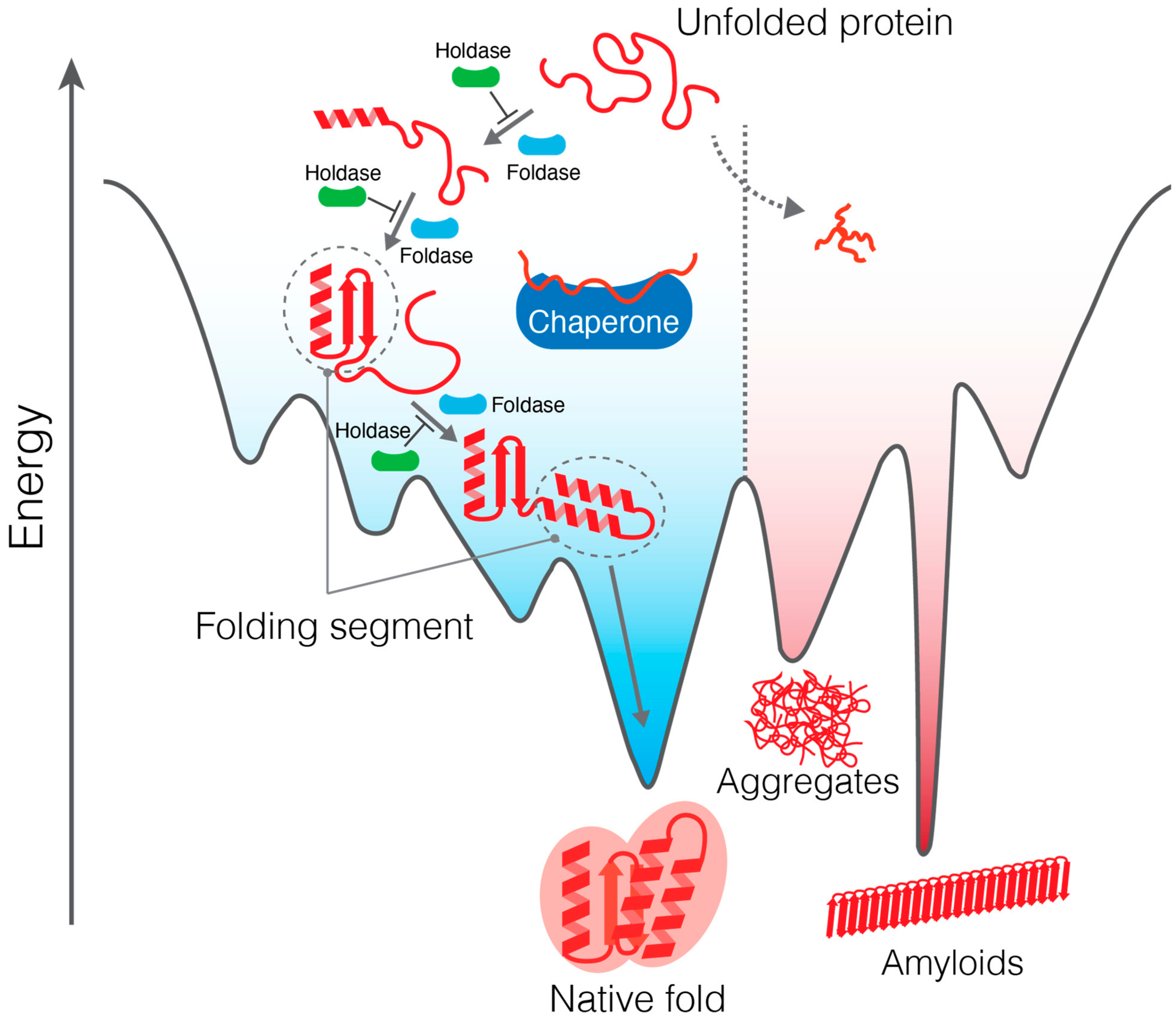
3. Effect of Molecular Chaperones on Protein Folding
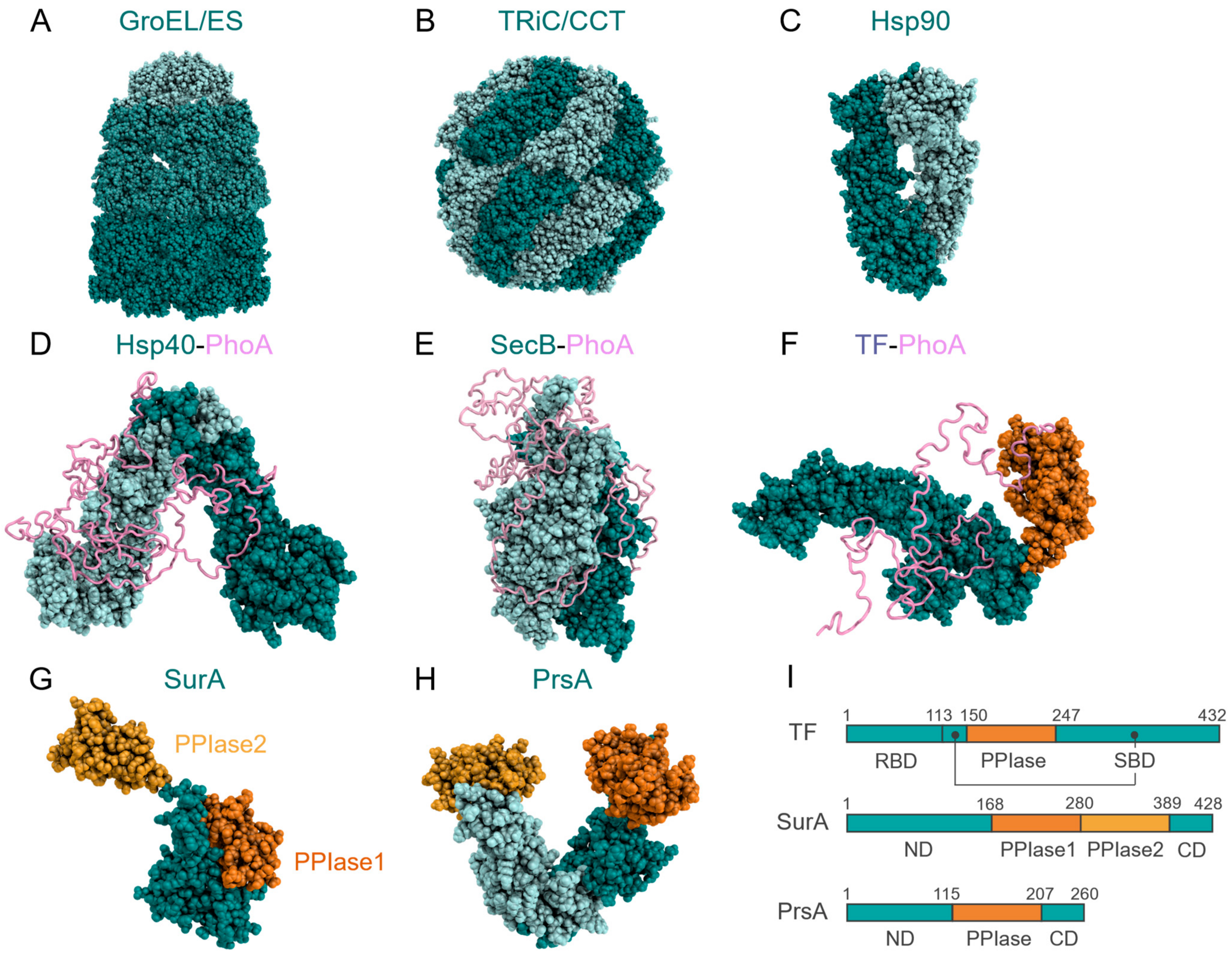
4. Structural Features of Molecular Chaperones
5. Structural Studies of Chaperone–Client Complexes
6. Kinetic Studies for Molecular Chaperones and Client Proteins
7. Catalytic Domains in Molecular Chaperones
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Xu, J. Distance-based protein folding powered by deep learning. Proc. Natl. Acad. Sci. USA 2019, 116, 16856–16865. [Google Scholar] [CrossRef] [PubMed]
- Zwanzig, R.; Szabo, A.; Bagchi, B. Levinthal’s paradox. Proc. Natl. Acad. Sci. USA 1992, 89, 20–22. [Google Scholar] [CrossRef]
- Šali, A.; Shakhnovich, E.; Karplus, M. How does a protein fold? Nature 1994, 369, 248–251. [Google Scholar] [PubMed]
- Levinthal’s Paradox. Available online: https://web.archive.org/web/20110523080407/http://www-miller.ch.cam.ac.uk/levinthal/levinthal.html (accessed on 7 January 2022).
- Jonathan, W.S.; Peter, K.S. Efficient catalysis of disulphide bond rearrangements by protein disulphide isomerase. Nature 1993, 365, 185–188. [Google Scholar]
- Sela, M.; Anfinsen, B. Some Spectrophotometric and Polarimetric Experiments With Ribonuclease. Biochem. Biophys. Acta 1957, 24, 229–235. [Google Scholar] [CrossRef]
- Roder, H.; Elöve, G.A.; Englander, S.W. Structural characterization of folding intermediates in cytochrome c by H-exchange labelling and proton NMR. Nature 1988, 335, 700–704. [Google Scholar] [CrossRef]
- Akiyama, S.; Takahashi, S.; Kimura, T.; Ishimori, K.; Morishima, I.; Nishikawa, Y.; Fujisawa, T. Conformational landscape of cytochrome c folding studied by microsecond-resolved small-angle x-ray scattering. Proc. Natl. Acad. Sci. USA 2002, 99, 1329–1334. [Google Scholar] [CrossRef]
- Levitt, M. Nature of the protein universe. Proc. Natl. Acad. Sci. USA 2009, 106, 11079–11084. [Google Scholar] [CrossRef]
- Han, J.H.; Batey, S.; Nickson, A.A.; Teichmann, S.A.; Clarke, J. The folding and evolution of multidomain proteins. Nat. Rev. Mol. Cell Biol. 2007, 8, 319–330. [Google Scholar] [CrossRef]
- Zimmerman, S.B.; Trach, S.O. Estimation of macromolecule concentrations and excluded volume effects for the cytoplasm of Escherichia coli. J. Mol. Biol. 1991, 222, 599–620. [Google Scholar] [CrossRef]
- Saio, T.; Kawagoe, S.; Ishimori, K.; Kalodimos, C.G. Oligomerization of a molecular chaperone modulates its activity. Elife 2018, 7, e35731. [Google Scholar] [CrossRef] [PubMed]
- Perales-Calvo, J.; Giganti, D.; Stirnemann, G.; Garcia-Manyes, S. The force-dependent mechanism of DnaK-mediated mechanical folding. Sci. Adv. 2018, 4, eaaq0243. [Google Scholar] [CrossRef]
- Stull, F.; Koldewey, P.; Humes, J.R.; Radford, S.E.; Bardwell, J.C.A. Substrate protein folds while it is bound to the ATP-independent chaperone Spy. Nat. Struct. Mol. Biol. 2016, 23, 53–58. [Google Scholar] [CrossRef]
- Huang, C.; Rossi, P.; Saio, T.; Kalodimos, C.G. Structural basis for the antifolding activity of a molecular chaperone. Nature 2016, 537, 202–206. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Rossi, P.; Kalodimos, C.G. Structural basis for client recognition and activity of Hsp40 chaperones. Science 2019, 365, 1313–1319. [Google Scholar] [CrossRef] [PubMed]
- Agashe, V.R.; Guha, S.; Chang, H.-C.; Genevaux, P.; Hayer-Hartl, M.; Stemp, M.; Georgopoulos, C.; Hartl, F.U.; Barral, J.M. Function of Trigger Factor and DnaK in Multidomain Protein Folding. Cell 2004, 117, 199–209. [Google Scholar] [CrossRef]
- Ye, X.; Mayne, L.; Kan, Z.Y.; Englander, S.W. Folding of maltose binding protein outside of and in GroEL. Proc. Natl. Acad. Sci. USA 2018, 115, 519–524. [Google Scholar] [CrossRef]
- Baker, T.A.; Sauer, R.T. ClpXP, an ATP-powered unfolding and protein-degradation machine. Biochim. Biophys. Acta Mol. Cell Res. 2012, 1823, 15–28. [Google Scholar] [CrossRef]
- Rizzolo, K.; Yu, A.Y.H.; Ologbenla, A.; Kim, S.R.; Zhu, H.; Ishimori, K.; Thibault, G.; Leung, E.; Zhang, Y.W.; Teng, M.; et al. Functional cooperativity between the trigger factor chaperone and the ClpXP proteolytic complex. Nat. Commun. 2021, 12, 1–18. [Google Scholar]
- Inanami, T.; Terada, T.P.; Sasai, M. Folding pathway of a multidomain protein depends on its topology of domain connectivity. Proc. Natl. Acad. Sci. USA 2014, 111, 15969–15974. [Google Scholar] [CrossRef] [PubMed]
- Rief, M.; Pascual, J.; Saraste, M.; Gaub, H.E. Single molecule force spectroscopy of spectrin repeats: Low unfolding forces in helix bundles. J. Mol. Biol. 1999, 286, 553–561. [Google Scholar] [CrossRef] [PubMed]
- Bechtluft, P.; van Leeuwen, R.G.H.; Tyreman, M.; Tomkiewicz, D.; Nouwen, N.; Tepper, H.L.; Driessen, A.J.M.; Tans, S.J. Direct observation of chaperone-induced changes in a protein folding pathway. Science 2007, 318, 1458–1461. [Google Scholar] [CrossRef] [PubMed]
- Peng, Q.; Li, H. Atomic force microscopy reveals parallel mechanical unfolding pathways of T4 lysozyme: Evidence for a kinetic partitioning mechanism. Proc. Natl. Acad. Sci. USA 2008, 105, 1885–1890. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Maciuba, K.; Kaiser, C.M. The Ribosome Cooperates with a Chaperone to Guide Multi-domain Protein Folding. Mol. Cell 2019, 74, 310–319.e7. [Google Scholar] [CrossRef] [PubMed]
- Bertz, M.; Rief, M. Mechanical Unfoldons as Building Blocks of Maltose-binding Protein. J. Mol. Biol. 2008, 378, 447–458. [Google Scholar] [CrossRef]
- Mashaghi, A.; Kramer, G.; Bechtluft, P.; Zachmann-Brand, B.; Driessen, A.J.M.; Bukau, B.; Tans, S.J. Reshaping of the conformational search of a protein by the chaperone trigger factor. Nature 2013, 500, 98–101. [Google Scholar] [CrossRef]
- Shilton, B.H.; Shuman, H.A.; Mowbray, S.L. Crystal Structures and Solution Conformations of a Dominant-negative Mutant of Escherichia coli Maltose-binding Protein. J. Mol. Biol. 1996, 264, 364–376. [Google Scholar] [CrossRef]
- Clementi, C.; Jennings, P.A.; Onuchic, J.N. How native-state topology affects the folding of dihydrofolate reductase and interleukin-1β. Proc. Natl. Acad. Sci. USA 2000, 97, 5871–5876. [Google Scholar] [CrossRef]
- Itoh, K.; Sasai, M. Cooperativity, connectivity, and folding pathways of multidomain proteins. Proc. Natl. Acad. Sci. USA 2008, 105, 13865–13870. [Google Scholar] [CrossRef]
- Krishna, M.M.G.; Hoang, L.; Lin, Y.; Englander, S.W. Hydrogen exchange methods to study protein folding. Methods 2004, 34, 51–64. [Google Scholar] [CrossRef] [PubMed]
- Mayne, L. Hydrogen Exchange Mass Spectrometry. Methods Enzymol. 2016, 566, 335–356. [Google Scholar] [PubMed]
- Englander, S.W.; Mayne, L.; Baldwin, R.L.; Goldenberg, D.P.; Woodside, M.T. The case for defined protein folding pathways. Proc. Natl. Acad. Sci. USA 2017, 114, 8253–8258. [Google Scholar] [CrossRef] [PubMed]
- Apetri, A.C.; Horwich, A.L. Chaperonin chamber accelerates protein folding through passive action of preventing aggregation. Proc. Natl. Acad. Sci. USA 2008, 105, 17351–17355. [Google Scholar] [CrossRef]
- Georgescauld, F.; Popova, K.; Gupta, A.J.; Bracher, A.; Engen, J.R.; Hayer-Hartl, M.; Hartl, F.U. GroEL/ES Chaperonin Modulates the Mechanism and Accelerates the Rate of TIM-Barrel Domain Folding. Cell 2014, 157, 922–934. [Google Scholar] [CrossRef]
- Fujiwara, K.; Ishihama, Y.; Nakahigashi, K.; Soga, T.; Taguchi, H. A systematic survey of in vivo obligate chaperonin-dependent substrates. EMBO J. 2010, 29, 1552–1564. [Google Scholar] [CrossRef]
- Kerner, M.J.; Naylor, D.J.; Ishihama, Y.; Maier, T.; Chang, H.C.; Stines, A.P.; Georgopoulos, C.; Frishman, D.; Hayer-Hartl, M.; Mann, M.; et al. Proteome-wide analysis of chaperonin-dependent protein folding in Escherichia coli. Cell 2005, 122, 209–220. [Google Scholar] [CrossRef]
- Martinez-Hackert, E.; Hendrickson, W.A. Promiscuous substrate recognition in folding and assembly activities of the trigger factor chaperone. Cell 2009, 138, 923–934. [Google Scholar] [CrossRef]
- Braig, K.; Otwinowski, Z.; Hegde, R.; Boisvert, D.C.; Joachimiak, A.; Horwich, A.L.; Sigler, P.B. The crystal structure of the bacterial chaperonln GroEL at 2.8 Å. Nature 1994, 371, 578–586. [Google Scholar] [CrossRef]
- Knowlton, J.J.; Gestaut, D.; Ma, B.; Taylor, G.; Seven, A.B.; Leitner, A.; Wilsonk, G.J.; Shanker, S.; Yates, N.A.; Prasad, B.V.V.; et al. Structural and functional dissection of reovirus capsid folding and assembly by the prefoldin-TRiC/CCT chaperone network. Proc. Natl. Acad. Sci. USA 2021, 118, e2018127118. [Google Scholar] [CrossRef]
- Ali, M.M.U.; Mark Roe, S.; Vaughan, C.K.; Meyer, P.; Panaretou, B.; Piper, P.W.; Prodromou, C.; Pearl, L.H. Crystal structure of an Hsp90–nucleotide–p23/Sba1 closed chaperone complex. Nature 2006, 440, 1013–1017. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Qian, X.; Sha, B. The Crystal Structure of the Yeast Hsp40 Ydj1 Complexed with Its Peptide Substrate. Structure 2003, 11, 1475–1483. [Google Scholar] [CrossRef] [PubMed]
- Dekker, C.; de Kruijff, B.; Gros, P. Crystal structure of SecB from Escherichia coli. J. Struct. Biol. 2003, 144, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Ferbitz, L.; Maier, T.; Patzelt, H.; Bukau, B.; Deuerling, E.; Ban, N. Trigger factor in complex with the ribosome forms a molecular cradle for nascent proteins. Nature 2004, 431, 590–596. [Google Scholar] [CrossRef] [PubMed]
- Reissmann, S.; Joachimiak, L.A.; Chen, B.; Meyer, A.S.; Nguyen, A.; Frydman, J. A Gradient of ATP Affinities Generates an Asymmetric Power Stroke Driving the Chaperonin TRIC/CCT Folding Cycle. Cell Rep. 2012, 2, 866–877. [Google Scholar] [CrossRef]
- Ranson, N.A.; Clare, D.K.; Farr, G.W.; Houldershaw, D.; Horwich, A.L.; Saibil, H.R. Allosteric signaling of ATP hydrolysis in GroEL–GroES complexes. Nat. Struct. Mol. Biol. 2006, 13, 147–152. [Google Scholar] [CrossRef]
- Leitner, A.; Joachimiak, L.A.; Bracher, A.; Mönkemeyer, L.; Walzthoeni, T.; Chen, B.; Pechmann, S.; Holmes, S.; Cong, Y.; Ma, B.; et al. The Molecular Architecture of the Eukaryotic Chaperonin TRiC/CCT. Structure 2012, 20, 814–825. [Google Scholar] [CrossRef]
- Joachimiak, L.A.; Walzthoeni, T.; Liu, C.W.; Aebersold, R.; Frydman, J. The Structural Basis of Substrate Recognition by the Eukaryotic Chaperonin TRiC/CCT. Cell 2014, 159, 1042–1055. [Google Scholar] [CrossRef]
- Shiau, A.K.; Harris, S.F.; Southworth, D.R.; Agard, D.A. Structural Analysis of E. coli hsp90 Reveals Dramatic Nucleotide-Dependent Conformational Rearrangements. Cell 2006, 127, 329–340. [Google Scholar] [CrossRef]
- Mickler, M.; Hessling, M.; Ratzke, C.; Buchner, J.; Hugel, T. The large conformational changes of Hsp90 are only weakly coupled to ATP hydrolysis. Nat. Struct. Mol. Biol. 2009, 16, 281–286. [Google Scholar] [CrossRef]
- Zhuravleva, A.; Clerico, E.M.; Gierasch, L.M. An Interdomain Energetic Tug-of-War Creates the Allosterically Active State in Hsp70 Molecular Chaperones. Cell 2012, 151, 1296–1307. [Google Scholar] [CrossRef] [PubMed]
- Lai, A.L.; Clerico, E.M.; Blackburn, M.E.; Patel, N.A.; Robinson, C.V.; Borbat, P.P.; Freed, J.H.; Gierasch, L.M. Key features of an Hsp70 chaperone allosteric landscape revealed by ion-mobility native mass spectrometry and double electron-electron resonance. J. Biol. Chem. 2017, 292, 8773–8785. [Google Scholar] [CrossRef] [PubMed]
- Saio, T.; Guan, X.; Rossi, P.; Economou, A.; Kalodimos, C.G. Structural basis for protein antiaggregation activity of the trigger factor chaperone. Science 2014, 344, 1250494. [Google Scholar] [CrossRef] [PubMed]
- Kawagoe, S.; Nakagawa, H.; Kumeta, H.; Ishimori, K.; Saio, T. Structural insight into proline cis/trans isomerization of unfolded proteins catalyzed by the trigger factor chaperone. J. Biol. Chem. 2018, 293, 15095–15106. [Google Scholar] [CrossRef]
- Sekhar, A.; Rosenzweig, R.; Bouvignies, G.; Kay, L.E. Mapping the conformation of a client protein through the Hsp70 functional cycle. Proc. Natl. Acad. Sci. USA 2015, 112, 10395–10400. [Google Scholar] [CrossRef]
- Calloni, G.; Chen, T.; Schermann, S.M.; Chang, H.-C.; Genevaux, P.; Agostini, F.; Tartaglia, G.G.; Hayer-Hartl, M.; Hartl, F.U. DnaK functions as a central hub in the E. coli chaperone network. Cell Rep. 2012, 1, 251–264. [Google Scholar] [CrossRef]
- Clerico, E.M.; Pozhidaeva, A.K.; Jansen, R.M.; Özden, C.; Tilitsky, J.M.; Gierasch, L.M. Selective promiscuity in the binding of E. coli Hsp70 to an unfolded protein. Proc. Natl. Acad. Sci. USA 2021, 118, e2016962118. [Google Scholar] [CrossRef]
- Meng, W.; Clerico, E.M.; McArthur, N.; Gierasch, L.M. Allosteric landscapes of eukaryotic cytoplasmic Hsp70s are shaped by evolutionary tuning of key interfaces. Proc. Natl. Acad. Sci. USA 2018, 115, 11970–11975. [Google Scholar] [CrossRef]
- Rosenzweig, R.; Sekhar, A.; Nagesh, J.; Kay, L.E. Promiscuous binding by Hsp70 results in conformational heterogeneity and fuzzy chaperone-substrate ensembles. Elife 2017, 6, e28030. [Google Scholar] [CrossRef]
- He, L.; Sharpe, T.; Mazur, A.; Hiller, S. A molecular mechanism of chaperone-client recognition. Sci. Adv. 2016, 2, e1601625. [Google Scholar] [CrossRef]
- Horowitz, S.; Salmon, L.; Koldewey, P.; Ahlstrom, L.S.; Martin, R.; Quan, S.; Afonine, P.V.; van den Bedem, H.; Wang, L.; Xu, Q.; et al. Visualizing chaperone-assisted protein folding. Nat. Struct. Mol. Biol. 2016, 23, 691–697. [Google Scholar] [CrossRef]
- Wu, K.; Stull, F.; Lee, C.; Bardwell, J.C.A. Protein folding while chaperone bound is dependent on weak interactions. Nat. Commun. 2019, 10, 1–10. [Google Scholar] [CrossRef]
- Mitra, R.; Gadkari, V.V.; Meinen, B.A.; van Mierlo, C.P.M.; Ruotolo, B.T.; Bardwell, J.C.A. Mechanism of the small ATP-independent chaperone Spy is substrate specific. Nat. Commun. 2021, 12, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Hardy, S.; Randall, L. A kinetic partitioning model of selective binding of nonnative proteins by the bacterial chaperone SecB. Science 1991, 251, 439–443. [Google Scholar] [CrossRef] [PubMed]
- Chiti, F.; Taddei, N.; Baroni, F.; Capanni, C.; Stefani, M.; Ramponi, G.; Dobson, C.M. Kinetic partitioning of protein folding and aggregation. Nat. Struct. Biol. 2002, 9, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Matsusaki, M.; Sugawara, T.; Ishimori, K.; Saio, T. Zinc-dependent oligomerization of thermus thermophilus trigger factor chaperone. Biology 2021, 10, 1106. [Google Scholar] [CrossRef]
- Imamoglu, R.; Balchin, D.; Hayer-Hartl, M.; Hartl, F.U. Bacterial Hsp70 resolves misfolded states and accelerates productive folding of a multi-domain protein. Nat. Commun. 2020, 11, 1–13. [Google Scholar] [CrossRef]
- Hoffmann, A.; Bukau, B.; Kramer, G. Structure and function of the molecular chaperone Trigger Factor. Biochim. Biophys. Acta 2010, 1803, 650–661. [Google Scholar] [CrossRef]
- Jakob, R.P.; Koch, J.R.; Burmann, B.M.; Schmidpeter, P.A.M.; Hunkeler, M.; Hiller, S.; Schmid, F.X.; Maier, T. Dimeric Structure of the Bacterial Extracellular Foldase PrsA. J. Biol. Chem. 2015, 290, 3278–3292. [Google Scholar] [CrossRef]
- Xu, X.; Wang, S.; Hu, Y.X.; McKay, D.B. The Periplasmic Bacterial Molecular Chaperone SurA Adapts its Structure to Bind Peptides in Different Conformations to Assert a Sequence Preference for Aromatic Residues. J. Mol. Biol. 2007, 373, 367–381. [Google Scholar] [CrossRef][Green Version]
- Okumura, M.; Noi, K.; Kanemura, S.; Kinoshita, M.; Saio, T.; Inoue, Y.; Hikima, T.; Akiyama, S.; Ogura, T.; Inaba, K. Dynamic assembly of protein disulfide isomerase in catalysis of oxidative folding. Nat. Chem. Biol. 2019, 15, 499–509. [Google Scholar] [CrossRef]
- Oroz, J.; Chang, B.J.; Wysoczanski, P.; Lee, C.T.; Pérez-Lara, Á.; Chakraborty, P.; Hofele, R.V.; Baker, J.D.; Blair, L.J.; Biernat, J.; et al. Structure and pro-toxic mechanism of the human Hsp90/PPIase/Tau complex. Nat. Commun. 2018, 9, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Guettouche, T.; Fenna, M.; Boellmann, F.; Pratt, W.B.; Toft, D.O.; Smith, D.F.; Voellmy, R. Evidence for a Mechanism of Repression of Heat Shock Factor 1 Transcriptional Activity by a Multichaperone Complex. J. Biol. Chem. 2001, 276, 45791–45799. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Thwin, A.C.; Nadel, C.M.; Tse, E.; Gates, S.N.; Gestwicki, J.E.; Southworth, D.R. The structure of an Hsp90-immunophilin complex reveals cochaperone recognition of the client maturation state. Mol. Cell 2021, 81, 3496–3508.e5. [Google Scholar] [CrossRef]
- Brown, N.R.; Noble, M.E.M.; Endicott, J.A.; Johnson, L.N. The structural basis for specificity of substrate and recruitment peptides for cyclin-dependent kinases. Nat. Cell Biol. 1999, 1, 438–443. [Google Scholar] [CrossRef] [PubMed]
- Weiwad, M.; Küllertz, G.; Schutkowski, M.; Fischer, G. Evidence that the substrate backbone conformation is critical to phosphorylation by p42 MAP kinase. FEBS Lett. 2000, 478, 39–42. [Google Scholar] [CrossRef]
- Zhou, X.Z.; Kops, O.; Werner, A.; Lu, P.J.; Shen, M.; Stoller, G.; Küllertz, G.; Stark, M.; Fischer, G.; Lu, K.P. Pin1-Dependent Prolyl Isomerization Regulates Dephosphorylation of Cdc25C and Tau Proteins. Mol. Cell 2000, 6, 873–883. [Google Scholar] [CrossRef]
- Piotukh, K.; Gu, W.; Kofler, M.; Labudde, D.; Helms, V.; Freund, C. Cyclophilin A binds to linear peptide motifs containing a consensus that is present in many human proteins. J. Biol. Chem. 2005, 280, 23668–23674. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kawagoe, S.; Ishimori, K.; Saio, T. Structural and Kinetic Views of Molecular Chaperones in Multidomain Protein Folding. Int. J. Mol. Sci. 2022, 23, 2485. https://doi.org/10.3390/ijms23052485
Kawagoe S, Ishimori K, Saio T. Structural and Kinetic Views of Molecular Chaperones in Multidomain Protein Folding. International Journal of Molecular Sciences. 2022; 23(5):2485. https://doi.org/10.3390/ijms23052485
Chicago/Turabian StyleKawagoe, Soichiro, Koichiro Ishimori, and Tomohide Saio. 2022. "Structural and Kinetic Views of Molecular Chaperones in Multidomain Protein Folding" International Journal of Molecular Sciences 23, no. 5: 2485. https://doi.org/10.3390/ijms23052485
APA StyleKawagoe, S., Ishimori, K., & Saio, T. (2022). Structural and Kinetic Views of Molecular Chaperones in Multidomain Protein Folding. International Journal of Molecular Sciences, 23(5), 2485. https://doi.org/10.3390/ijms23052485





