Plant Peroxisome-Targeting Effector MoPtep1 Is Required for the Virulence of Magnaporthe oryzae
Abstract
:1. Introduction
2. Results
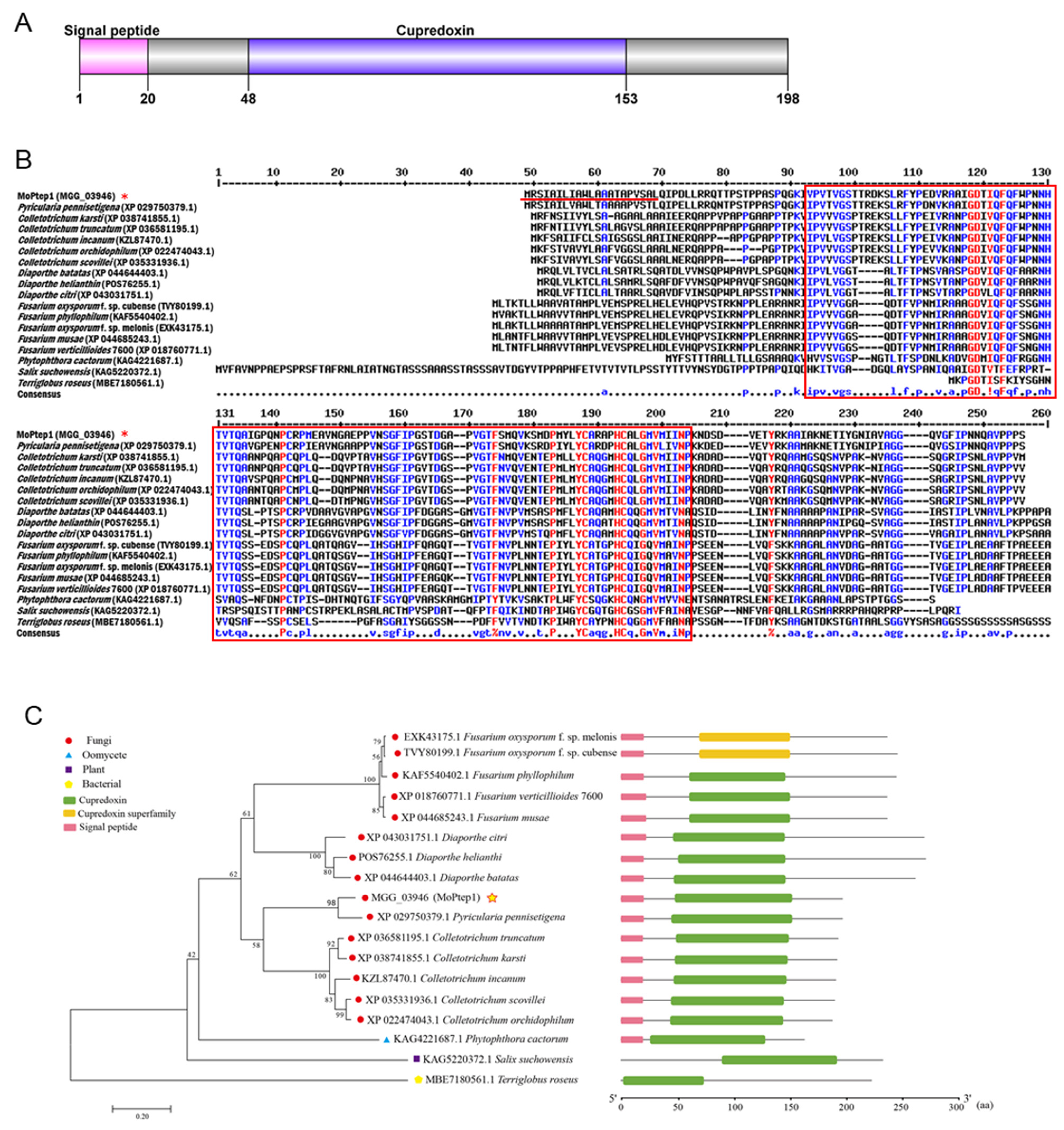
2.1. Amino Acid Sequence Alignment and Phylogenetic Analysis of MoPtep1 in Different Species
2.2. Expression of the MoPtep1 Gene Was Induced during Infection
2.3. MoPtep1 Is a Secreted Protein in M. oryzae
2.4. MoPtep1 Is Important for Pathogenicity, but Not Growth
2.5. Sensitivity Analysis of Vegetative Growth of Δmoptep1 Mutant Strains under Multiple Stresses
2.6. Subcellular Localization of MoPtep1 in Plants
2.7. MoPtep1 Suppressed Programmed Cell Death in N. benthamiana
2.8. Screening and Identification of MoPtep1-Interacting Proteins
3. Discussion
4. Materials and Methods
4.1. Fungal Strains and Plant Growth Conditions
4.2. Bioinformatic and Phylogenetic Analysis of MoPtep1 Homologous Proteins
4.3. Secretion Characterization of MoPtep1
4.4. Subcellular Localization and Cell Death Inhibition Assay
4.5. Construction of MoPtep1 ko Mutant and Complemented Strains
4.6. Pathogenicity Analysis
4.7. MoPtep1 Gene Expression Analysis
4.8. Response of the Δmoptep1 Mutants under Different Stress Conditions
4.9. Yeast Two-Hybrid Assay
4.10. Statistical Assays
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Peng, Y.; Wersch, R.V.; Zhang, Y. Convergent and divergent signaling in PAMP-triggered immunity and Effector-triggered immunity. Mol. Plant Microbe Interact. 2017, 31, 403–409. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Kapos, P.; Zhang, Y. NLRs in plants. Curr. Opin. Immunol. 2015, 32, 114–121. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.; Vance, R.E.; Dangl, J.L. Intracellular innate immune surveillance devices in plants and animals. Science 2016, 354, 6316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.; Cui, D.; Zhao, J.; Liu, N.; Wang, B.; Liu, J.; Xu, E.; Hu, Z.; Ren, D.; Tang, D.; et al. Two Arabidopsis Receptor-like Cytoplasmic Kinases SZE1 and SZE2 Associate with the ZAR1-ZED1 Complex and Are Required for Effector-Triggered Immunity. Mol. Plant 2019, 12, 967–983. [Google Scholar] [CrossRef] [PubMed]
- Naveed, Z.A.; Bibi, S.; Ali, G.S. The Phytophthora RXLR Effector Avrblb2 Modulates Plant Immunity by Interfering With Ca2+ Signaling Pathway. Front. Plant Sci. 2019, 10, 374. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Shang, B.; Dou, M.; Liu, R.; Chen, T.; Xiang, G.; Li, Y.; Liu, G.; Xu, Y. The Nuclear-Localized RxLR Effector PvAvh74 From Plasmopara viticola Induces Cell Death and Immunity Responses in Nicotiana benthamiana. Front. Microbiol. 2019, 10, 1531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.; Kim, C.Y.; Park, S.Y.; Kim, K.T.; Jeon, J.; Chung, H.; Choi, G.; Kwon, S.; Choi, J.; Jeon, J.; et al. Two nuclear effectors of the rice blast fungus modulate host immunity via transcriptional reprogramming. Nat. Commun. 2020, 11, 5845. [Google Scholar] [CrossRef]
- Xu, G.J.; Zhong, X.H.; Shi, Y.L.; Liu, Z.; Wang, X.L. A fungal effector targets a heat shock-dynamin protein complex to modulate mitochondrial dynamics and reduce plant immunity. Sci. Adv. 2020, 6, 48. [Google Scholar] [CrossRef]
- Kao, Y.T.; Gonzalez, K.L.; Bartel, B. Peroxisome Function, Biogenesis, and Dynamics in Plants. Plant Physiol. 2018, 176, 162–177. [Google Scholar] [CrossRef] [Green Version]
- Cook, K.C.; Moreno, J.A.; Beltran, P.; Cristea, I.M. Peroxisome Plasticity at the Virus–Host Interface. Trends Microbiol. 2019, 27, 906–914. [Google Scholar] [CrossRef]
- Hu, J.; Baker, A.; Bartel, B.; Linka, N.; Mullen, R.T.; Reumann, S.; Zolman, B.K. Plant peroxisomes: Biogenesis and function. Plant Cell 2012, 24, 2279–2303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.; Schilmiller, A.L.; Liu, G.; Lee, G.I.; Jayanty, S.; Sageman, C.; Vrebalov, J.; Giovannoni, J.J.; Yagi, K.; Kobayashi, Y.; et al. Role of beta-oxidation in jasmonate biosynthesis and systemic wound signaling in tomato. Plant Cell 2005, 17, 971–986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mhamdi, A.; Noctor, G.; Baker, A. Plant catalases: Peroxisomal redox guardians. Arch. Biochem. Biophys. 2012, 525, 181–194. [Google Scholar] [CrossRef] [PubMed]
- Siamer, S.; Gaubert, S.; Boureau, T.; Brisset, M.N.; Barny, M.A. Mutational analysis of a predicted double β-propeller domain of the DspA/E effector of Erwinia amylovora. FEMS Microbiol. Lett. 2013, 342, 54–61. [Google Scholar] [CrossRef]
- Robin, G.P.; Jochen, K.; Ulla, N.; Lisa, C.; Jean-Félix, D.; Nicolas, L.; O’Connell, R.J. Subcellular Localization Screening of Colletotrichum higginsianum Effector Candidates Identifies Fungal Proteins Targeted to Plant Peroxisomes, Golgi Bodies, and Microtubules. Front. Plant Sci. 2018, 9, 562. [Google Scholar] [CrossRef] [Green Version]
- Reumann, S.; Bartel, B. Plant peroxisomes: Recent discoveries in functional complexity, organelle homeostasis, and morphological dynamics. Curr. Opin. Plant Biol. 2016, 34, 17–26. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.B.; Songkumarn, P.; Venu, R.C.; Gowda, M.; Bellizzi, M.; Hu, J.N.; Liu, W.D.; Ebbole, D.; Meyers, B.; Mitchell, T.; et al. Identification and Characterization of In planta-Expressed Secreted Effector Proteins from Magnaporthe oryzae That Induce Cell Death in Rice. Mol. Plant Microbe Interact. 2013, 26, 191–202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dai, M.D.; Wu, M.; Li, Y.; Su, Z.Z.; Lin, F.C.; Liu, X.H. The chitin deacetylase PoCda7 is involved in the pathogenicity of Pyricularia oryzae. Microbiol. Res. 2021, 248, 126749. [Google Scholar] [CrossRef]
- Guo, X.; Zhong, D.; Xie, W.; He, Y.; Zheng, Y.; Lin, Y.; Chen, Z.; Han, Y.; Tian, D.; Liu, W.; et al. Functional Identification of Novel Cell Death-inducing Effector Proteins from Magnaporthe oryzae. Rice 2019, 12, 59. [Google Scholar] [CrossRef] [PubMed]
- Hogenhout, S.A.; Van der Hoorn, R.A.; Terauchi, R.; Kamoun, S. Emerging concepts in effector biology of plant-associated organisms. Mol. Plant Microbe Interact. 2009, 22, 115–122. [Google Scholar] [CrossRef] [Green Version]
- Tian, L.; Huang, C.M.; Zhang, D.D.; Ran, L.I.; Chen, J.Y.; Sun, W.X.; Qiu, N.W.; Dai, X.F. Extracellular superoxide dismutase VdSOD5 is required for virulence in Verticillium dahliae. J. Integr. Agric. 2021, 20, 1858–1870. [Google Scholar] [CrossRef]
- Li, Q.; Ai, G.; Shen, D.Y.; Zou, F.; Wang, J.; Bai, T.; Chen, Y.Y.; Li, S.T.; Zhang, M.X.; Jing, M.F.; et al. A Phytophthora capsici Effector Targets ACD11 Binding Partners that Regulate ROS-Mediated Defense Response in Arabidopsis. Mol. Plant 2019, 12, 565–581. [Google Scholar] [CrossRef] [Green Version]
- Dixit, E.; Boulant, S.; Zhang, Y.; Lee, A.S.; Odendall, C.; Shum, B.; Hacohen, N.; Chen, Z.J.; Whelan, S.P.; Fransen, M.; et al. Peroxisomes are signaling platforms for antiviral innate immunity. Cell 2010, 141, 668–681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peter, T.; Sophie, M.; Peter, J.C.; Vivian, C.B.; Mirela, C.C.; Sebastian, E.A.; Elena, G.; Catherine, J.L.; Geert, S.; Adam, J.R.; et al. Genomic characterisation of the effector complement of the potato cyst nematode Globodera pallida. BMC Genomics 2014, 15, 923. [Google Scholar] [CrossRef] [Green Version]
- Pan, R.H.; Liu, J.; Wang, S.S.; Hu, J.P. Peroxisomes: Versatile organelles with diverse roles in plants. New Phytol. 2020, 225, 1410–1427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, Z.; Gang, W.; Deng, W.; Pu, Y.; Wei, C.; Yi, L. Interaction of rice dwarf virus outer capsid P8 protein with rice glycolate oxidase mediates relocalization of P8. FEBS Lett. 2007, 581, 34–40. [Google Scholar] [CrossRef] [Green Version]
- Yang, M.; Li, Z.; Zhang, K.; Zhang, X.; Zhang, Y.; Wang, X.; Han, C.; Yu, J.; Xu, K.; Li, D. Barley Stripe Mosaic Virus γb Interacts with Glycolate Oxidase and Inhibits Peroxisomal ROS Production to Facilitate Virus Infection. Mol. Plant 2017, 11, 338–341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hasegawa, M.; Mitsuhara, I.; Seo, S.; Imai, T.; Koga, J.; Okada, K.; Yamane, H.; Ohashi, Y. Phytoalexin accumulation in the interaction between rice and the blast fungus. Mol. Plant. Microbe Interact. 2010, 23, 1000–1011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grayer, R.J.; Kokubun, T. Plant–fungal interactions: The search for phytoalexins and other antifungal compounds from higher plants. Phytochemistry 2001, 56, 253–263. [Google Scholar] [CrossRef]
- Cui, Z.; Liang, F.; Zhang, J.; Wang, F.; Liu, D.; Wang, H. Transgenic expression of TaTLP1, a thaumatin-like protein gene, reduces susceptibility to common root rot and leaf rust in wheat. Crop J. 2021, 9, 1214–1218. [Google Scholar] [CrossRef]
- Xu, Q.; Tang, C.L.; Wang, X.D.; Sun, S.T.; Wang, X.R. An effector protein of the wheat stripe rust fungus targets chloroplasts and suppresses chloroplast function. Nat. Commun. 2019, 10, 5571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goverse, A.; Smant, G. The activation and suppression of plant innate immunity by parasitic nematodes. Annu. Rev. Phytopathol. 2014, 52, 243–265. [Google Scholar] [CrossRef] [PubMed]
- Kud, J.; Wang, W.; Gross, R.; Fan, Y.; Huang, L.; Yuan, Y.; Gray, A.; Duarte, A.; Kuhl, J.C.; Caplan, A. The potato cyst nematode effector RHA1B is a ubiquitin ligase and uses two distinct mechanisms to suppress plant immune signaling. PLoS Pathog. 2019, 15, e1007720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Irieda, H.; Inoue, Y.; Mori, M.; Yamada, K.; Oshikawa, Y.; Saitoh, H.; Uemura, A.; Terauchi, R.; Kitakura, S.; Kosaka, A.; et al. Conserved fungal effector suppresses PAMP-triggered immunity by targeting plant immune kinases. Proc. Natl. Acad. Sci. USA 2019, 116, 496–505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mur, L.A.; Kenton, P.; Lloyd, A.J.; Ougham, H.; Prats, E. The hypersensitive response; the centenary is upon us but how much do we know? J. Exp. Bot. 2008, 59, 501–520. [Google Scholar] [CrossRef] [Green Version]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; Von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef]
- Yin, W.; Wang, Y.; Chen, T.; Lin, Y.; Luo, C. Functional Evaluation of the Signal Peptides of Secreted Proteins. Bio-Protocol 2018, 8, e2839. [Google Scholar] [CrossRef]
- Oh, S.K.; Young, C.; Lee, M.; Oliva, R.; Bozkurt, T.O.; Cano, L.M.; Win, J.; Bos, J.I.B.; Liu, H.Y.; Van Damme, M. In planta expression screens of Phytophthora infestans RXLR effectors reveal diverse phenotypes, including activation of the Solanum bulbocastanum disease resistance protein Rpi-blb2. Plant Cell 2009, 21, 2928–2947. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Zheng, W.; Chen, L.; Li, C.; Liang, T.; Chen, Z.; Xu, H.; Han, Y.; Kong, L.; Zhao, X.; et al. Green Fluorescent Protein- and Discosoma sp. Red Fluorescent Protein-Tagged Organelle Marker Lines for Protein Subcellular Localization in Rice. Front. Plant Sci. 2019, 10, 1421. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Ding, B.; Zhou, X.; Wang, G.L. The Rice Dynamin-Related Protein OsDRP1E Negatively Regulates Programmed Cell Death by Controlling the Release of Cytochrome c from Mitochondria. PLoS Pathog. 2017, 13, e1006157. [Google Scholar] [CrossRef]
- Li, Z.Q.; Wu, L.Y.; Wu, H.; Zhang, X.X.; Mei, J.; Zhou, X.P.; Wang, G.; Liu, W. Arginine methylation is required for remodelling pre-mRNA splicing and induction of autophagy in rice blast fungus. New Phytol. 2020, 225, 413–429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Son, H.; Seo, Y.S.; Min, K.; Park, A.R.; Lee, J.; Jin, J.M.; Lin, Y.; Cao, P.; Hong, S.Y.; Kim, E.K.; et al. A phenome-based functional analysis of transcription factors in the cereal head blight fungus, Fusarium graminearum. PLoS Pathog. 2011, 7, e1002310. [Google Scholar] [CrossRef]
- Shi, D.Y.; Ren, W.C.; Wang, J.; Zhang, J.; Mbadianya, J.I.; Mao, X.W.; Chen, C.J. The transcription factor FgNsf1 regulates fungal development, virulence and stress responses in Fusarium graminearum. J. Integr. Agric. 2021, 20, 2156–2169. [Google Scholar] [CrossRef]
- Khan, I.A.; Wang, Y.; Li, H.J.; Lu, J.P.; Liu, X.H.; Lin, F.C. Disruption and molecular characterization of calpains-related (MoCAPN1, MoCAPN3 and MoCAPN4) genes in Magnaporthe oryzae. Microbiol. Res. 2014, 169, 844–854. [Google Scholar] [CrossRef] [PubMed]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ning, N.; Xie, X.; Yu, H.; Mei, J.; Li, Q.; Zuo, S.; Wu, H.; Liu, W.; Li, Z. Plant Peroxisome-Targeting Effector MoPtep1 Is Required for the Virulence of Magnaporthe oryzae. Int. J. Mol. Sci. 2022, 23, 2515. https://doi.org/10.3390/ijms23052515
Ning N, Xie X, Yu H, Mei J, Li Q, Zuo S, Wu H, Liu W, Li Z. Plant Peroxisome-Targeting Effector MoPtep1 Is Required for the Virulence of Magnaporthe oryzae. International Journal of Molecular Sciences. 2022; 23(5):2515. https://doi.org/10.3390/ijms23052515
Chicago/Turabian StyleNing, Na, Xin Xie, Haiyue Yu, Jie Mei, Qianqian Li, Shimin Zuo, Hanxiang Wu, Wende Liu, and Zhiqiang Li. 2022. "Plant Peroxisome-Targeting Effector MoPtep1 Is Required for the Virulence of Magnaporthe oryzae" International Journal of Molecular Sciences 23, no. 5: 2515. https://doi.org/10.3390/ijms23052515
APA StyleNing, N., Xie, X., Yu, H., Mei, J., Li, Q., Zuo, S., Wu, H., Liu, W., & Li, Z. (2022). Plant Peroxisome-Targeting Effector MoPtep1 Is Required for the Virulence of Magnaporthe oryzae. International Journal of Molecular Sciences, 23(5), 2515. https://doi.org/10.3390/ijms23052515






