Identification of Surface Antigens That Define Human Pluripotent Stem Cell-Derived PRRX1+Limb-Bud-like Mesenchymal Cells
Abstract
:1. Introduction
2. Results
2.1. Effect of Pluripotent Cell Density before LBM-like Induction on PRRX1 Expression
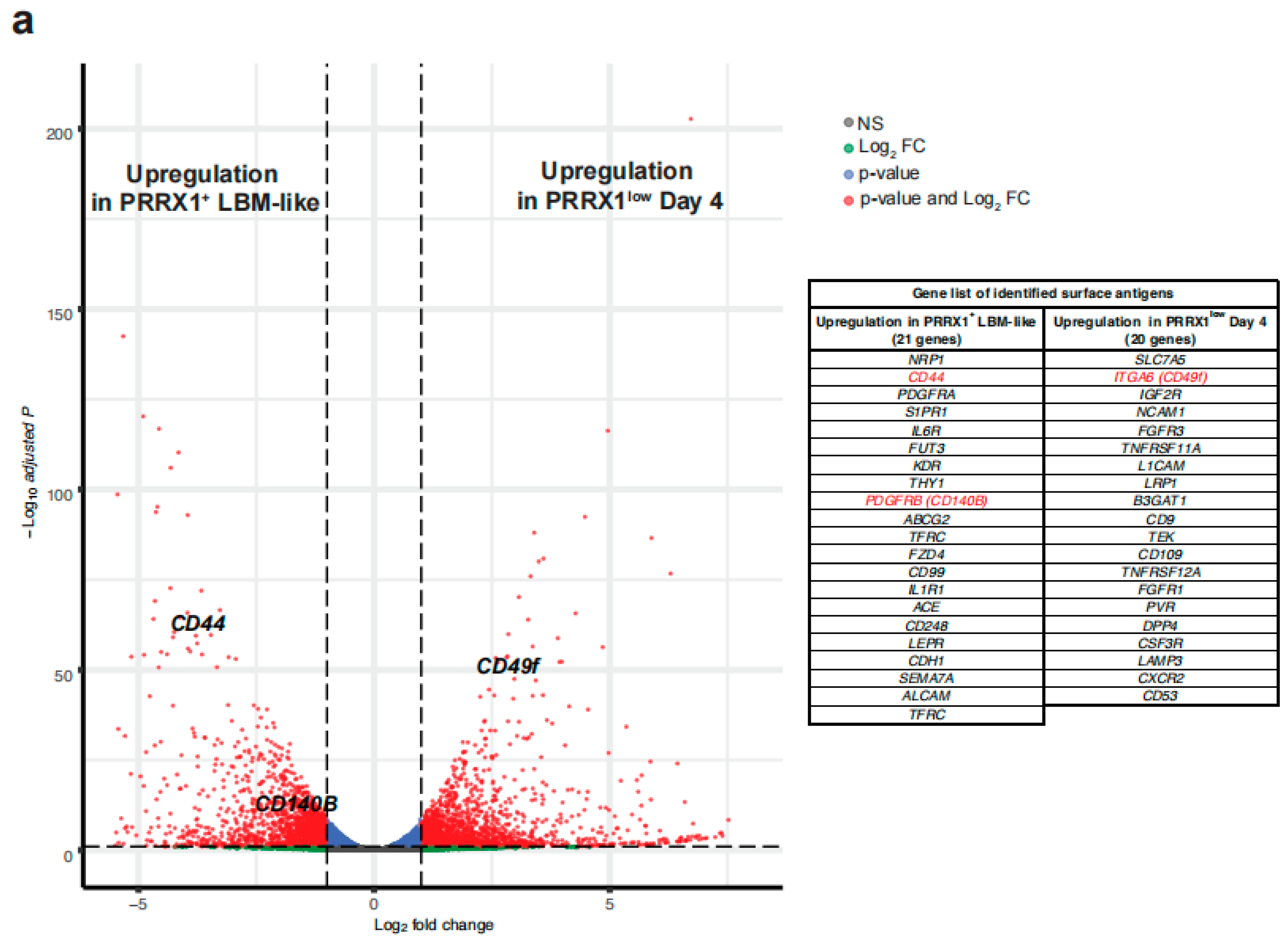
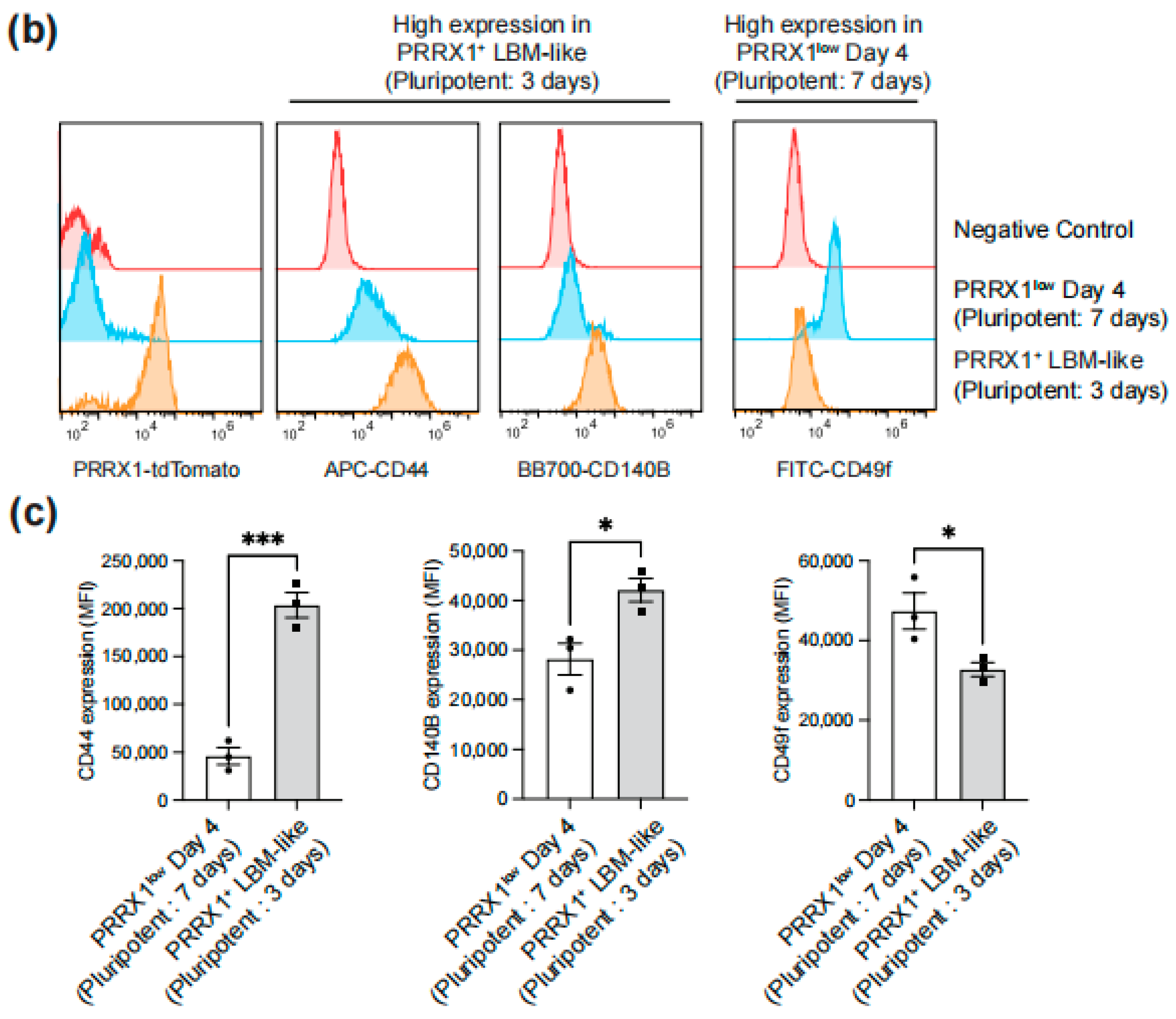
2.2. Identification of Surface Antigens Defining PRRX1+ LBM-like Cells
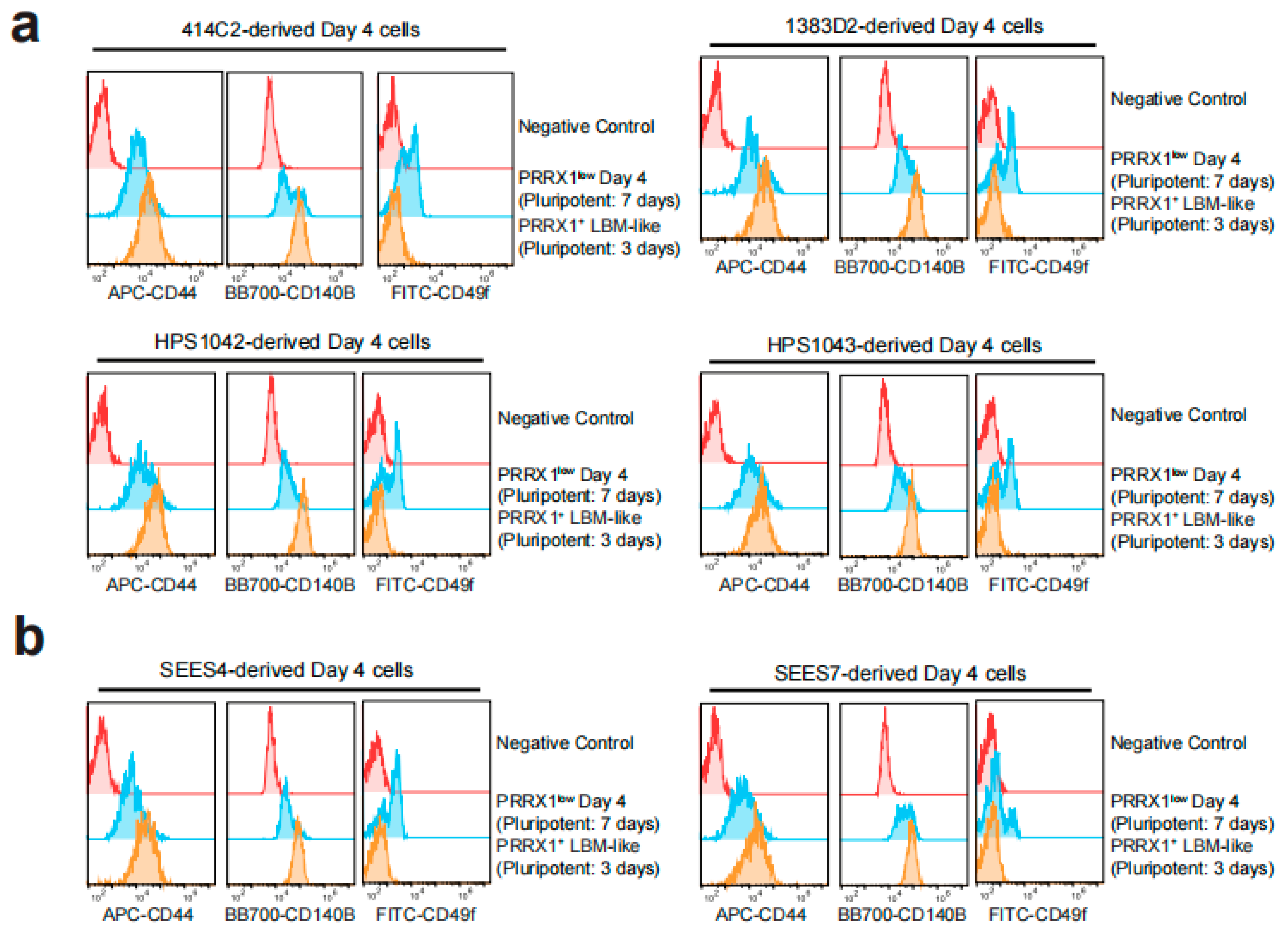
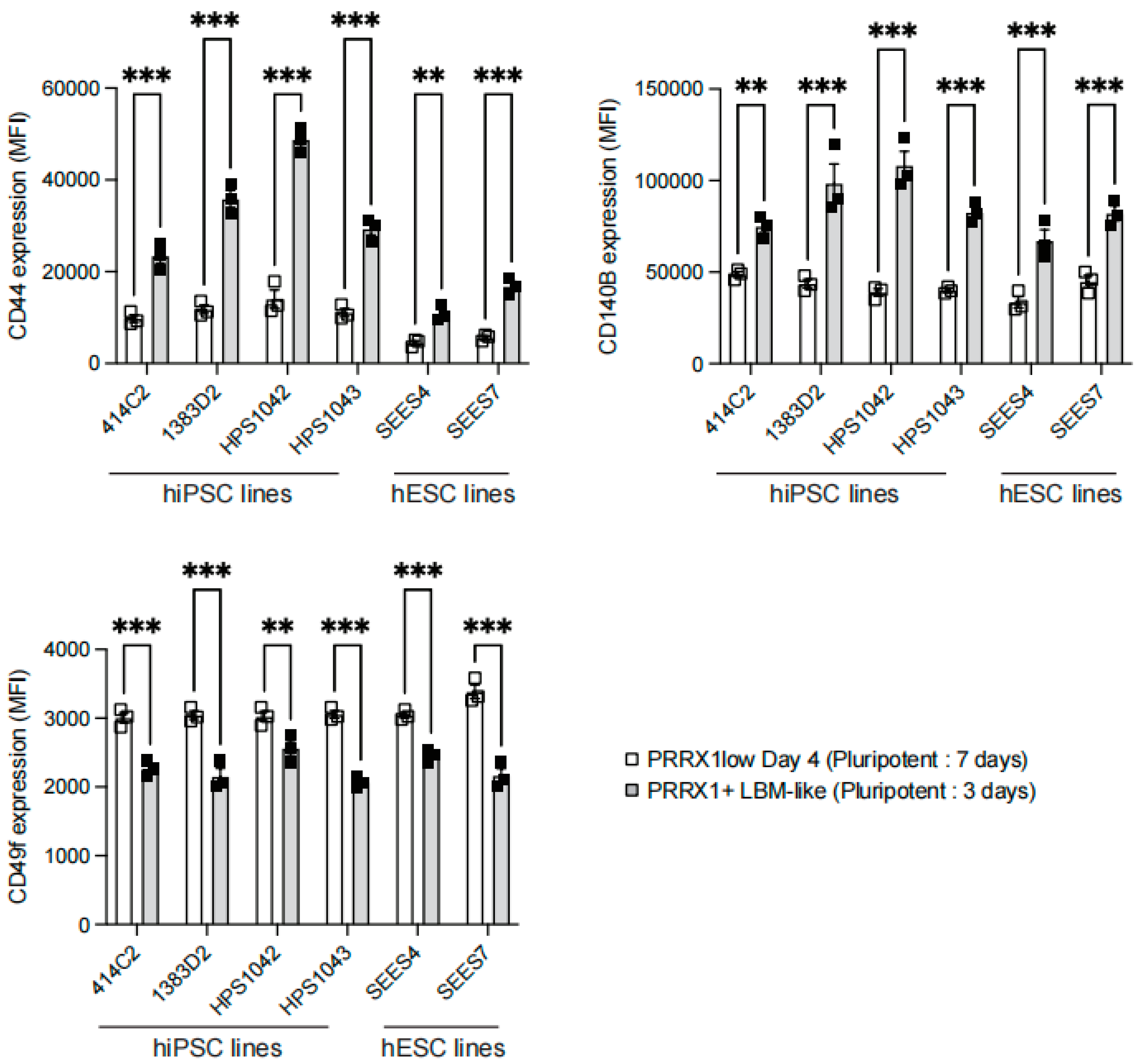
2.3. Detection of CD44high CD140Bhigh CD49f− LBM-like Cells Derived from hiPSC Lines
2.4. Detection of CD44high CD140Bhigh CD49f− LBM-like Cells Derived from hESC Lines
3. Discussion
4. Materials and Methods
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pap, T.; Korb-Pap, A. Cartilage damage in osteoarthritis and rheumatoid arthritis—Two unequal siblings. Nat. Rev. Rheumatol. 2015, 11, 606–615. [Google Scholar] [CrossRef] [PubMed]
- Redondo, M.; Beer, A.; Yanke, A. Cartilage Restoration: Microfracture and Osteochondral Autograft Transplantation. J. Knee Surg. 2018, 31, 231–238. [Google Scholar] [CrossRef] [PubMed]
- Tam, W.L.; Luyten, F.P.; Roberts, S.J. From skeletal development to the creation of pluripotent stem cell-derived bone-forming progenitors. Philos. Trans. R. Soc. B: Biol. Sci. 2018, 373, 20170218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berendsen, A.D.; Olsen, B.R. Bone development. Bone 2015, 80, 14–18. [Google Scholar] [CrossRef] [Green Version]
- Yamanaka, S. Pluripotent Stem Cell-Based Cell Therapy—Promise and Challenges. Cell Stem Cell 2020, 27, 523–531. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, T.; Shibata, M.; Nishio, M.; Nagata, S.; Alev, C.; Sakurai, H.; Toguchida, J.; Ikeya, M. Modeling human somite development and fibrodysplasia ossificans progressiva with induced pluripotent stem cells. Development 2018, 145, dev165431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamashita, A.; Morioka, M.; Yahara, Y.; Okada, M.; Kobayashi, T.; Kuriyama, S.; Matsuda, S.; Tsumaki, N. Generation of Scaffoldless Hyaline Cartilaginous Tissue from Human iPSCs. Stem Cell Rep. 2015, 4, 404–418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Craft, A.M.; Rockel, J.S.; Nartiss, Y.; Kandel, R.A.; Alman, B.A.; Keller, G. Generation of articular chondrocytes from human pluripotent stem cells. Nat. Biotechnol. 2015, 33, 638–645. [Google Scholar] [CrossRef] [PubMed]
- Kawata, M.; Mori, D.; Kanke, K.; Hojo, H.; Ohba, S.; Chung, U.-I.; Yano, F.; Masaki, H.; Otsu, M.; Nakauchi, H.; et al. Simple and Robust Differentiation of Human Pluripotent Stem Cells toward Chondrocytes by Two Small-Molecule Compounds. Stem Cell Rep. 2019, 13, 530–544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Umeda, K.; Oda, H.; Yan, Q.; Matthias, N.; Zhao, J.; Davis, B.R.; Nakayama, N. Long-term expandable SOX9+ chondrogenic ectomesenchymal cells from human pluripotent stem cells. Stem Cell Rep. 2015, 4, 712–726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Humphreys, P.A.; Mancini, F.E.; Ferreira, M.J.; Woods, S.; Ogene, L.; Kimber, S.J. Developmental principles informing human pluripotent stem cell differentiation to cartilage and bone. Semin. Cell Dev. Biol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Prummel, K.D.; Nieuwenhuize, S.; Mosimann, C. The lateral plate mesoderm. Development 2020, 147, dev175059. [Google Scholar] [CrossRef] [PubMed]
- Akiyama, H.; Kim, J.-E.; Nakashima, K.; Balmes, G.; Iwai, N.; Deng, J.M.; Zhang, Z.; Martin, J.F.; Behringer, R.R.; Nakamura, T.; et al. Osteo-chondroprogenitor cells are derived from Sox9 expressing precursors. Proc. Natl. Acad. Sci. 2005, 102, 14665–14670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamada, D.; Nakamura, M.; Takao, T.; Takihira, S.; Yoshida, A.; Kawai, S.; Miura, A.; Ming, L.; Yoshitomi, H.; Gozu, M.; et al. Induction and expansion of human PRRX1+ limb-bud-like mesenchymal cells from pluripotent stem cells. Nat. Biomed. Eng. 2021, 5, 926–940. [Google Scholar] [CrossRef]
- Karamboulas, K.; Dranse, H.J.; Underhill, T.M. Regulation of BMP-dependent chondrogenesis in early limb mesenchyme by TGFbeta signals. J. Cell Sci. 2010, 123, 2068–2076. [Google Scholar] [CrossRef] [Green Version]
- Mori, S.; Sakakura, E.; Tsunekawa, Y.; Hagiwara, M.; Suzuki, T.; Eiraku, M. Self-organized formation of developing appendages from murine pluripotent stem cells. Nat. Commun. 2019, 10, 3802. [Google Scholar] [CrossRef] [Green Version]
- Marín-Llera, J.C.; Chimal-Monroy, J. A small population of resident limb bud mesenchymal cells express few MSC-associated markers, but the expression of these markers is increased immediately after cell culture. Cell Biol. Int. 2018, 42, 570–579. [Google Scholar] [CrossRef]
- Loh, K.M.; Chen, A.; Koh, P.W.; Deng, T.Z.; Sinha, R.; Tsai, J.M.; Barkal, A.A.; Shen, K.Y.; Jain, R.; Morganti, R.M.; et al. Mapping the Pairwise Choices Leading from Pluripotency to Human Bone, Heart, and Other Mesoderm Cell Types. Cell 2016, 166, 451–467. [Google Scholar] [CrossRef] [Green Version]
- Wright, A.J.; Andrews, P.W. Surface marker antigens in the characterization of human embryonic stem cells. Stem Cell Res. 2009, 3, 3–11. [Google Scholar] [CrossRef] [Green Version]
- Lv, F.-J.; Tuan, R.S.; Cheung, K.M.C.; Leung, V.Y.L. Concise Review: The Surface Markers and Identity of Human Mesenchymal Stem Cells. Stem Cells 2014, 32, 1408–1419. [Google Scholar] [CrossRef]
- Ambrosi, T.H.; Longaker, M.T.; Chan, C.K.F. A Revised Perspective of Skeletal Stem Cell Biology. Front. Cell Dev. Biol. 2019, 7, 189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haas, L.; Obenauf, A.C. Allies or Enemies—The Multifaceted Role of Myeloid Cells in the Tumor Microenvironment. Front. Immunol. 2019, 10, 2746. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takarada, T.; Nakazato, R.; Tsuchikane, A.; Fujikawa, K.; Iezaki, T.; Yoneda, Y.; Hinoi, E. Genetic analysis of Runx2 function during intramembranous ossification. Development 2016, 143, 211–218. [Google Scholar] [CrossRef] [Green Version]
- Nakagawa, M.; Taniguchi, Y.; Senda, S.; Takizawa, N.; Ichisaka, T.; Asano, K.; Morizane, A.; Doi, D.; Takahashi, J.; Nishizawa, M.; et al. A novel efficient feeder-free culture system for the derivation of human induced pluripotent stem cells. Sci. Rep. 2014, 4, 3594. [Google Scholar] [CrossRef] [Green Version]
- Gu, W.; Gaeta, X.; Sahakyan, A.; Chan, A.B.; Hong, C.S.; Kim, R.; Braas, D.; Plath, K.; Lowry, W.E.; Christofk, H.R. Glycolytic Metabolism Plays a Functional Role in Regulating Human Pluripotent Stem Cell State. Cell Stem Cell 2016, 19, 476–490. [Google Scholar] [CrossRef] [Green Version]
- Tsogtbaatar, E.; Landin, C.; Minter-Dykhouse, K.; Folmes, C.D.L. Energy Metabolism Regulates Stem Cell Pluripotency. Front. Cell Dev. Biol. 2020, 8, 87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanosaki, S.; Tohyama, S.; Kishino, Y.; Fujita, J.; Fukuda, K. Metabolism of human pluripotent stem cells and differentiated cells for regenerative therapy: A focus on cardiomyocytes. Inflamm. Regen. 2021, 41, 5. [Google Scholar] [CrossRef] [PubMed]
- Lin, B.; Lin, X.; Stachel, M.; Wang, E.; Luo, Y.; Lader, J.; Sun, X.; Delmar, M.; Bu, L. Culture in Glucose-Depleted Medium Supplemented with Fatty Acid and 3,3′,5-Triiodo-l-Thyronine Facilitates Purification and Maturation of Human Pluripotent Stem Cell-Derived Cardiomyocytes. Front. Endocrinol. 2017, 8, 253. [Google Scholar] [CrossRef] [Green Version]
- Römer, P.S.; Berr, S.; Avota, E.; Na, S.-Y.; Battaglia, M.; ten Berge, I.; Einsele, H.; Hünig, T. Preculture of PBMCs at high cell density increases sensitivity of T-cell responses, revealing cytokine release by CD28 superagonist TGN1412. Blood 2011, 118, 6772–6782. [Google Scholar] [CrossRef]
- Sukho, P.; Kirpensteijn, J.; Hesselink, J.W.; van Osch, G.J.V.M.; Verseijden, F.; Bastiaansen-Jenniskens, Y.M. Effect of Cell Seeding Density and Inflammatory Cytokines on Adipose Tissue-Derived Stem Cells: An in Vitro Study. Stem Cell Rev. Rep. 2017, 13, 267–277. [Google Scholar] [CrossRef]
- Najafabadi, M.M.; Bayati, V.; Orazizadeh, M.; Hashemitabar, M.; Absalan, F. Impact of Cell Density on Differentiation Efficiency of Rat Adipose-derived Stem Cells into Schwann-like Cells. Int. J. Stem Cells 2016, 9, 213–220. [Google Scholar] [CrossRef] [Green Version]
- Chen, Q.; Shou, P.; Zheng, C.; Jiang, M.; Cao, G.; Yang, Q.; Cao, J.; Xie, N.; Velletri, T.; Zhang, X.; et al. Fate decision of mesenchymal stem cells: Adipocytes or osteoblasts? Cell Death Differ. 2016, 23, 1128–1139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, H.; Niu, M.; Yuan, X.; Wu, K.; Liu, A. CD44 as a tumor biomarker and therapeutic target. Exp. Hematol. Oncol. 2020, 9, 36. [Google Scholar] [CrossRef]
- Jiang, Y.; Wells, A.; Sylakowski, K.; Clark, A.M.; Ma, B. Adult Stem Cell Functioning in the Tumor Micro-Environment. Int. J. Mol. Sci. 2019, 20, 2566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sherman, L.; Wainwright, D.; Ponta, H.; Herrlich, P. A splice variant of CD44 expressed in the apical ectodermal ridge presents fibroblast growth factors to limb mesenchyme and is required for limb outgrowth. Genes Dev. 1998, 12, 1058–1071. [Google Scholar] [CrossRef]
- Schmitt, M.; Metzger, M.; Gradl, D.; Davidson, G.; Orian-Rousseau, V. CD44 functions in Wnt signaling by regulating LRP6 localization and activation. Cell Death Differ. 2015, 22, 677–689. [Google Scholar] [CrossRef] [Green Version]
- Krebsbach, P.H.; Villa-Diaz, L.G. The Role of Integrin α6 (CD49f) in Stem Cells: More than a Conserved Biomarker. Stem Cells Dev. 2017, 26, 1090–1099. [Google Scholar] [CrossRef] [PubMed]
- Le, M.N.T.; Takahi, M.; Ohnuma, K. Auto/paracrine factors and early Wnt inhibition promote cardiomyocyte differentiation from human induced pluripotent stem cells at initial low cell density. Sci. Rep. 2021, 11, 21426. [Google Scholar] [CrossRef]
- Conway, S.J.; Henderson, D.J.; Copp, A.J. Pax3 is required for cardiac neural crest migration in the mouse: Evidence from the splotch (Sp2H) mutant. Development 1997, 124, 505–514. [Google Scholar] [CrossRef] [PubMed]
- Reinisch, A.; Etchart, N.; Thomas, D.; Hofmann, N.A.; Fruehwirth, M.; Sinha, S.; Chan, C.K.; Senarath-Yapa, K.; Seo, E.-Y.; Wearda, T.; et al. Epigenetic and in vivo comparison of diverse MSC sources reveals an endochondral signature for human hematopoietic niche formation. Blood 2015, 125, 249–260. [Google Scholar] [CrossRef] [PubMed]
- Chamberlain, G.; Fox, J.; Ashton, B.; Middleton, J. Concise Review: Mesenchymal Stem Cells: Their Phenotype, Differentiation Capacity, Immunological Features, and Potential for Homing. Stem Cells 2007, 25, 2739–2749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, M.; Erickson, I.E.; Huang, A.H.; Garrity, S.T.; Mauck, R.L.; Steinberg, D.R. Donor Variation and Optimization of Human Mesenchymal Stem Cell Chondrogenesis in Hyaluronic Acid. Tissue Eng. Part A 2018, 24, 1693–1703. [Google Scholar] [CrossRef] [PubMed]
- Garcia, J.; Mennan, C.; McCarthy, H.S.; Roberts, S.; Richardson, J.B.; Wright, K.T. Chondrogenic Potency Analyses of Donor-Matched Chondrocytes and Mesenchymal Stem Cells Derived from Bone Marrow, Infrapatellar Fat Pad, and Subcutaneous Fat. Stem Cells Int. 2016, 2016, 6969726. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yamada, D.; Takao, T.; Nakamura, M.; Kitano, T.; Nakata, E.; Takarada, T. Identification of Surface Antigens That Define Human Pluripotent Stem Cell-Derived PRRX1+Limb-Bud-like Mesenchymal Cells. Int. J. Mol. Sci. 2022, 23, 2661. https://doi.org/10.3390/ijms23052661
Yamada D, Takao T, Nakamura M, Kitano T, Nakata E, Takarada T. Identification of Surface Antigens That Define Human Pluripotent Stem Cell-Derived PRRX1+Limb-Bud-like Mesenchymal Cells. International Journal of Molecular Sciences. 2022; 23(5):2661. https://doi.org/10.3390/ijms23052661
Chicago/Turabian StyleYamada, Daisuke, Tomoka Takao, Masahiro Nakamura, Toki Kitano, Eiji Nakata, and Takeshi Takarada. 2022. "Identification of Surface Antigens That Define Human Pluripotent Stem Cell-Derived PRRX1+Limb-Bud-like Mesenchymal Cells" International Journal of Molecular Sciences 23, no. 5: 2661. https://doi.org/10.3390/ijms23052661
APA StyleYamada, D., Takao, T., Nakamura, M., Kitano, T., Nakata, E., & Takarada, T. (2022). Identification of Surface Antigens That Define Human Pluripotent Stem Cell-Derived PRRX1+Limb-Bud-like Mesenchymal Cells. International Journal of Molecular Sciences, 23(5), 2661. https://doi.org/10.3390/ijms23052661






