Genome-Wide Identification and Expression Analysis of Pseudouridine Synthase Family in Arabidopsis and Maize
Abstract
:1. Introduction
2. Results
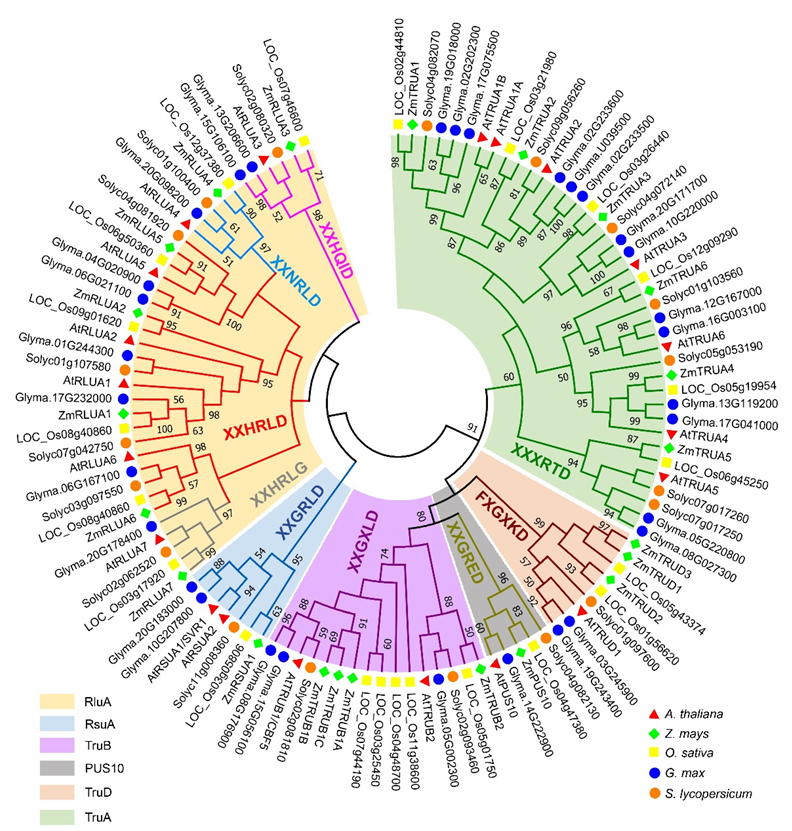
2.1. Identification and Phylogenetic Analysis of the PUS Genes in Arabidopsis thaliana and Zea mays
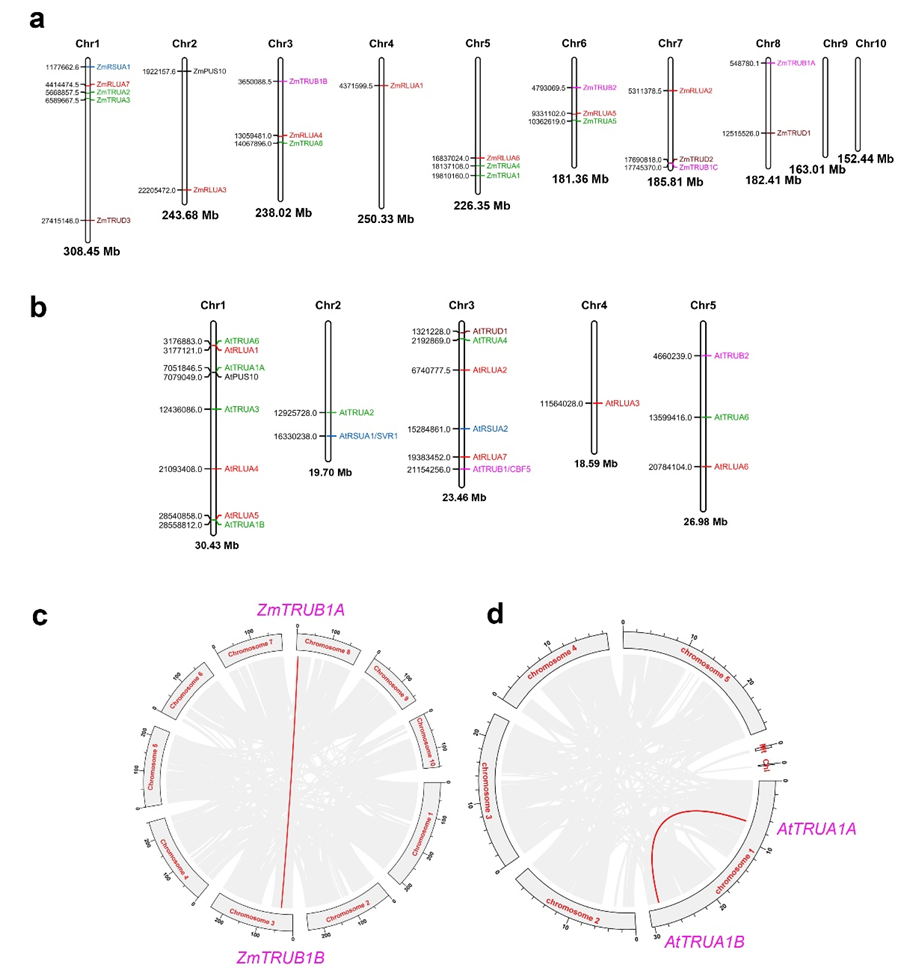
2.2. Chromosomal Distribution and Gene Synteny of AtPUS and ZmPUS Genes
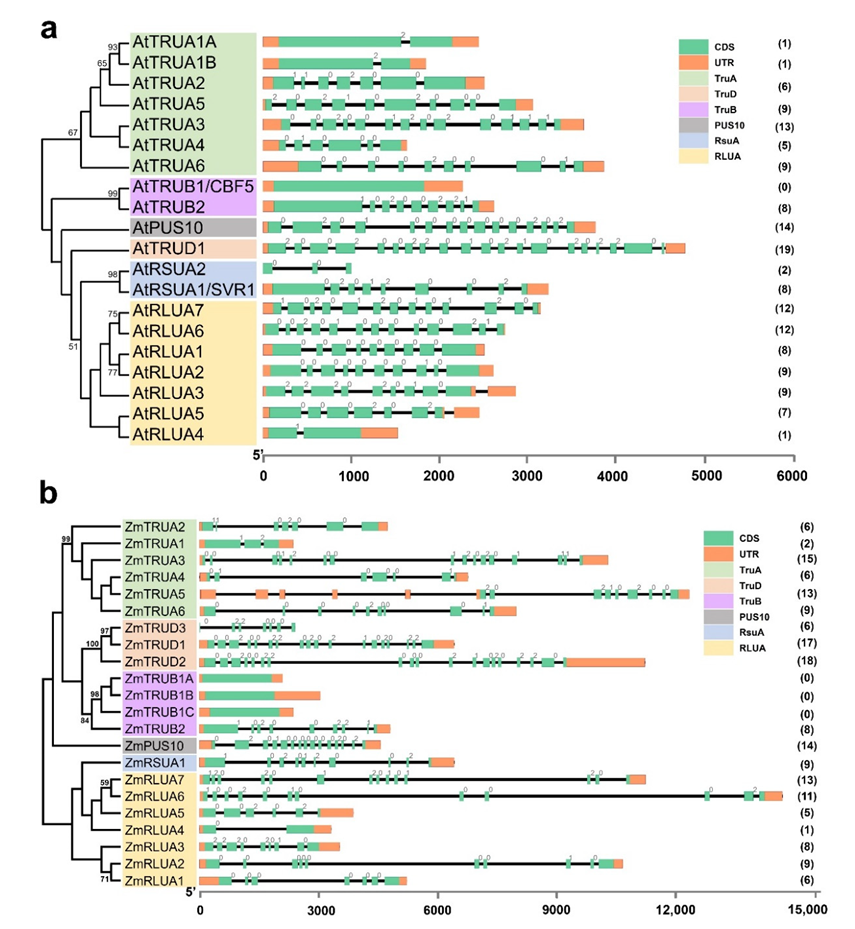
2.3. Gene Structure of AtPUS and ZmPUS Genes
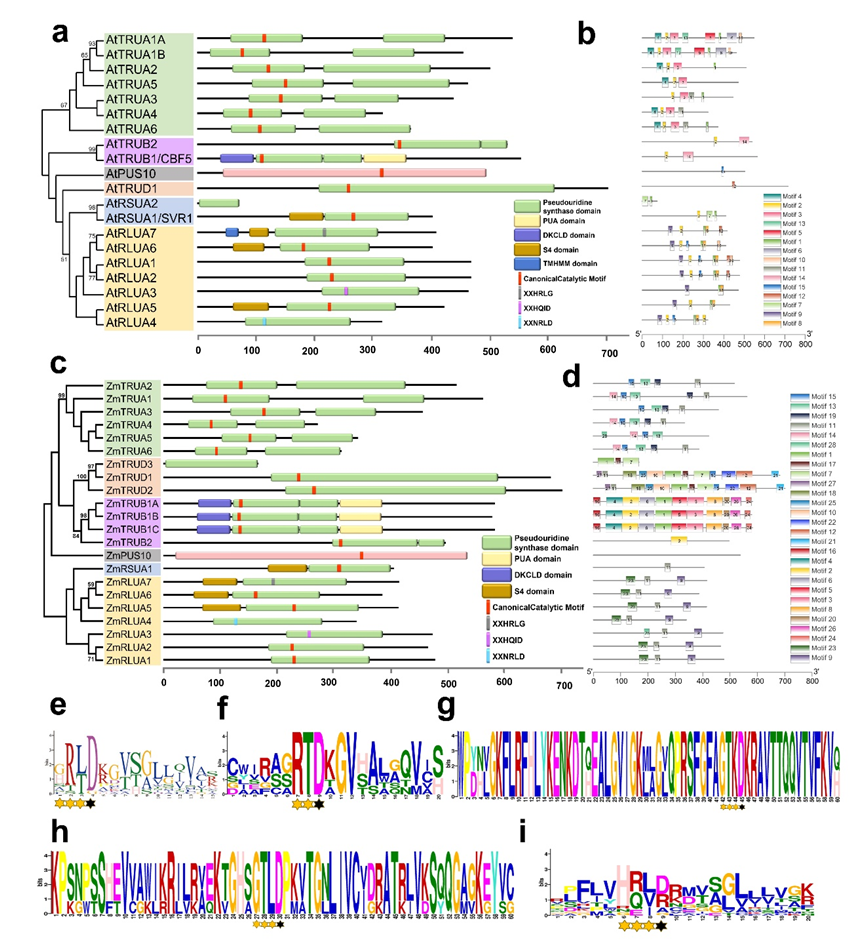
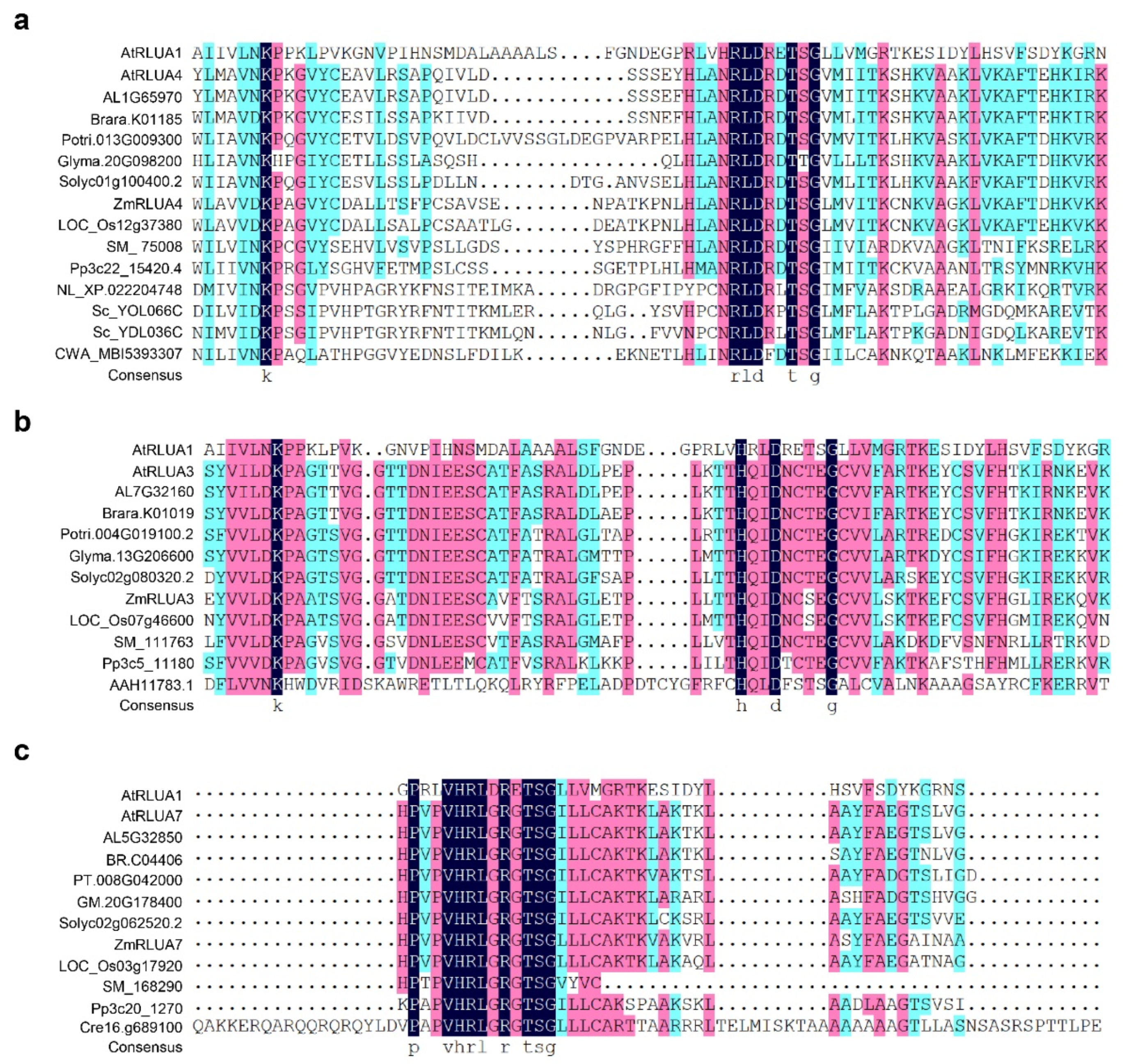
2.4. Protein Features of AtPUSs and ZmPUSs
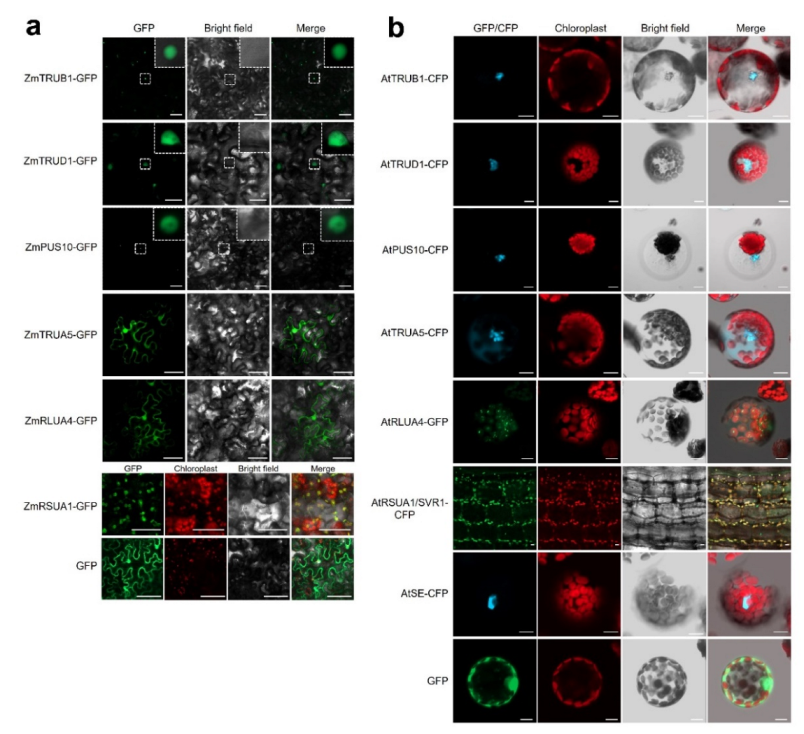
2.5. Subcellular Localization of AtPUSs and ZmPUSs
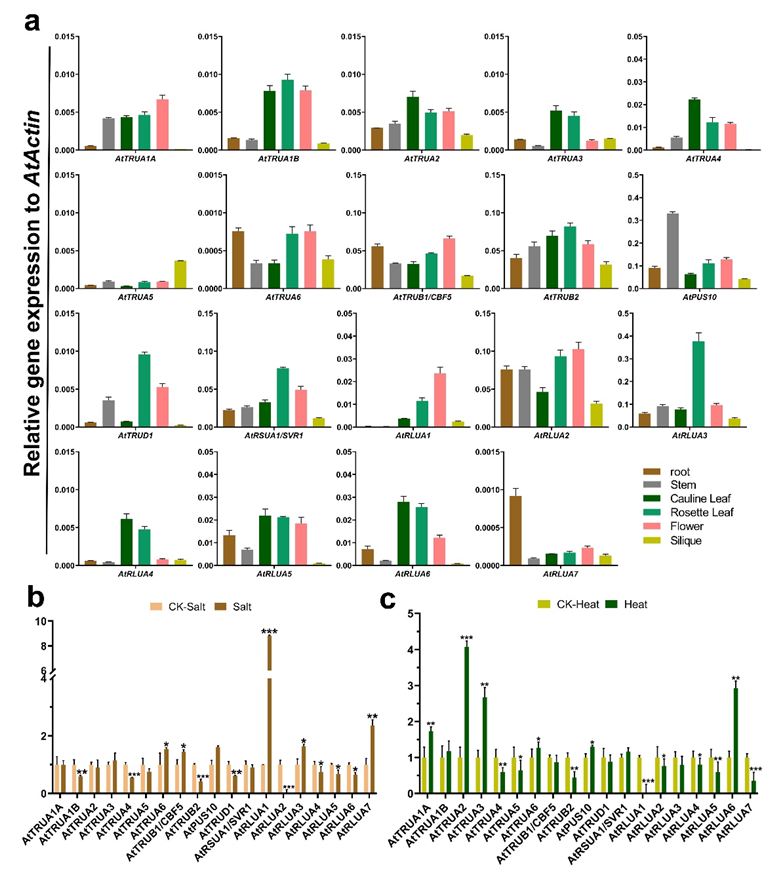
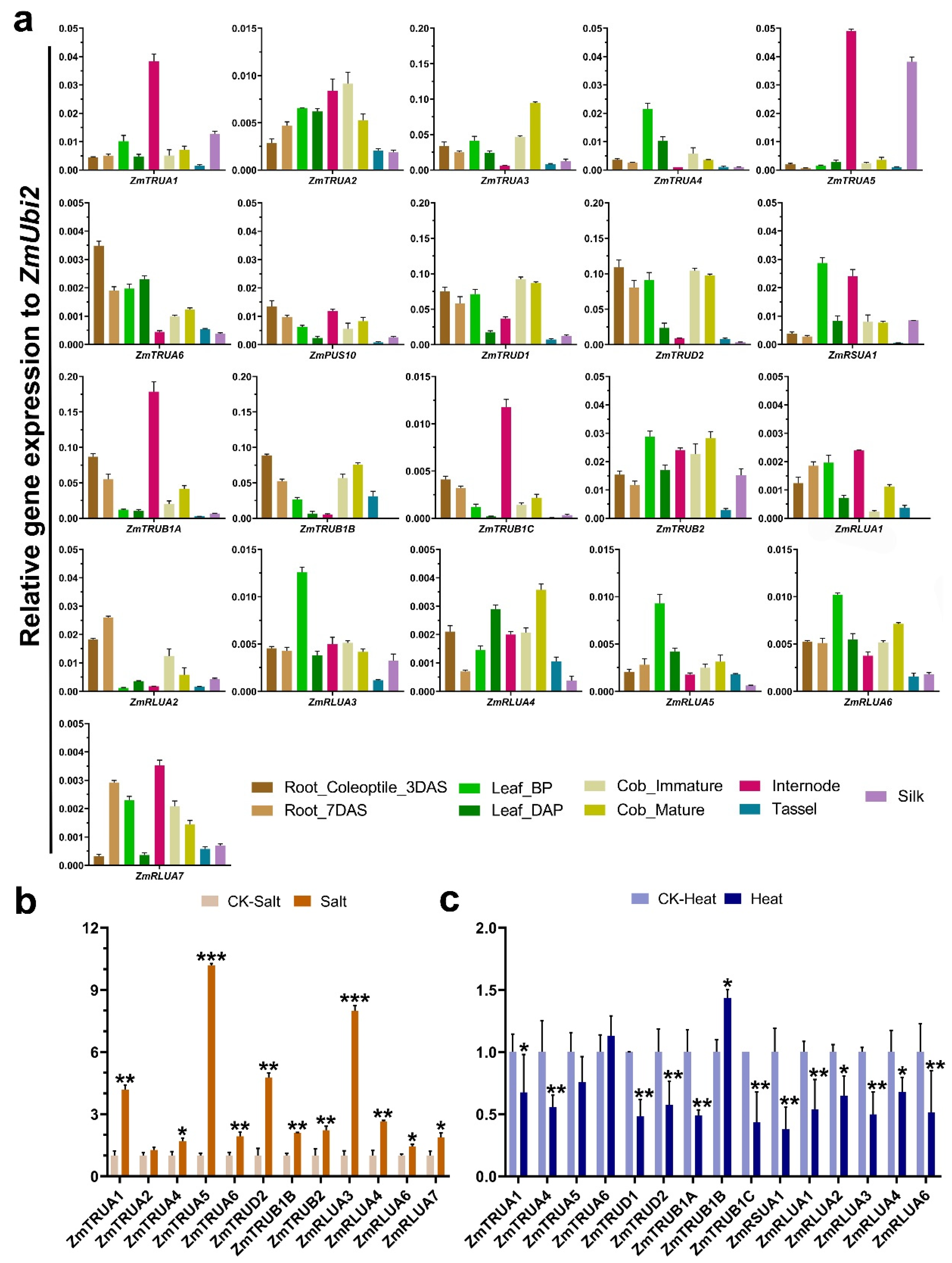
2.6. Expression Analysis of the AtPUS and ZmPUS Genes in Different Tissues and in Response to Abiotic Stresses
3. Discussion
4. Materials and Methods
4.1. Identification of the PUS Genes in Arabidopsis and Maize
4.2. Phylogenetic Analysis and Gene Structure
4.3. Amino Acid Sequence Analysis
4.4. Plant Materials and Growth Conditions
4.5. Expression of AtPUS and ZmPUS Genes Analyzed by Quantitative Real-Time PCR
4.6. Subcellular Localization of ZmPUS and AtPUS Proteins by Confocal Imaging Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Frye, M.; Harada, B.T.; Behm, M.; He, C. RNA modifications modulate gene expression during development. Science 2018, 361, 1346–1349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cohn, W.E.; Volkin, E. Nucleoside-5’-Phosphates from Ribonucleic Acid. Nature 1951, 167, 483–484. [Google Scholar] [CrossRef]
- Davis, F.F.; Allen, F.W. Ribonucleic acids from yeast which contain a fifth nucleotide. J. Biol. Chem. 1957, 227, 907–915. [Google Scholar] [CrossRef]
- Charette, M.; Gray, M.W. Pseudouridine in RNA: What, where, how, and why. IUBMB Life 2000, 49, 341–351. [Google Scholar]
- Carlile, T.M.; Rojas-Duran, M.F.; Gilbert, W.V. Transcriptome-Wide Identification of Pseudouridine Modifications Using Pseudo-seq. Curr. Protoc. Mol. Biol. 2015, 112, 4.25.1–4.25.24. [Google Scholar] [CrossRef]
- Schwartz, S.; Bernstein, D.A.; Mumbach, M.R.; Jovanovic, M.; Herbst, R.H.; Leon-Ricardo, B.X.; Engreitz, J.M.; Guttman, M.; Satija, R.; Lander, E.S.; et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell 2014, 159, 148–162. [Google Scholar] [CrossRef] [Green Version]
- Lovejoy, A.F.; Riordan, D.P.; Brown, P.O. Transcriptome-wide mapping of pseudouridines: Pseudouridine synthases modify specific mRNAs in S. cerevisiae. PLoS ONE 2014, 9, e110799. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Zhu, P.; Ma, S.; Song, J.; Bai, J.; Sun, F.; Yi, C. Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nat. Chem. Biol. 2015, 11, 592–597. [Google Scholar] [CrossRef]
- Spenkuch, F.; Motorin, Y.; Helm, M. Pseudouridine: Still mysterious, but never a fake (uridine)! RNA Biol. 2014, 11, 1540–1554. [Google Scholar] [CrossRef] [Green Version]
- Hamma, T.; Ferre-D’Amare, A.R. Pseudouridine synthases. Chem. Biol. 2006, 13, 1125–1135. [Google Scholar] [CrossRef] [Green Version]
- Fitzek, E.; Joardar, A.; Gupta, R.; Geisler, M. Evolution of Eukaryal and Archaeal Pseudouridine Synthase Pus10. Mol. Biol. Evol. 2018, 86, 77–89. [Google Scholar] [CrossRef] [PubMed]
- Carlile, T.M.; Rojas-Duran, M.F.; Zinshteyn, B.; Shin, H.; Bartoli, K.M.; Gilbert, W.V. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 2014, 515, 143–146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Basak, A.; Query, C.C. A pseudouridine residue in the spliceosome core is part of the filamentous growth program in yeast. Cell Rep. 2014, 8, 966–973. [Google Scholar] [CrossRef] [Green Version]
- Nakamoto, M.A.; Lovejoy, A.F.; Cygan, A.M.; Boothroyd, J.C. mRNA pseudouridylation affects RNA metabolism in the parasite Toxoplasma gondii. RNA 2017, 23, 1834–1849. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, F.; Liu, X.; Alsheikh, M.; Park, S.; Rodermel, S. Mutations in SUPPRESSOR OF VARIEGATION1, a factor required for normal chloroplast translation, suppress var2-mediated leaf variegation in Arabidopsis. Plant Cell 2008, 20, 1786–1804. [Google Scholar] [CrossRef] [Green Version]
- Lu, S.; Li, C.; Zhang, Y.; Zheng, Z.; Liu, D. Functional Disruption of a Chloroplast Pseudouridine Synthase Desensitizes Arabidopsis Plants to Phosphate Starvation. Front. Plant Sci. 2017, 8, 1421. [Google Scholar] [CrossRef] [Green Version]
- Sun, L.; Xu, Y.; Bai, S.; Bai, X.; Zhu, H.; Dong, H.; Wang, W.; Zhu, X.; Hao, F.; Song, C.P. Transcriptome-wide analysis of pseudouridylation of mRNA and non-coding RNAs in Arabidopsis. J. Exp. Bot. 2019, 70, 5089–5600. [Google Scholar] [CrossRef] [Green Version]
- Lin, D.; Kong, R.; Chen, L.; Wang, Y.; Wu, L.; Xu, J.; Piao, Z.; Lee, G.; Dong, Y. Chloroplast development at low temperature requires the pseudouridine synthase gene TCD3 in rice. Sci. Rep. 2020, 10, 8518. [Google Scholar] [CrossRef]
- Lermontova, I.; Schubert, V.; Bornke, F.; Macas, J.; Schubert, I. Arabidopsis CBF5 interacts with the H/ACA snoRNP assembly factor NAF1. Plant Mol. Biol. 2007, 65, 615–626. [Google Scholar] [CrossRef]
- Kannan, K.; Nelson, A.D.; Shippen, D.E. Dyskerin is a component of the Arabidopsis telomerase RNP required for telomere maintenance. Mol. Cell. Biol. 2008, 28, 2241–2332. [Google Scholar] [CrossRef] [Green Version]
- Schnable, P.S.; Ware, D.; Fulton, R.S.; Stein, J.C.; Wei, F.; Pasternak, S.; Liang, C.; Zhang, J.; Fulton, L.; Graves, T.A. The B73 Maize Genome: Complexity, Diversity, and Dynamics. Science 2009, 326, 1112–1115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, S.; Zhou, Y.; Chen, J.; Shi, J.; Zhao, H.; Zhao, H.; Song, W.; Zhang, M.; Cui, Y.; Dong, X.; et al. Extensive intraspecific gene order and gene structural variations between Mo17 and other maize genomes. Nat. Genet. 2018, 50, 1289–1295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Springer, N.M.; Anderson, S.N.; Andorf, C.M.; Ahern, K.R.; Bai, F.; Barad, O.; Barbazuk, W.B.; Bass, H.W.; Baruch, K.; Ben-Zvi, G.; et al. The maize W22 genome provides a foundation for functional genomics and transposon biology. Nat. Genet. 2018, 50, 1282–1288. [Google Scholar] [CrossRef] [Green Version]
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef] [Green Version]
- Letunic, I.; Bork, P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018, 46, D493–D496. [Google Scholar] [CrossRef]
- Wang, X.; Li, G.Z.; Lu, W.C. Virus-ECC-mPLoc: A Multi-Label Predictor for Predicting the Subcellular Localization of Virus Proteins with Both Single and Multiple Sites Based on a General Form of Chou’s Pseudo Amino Acid Composition. Protein Pept. Lett. 2013, 20, 309–317. [Google Scholar]
- Chou, K.C.; Shen, H.B. Cell-PLoc: A package of Web servers for predicting subcellular localization of proteins in various organisms. Nat. Protoc. 2008, 3, 153–162. [Google Scholar] [CrossRef]
- Chou, K.C.; Shen, H.B. Plant-mPLoc: A Top-Down Strategy to Augment the Power for Predicting Plant Protein Subcellular Localization. PLoS ONE 2010, 5, e11335. [Google Scholar] [CrossRef] [Green Version]
- Chou, K.C.; Shen, H.B. Large-scale plant protein subcellular location prediction. J. Cell. Chem. 2007, 100, 665–678. [Google Scholar] [CrossRef]
- Behm-Ansmant, I.; Grosjean, H.; Massenet, S.; Motorin, Y.; Branlant, C. Pseudouridylation at position 32 of mitochondrial and cytoplasmic tRNAs requires two distinct enzymes in Saccharomyces cerevisiae. J. Biol. Chem. 2004, 279, 52998–53006. [Google Scholar] [CrossRef] [Green Version]
- Perron, K.; Goldschmidt-Clermont, M.; Rochaix, J.D. A factor related to pseudouridine synthases is required for chloroplast group II intron trans-splicing in Chlamydomonas reinhardtii. EMBO J. 1999, 18, 6481–6490. [Google Scholar] [CrossRef] [Green Version]
- Gutgsell, N.S.; Del Campo, M.; Raychaudhuri, S.; Ofengand, J. A second function for pseudouridine synthases: A point mutant of RluD unable to form pseudouridines 1911, 1915, and 1917 in Escherichia coli 23S ribosomal RNA restores normal growth to an RluD-minus strain. RNA 2001, 7, 990–998. [Google Scholar] [CrossRef] [Green Version]
- Gutgsell, N.; Englund, N.; Niu, L.; Kaya, Y.; Lane, B.G.; Ofengand, J. Deletion of the Escherichia coli pseudouridine synthase gene truB blocks formation of pseudouridine 55 in tRNA in vivo, does not affect exponential growth, but confers a strong selective disadvantage in competition with wild-type cells. RNA 2000, 6, 1870–1881. [Google Scholar] [CrossRef] [Green Version]
- Lafontaine, D.L.; Bousquet-Antonelli, C.; Henry, Y.; Caizergues-Ferrer, M.; Tollervey, D. The box H + ACA snoRNAs carry Cbf5p, the putative rRNA pseudouridine synthase. Genes Dev. 1998, 12, 527–537. [Google Scholar] [CrossRef] [Green Version]
- Ma, X.; Zhao, X.; Yu, Y.T. Pseudouridylation (Psi) of U2 snRNA in S. cerevisiae is catalyzed by an RNA-independent mechanism. EMBO J. 2003, 22, 1889–1897. [Google Scholar] [CrossRef] [Green Version]
- Wu, G.; Xiao, M.; Yang, C.; Yu, Y.T. U2 snRNA is inducibly pseudouridylated at novel sites by Pus7p and snR81 RNP. EMBO J. 2011, 30, 79–89. [Google Scholar] [CrossRef] [Green Version]
- Behm-Ansmant, I.; Urban, A.; Ma, X.; Yu, Y.T.; Motorin, Y.; Branlant, C. The Saccharomyces cerevisiae U2 snRNA:pseudouridine-synthase Pus7p is a novel multisite-multisubstrate RNA:Psi-synthase also acting on tRNAs. RNA 2003, 9, 1371–1382. [Google Scholar] [CrossRef] [Green Version]
- Decatur, W.A.; Schnare, M.N. Different mechanisms for pseudouridine formation in yeast 5S and 5.8S rRNAs. Mol. Cell. Biol. 2008, 28, 3089–3100. [Google Scholar] [CrossRef] [Green Version]
- Guegueniat, J.; Halabelian, L.; Zeng, H.; Dong, A.; Li, Y.; Wu, H.; Arrowsmith, C.H.; Kothe, U. The human pseudouridine synthase PUS7 recognizes RNA with an extended multi-domain binding surface. Nucleic Acids Res. 2021, 49, 11810–11822. [Google Scholar] [CrossRef]
- Guzzi, N.; Ciesla, M.; Ngoc, P.C.T.; Lang, S.; Arora, S.; Dimitriou, M.; Pimkova, K.; Sommarin, M.N.E.; Munita, R.; Lubas, M.; et al. Pseudouridylation of tRNA-Derived Fragments Steers Translational Control in Stem Cells. Cell 2018, 173, 1204–1216. e26. [Google Scholar] [CrossRef] [Green Version]
- Mukhopadhyay, S.; Deogharia, M.; Gupta, R. Mammalian nuclear TRUB1, mitochondrial TRUB2, and cytoplasmic PUS10 produce conserved pseudouridine 55 in different sets of tRNA. RNA 2021, 27, 66–79. [Google Scholar] [CrossRef]
- Song, J.; Zhuang, Y.; Zhu, C.; Meng, H.; Lu, B.; Xie, B.; Peng, J.; Li, M.; Yi, C. Differential roles of human PUS10 in miRNA processing and tRNA pseudouridylation. Nat. Chem. Biol. 2020, 16, 160–169. [Google Scholar] [CrossRef]
- Deogharia, M.; Mukhopadhyay, S.; Joardar, A.; Gupta, R. The human ortholog of archaeal Pus10 produces pseudouridine 54 in select tRNAs where its recognition sequence contains a modified residue. RNA 2019, 25, 336–351. [Google Scholar] [CrossRef] [Green Version]
- Gurha, P.; Gupta, R. Archaeal Pus10 proteins can produce both pseudouridine 54 and 55 in tRNA. RNA 2008, 14, 2521–2527. [Google Scholar] [CrossRef] [Green Version]
- Roovers, M.; Hale, C.; Tricot, C.; Terns, M.P.; Terns, R.M.; Grosjean, H.; Droogmans, L. Formation of the conserved pseudouridine at position 55 in archaeal tRNA. Nucleic Acids Res. 2006, 34, 4293–4301. [Google Scholar] [CrossRef]
- Whelan, S.; Goldman, N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol Evol. 2001, 18, 691–699. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Ludwig, Y.; Zhang, Y.X.; Hochholdinger, F. The Maize (Zea mays L.) AUXIN/INDOLE-3-ACETIC ACID Gene Family: Phylogeny, Synteny, and Unique Root-Type and Tissue-Specific Expression Patterns during Development. PLoS ONE 2013, 8, e78859. [Google Scholar] [CrossRef] [Green Version]
- Walley, J.W.; Sartor, R.C.; Shen, Z.; Schmitz, R.J.; Wu, K.J.; Urich, M.A.; Nery, J.R.; Smith, L.G.; Schnable, J.C.; Ecker, J.R.; et al. Integration of omic networks in a developmental atlas of maize. Science 2016, 353, 814–818. [Google Scholar] [CrossRef] [Green Version]
- Makarevitch, I.; Waters, A.J.; West, P.T.; Stitzer, M.; Hirsch, C.N.; Ross-Ibarra, J.; Springer, N.M. Transposable Elements Contribute to Activation of Maize Genes in Response to Abiotic Stress. PLoS Genet. 2015, 11, e1004915. [Google Scholar]
- Nguyen, T.X.; Sticklen, M. Barley HVA1 Gene Confers Drought and Salt Tolerance in Transgenic Maize (Zea Mays, L.). Adv. Crop Sci. Tech 2013, 1, 105. [Google Scholar] [CrossRef] [Green Version]
- Yoo, S.D.; Cho, Y.H.; Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Group | Name | Transcript ID | Chromosome Location | F(+)/R(−) | AA a | pI b | MW c (KDa) | Predicted Subcellular Localization |
|---|---|---|---|---|---|---|---|---|
| TruA | AtTRUA1A | AT1G20370.1 | Chr1:7051846-7053588 | − | 549 | 5.22 | 61.5 | Nc, Cp |
| AtTRUA1B | AT1G76120.1 | Chr1:28558813-28560294 | − | 463 | 7.23 | 51.8 | Cp, Cm | |
| AtTRUA2 | AT2G30320.1 | Chr2:12925728-12927896 | − | 510 | 6.02 | 59.0 | Chl, Mt | |
| AtTRUA5 | AT5G35400.2 | Chr5:13599416-13602240 | − | 471 | 6.19 | 53.3 | Chl,ER, Mt | |
| AtTRUA3 | AT1G34150.1 | Chr1:12436086-12439237 | + | 565 | 9.19 | 63.0 | Nc, Cp | |
| AtTRUA4 | AT3G06950.1 | Chr3:2192869-2194254 | + | 323 | 8.96 | 36.2 | Chl | |
| AtTRUA6 | AT1G09800.1 | Chr1:3177121-3180336 | − | 372 | 7.27 | 41.5 | Nc | |
| TruB | AtTRUB2 | AT5G14460.1 | Chr5:4660239-4662543 | − | 540 | 8.93 | 61.5 | Chl |
| AtTRUB1/CBF5 | AT3G57150.1 | Chr3:21154255-21155952 | − | 446 | 8.84 | 51.0 | Nc | |
| Pus10 | AtPUS10 | AT1G20410.1 | Chr1:7079049-7082504 | − | 504 | 6.55 | 56.5 | Nc |
| TruD | AtTRUD1 | AT3G04820.1 | Chr3:1321228-1325953 | − | 715 | 5.76 | 79.4 | Chl, Mt |
| RsuA | AtRSUA2 | AT3G43340.1 | Chr3:15284861-15285851 | − | 74 | 6.46 | 8.3 | Chl, Cp |
| AtRSUA1/SVR1 | AT2G39140.1 | Chr2:16330238-16333153 | + | 410 | 9.95 | 45.1 | Chl | |
| RluA | AtRLUA7 | AT3G52260.3 | Chr3:19383452-19386440 | − | 416 | 6.24 | 45.8 | Chl, Cp, Nc |
| AtRLUA6 | AT5G51140.2 | Chr5:20784103-20786793 | − | 410 | 6.29 | 46.5 | Cp | |
| AtRLUA1 | AT1G78910.1 | Chr1:3177121-3180336 | − | 478 | 9.48 | 53.8 | Chl, Nc, Cp | |
| AtRLUA2 | AT3G19440.1 | Chr3:6740778-6743132 | + | 477 | 9.46 | 53.0 | Mt, Chl | |
| AtRLUA3 | AT4G21770.1 | Chr4:11564028-11566345 | − | 472 | 8.13 | 52.9 | Chl | |
| AtRLUA5 | AT1G76050.2 | Chr1:28540858-28542826 | + | 430 | 7.04 | 46.7 | Chl, Mt | |
| AtRLUA4 | AT1G56345.1 | Chr1:21093409-21094454 | − | 322 | 6.94 | 35.9 | Chl, Nc |
| Group | Name | Transcript ID | Chromosome Location | F(+)/R(−) | AA a | pI b | MW c (KDa) | Predicted Subcellular Localization |
|---|---|---|---|---|---|---|---|---|
| TruA | ZmTRUA2 | Zm00001eb016190_T001 | 1:56688574-56693320 | + | 516 | 8.66 | 57.4 | Chl |
| ZmTRUA1 | Zm00001eb250100_T001 | 5:198101606-198103976 | + | 562 | 5.67 | 61.6 | Nc, Chl, Mt | |
| ZmTRUA3 | Zm00001eb018490_T001 | 1:65896675-65906968 | + | 457 | 8.42 | 51.7 | Chl, Nc | |
| ZmTRUA4 | Zm00001eb245280_T002 | 5:181371080-181377861 | + | 334 | 9.00 | 37.1 | Mt, Chl | |
| ZmTRUA5 | Zm00001eb274430_T001 | 6:103626192-103638522 | + | 422 | 8.65 | 46.7 | Mt, Chl | |
| ZmTRUA6 | Zm00001eb138750_T005 | 3:140678961-140686947 | + | 386 | 8.05 | 42.3 | Nc,Cp,Chl | |
| TruD | ZmTRUD3 | Zm00001eb054900_T001 | 1:274151464-274153881 | − | 167 | 9.92 | 18.8 | Chl, Cp |
| ZmTRUD1 | Zm00001eb352870_T005 | 8:125155260-125161684 | − | 682 | 6.82 | 75.2 | Nc,Cp,Chl | |
| ZmTRUD2 | Zm00001eb328420_T002 | 7:176908181-176919404 | − | 701 | 6.22 | 77.4 | Chl, Cp | |
| TruB | ZmTRUB1A | Zm00001eb333520_T001 | 8:5487801-5489896 | + | 583 | 9.19 | 64.2 | Nc |
| ZmTRUB1B | Zm00001eb127780_T001 | 3:36500885-36503926 | − | 579 | 9.17 | 63.8 | Nc | |
| ZmTRUB1C | Zm00001eb328640_T001 | 7:177453702-177456063 | − | 583 | 9.08 | 63.9 | Nc | |
| ZmTRUB2 | Zm00001eb267110_T001 | 6:47930697-47935508 | − | 497 | 9.25 | 56.1 | Chl, Mt | |
| Pus10 | ZmPUS10 | Zm00001eb074060_T004 | 2:19221575-19226195 | + | 536 | 6.34 | 60.6 | Nc |
| RsuA | ZmRSUA1 | Zm00001eb004240_T001 | 1:11776626-11783053 | + | 406 | 9.93 | 43.7 | Chl |
| RluA | ZmRLUA7 | Zm00001eb013230_T001 | 1:44144745-44155879 | + | 415 | 7.04 | 45.6 | Chl, Nc |
| ZmRLUA6 | Zm00001eb241970_T001 | 5:168370244-168384931 | − | 386 | 7.69 | 42.9 | Cp | |
| ZmRLUA5 | Zm00001eb272250_T003 | 6:93311018-93314911 | + | 414 | 9.52 | 44.6 | Chl, Mt | |
| ZmRLUA4 | Zm00001eb137040_T002 | 3:130594814-130598135 | − | 348 | 8.66 | 37.5 | Chl, Mt | |
| ZmRLUA3 | Zm00001eb110780_T001 | 2:222054727-222058271 | + | 474 | 8.82 | 52.6 | Chl | |
| ZmRLUA2 | Zm00001eb307420_T004 | 7:53113783-53124431 | − | 466 | 9.55 | 51.6 | Chl, Mt | |
| ZmRLUA1 | Zm00001eb174620_T001 | 4:43715995-43721226 | − | 478 | 9.90 | 54.0 | Chl, Mt |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, Y.; Gu, Y.; Shi, G.; He, J.; Hu, W.; Zhang, Z. Genome-Wide Identification and Expression Analysis of Pseudouridine Synthase Family in Arabidopsis and Maize. Int. J. Mol. Sci. 2022, 23, 2680. https://doi.org/10.3390/ijms23052680
Xie Y, Gu Y, Shi G, He J, Hu W, Zhang Z. Genome-Wide Identification and Expression Analysis of Pseudouridine Synthase Family in Arabidopsis and Maize. International Journal of Molecular Sciences. 2022; 23(5):2680. https://doi.org/10.3390/ijms23052680
Chicago/Turabian StyleXie, Yuting, Yeting Gu, Guangping Shi, Jianliang He, Wenjing Hu, and Zhonghui Zhang. 2022. "Genome-Wide Identification and Expression Analysis of Pseudouridine Synthase Family in Arabidopsis and Maize" International Journal of Molecular Sciences 23, no. 5: 2680. https://doi.org/10.3390/ijms23052680







