Droplet Digital PCR for Non-Invasive Prenatal Detection of Fetal Single-Gene Point Mutations in Maternal Plasma
Abstract
:1. Introduction
2. Materials and Methods
2.1. Collection of Samples
2.2. Preparation of Plasma
2.3. Extraction of Genomic DNA
2.4. Extraction of Circulating Cell-Free DNA
2.5. Synthetic Oligonucleotides
2.6. Sequencing Reaction
2.7. Droplet Digital™ PCR (ddPCR)
2.8. Statistical Analysis
3. Results
3.1. Validation of β+IVSI-110 and β039 Thalassemia ddPCR Assays by Genomic DNA Mixtures Mimicking Circulating Cell-Free Fetal DNA and Maternal DNA
3.2. NIPT of β+IVSI-110 or β039 Thalassemia Mutations Using ddPCR Assays
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lo, Y.M.; Corbetta, N.; Chamberlain, P.F.; Rai, V.; Sargent, I.L.; Redman, C.W.; Wainscoat, J.S. Presence of fetal DNA in maternal plasma and serum. Lancet 1997, 350, 485–487. [Google Scholar] [CrossRef]
- Lun, F.M.; Tsui, N.B.; Chan, K.C.; Leung, T.Y.; Lau, T.K.; Charoenkwan, P.; Chow, K.C.; Lo, W.Y.; Wanapirak, C.; Sanguansermsri, T.; et al. Noninvasive prenatal diagnosis of monogenic diseases by digital size selection and relative mutation dosage on DNA in maternal plasma. Proc. Natl. Acad. Sci. USA 2008, 105, 19920–19925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Illanes, S.; Denbow, M.; Kailasam, C.; Finning, K.; Soothill, P.W. Early detection of cell-free fetal DNA in maternal plasma. Early Hum. Dev. 2007, 83, 563–566. [Google Scholar] [CrossRef] [PubMed]
- Drury, S.; Hill, M.; Chitty, L.S. Cell-Free Fetal DNA Testing for Prenatal Diagnosis. Adv. Clin. Chem. 2016, 76, 1–35. [Google Scholar] [PubMed]
- Zhou, Y.; Zhu, Z.; Gao, Y.; Yuan, Y.; Guo, Y.; Zhou, L.; Liao, K.; Wang, J.; Du, B.; Hou, Y.; et al. Effects of maternal and fetal characteristics on cell-free fetal DNA fraction in maternal plasma. Reprod. Sci. 2015, 22, 1429–1435. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Kim, H.J.; Park, S.Y.; Han, Y.J.; Choi, J.S.; Ryu, H.M. Early Prediction of Hypertensive Disorders of Pregnancy Using Cell-Free Fetal DNA, Cell-Free Total DNA, and Biochemical Markers. Fetal Diagn. Ther. 2016, 40, 255–262. [Google Scholar] [CrossRef]
- Vora, N.L.; Johnson, K.L.; Basu, S.; Catalano, P.M.; Hauguel-De Mouzon, S.; Bianchi, D.W. A multifactorial relationship exists between total circulating cell-free DNA levels and maternal BMI. Prenat. Diagn. 2012, 32, 912–914. [Google Scholar] [CrossRef]
- Attilakos, G.; Maddocks, D.G.; Davies, T.; Hunt, L.P.; Avent, N.D.; Soothill, P.W.; Grant, S.R. Quantification of free fetal DNA in multiple pregnancies and relationship with chorionicity. Prenat. Diagn. 2011, 31, 967–972. [Google Scholar] [CrossRef]
- Wright, C.F.; Wei, Y.; Higgins, J.P.; Sagoo, G.S. Non-invasive prenatal diagnostic test accuracy for fetal sex using cell-free DNA a review and meta-analysis. BMC Res. Notes 2012, 5, 476. [Google Scholar] [CrossRef] [Green Version]
- Perlado-Marina, S.; Bustamante-Aragones, A.; Horcajada, L.; Trujillo-Tiebas, M.J.; Lorda-Sanchez, I.; Ruiz Ramos, M.; Plaza, J.; Rodriguez de Alba, M. Overview of Five-Years of Experience Performing Non-Invasive Fetal Sex Assessment in Maternal Blood. Diagnostics 2013, 3, 283–290. [Google Scholar] [CrossRef] [Green Version]
- Clausen, F.B.; Damkjær, M.B.; Dziegiel, M.H. Noninvasive fetal RhD genotyping. Transfus. Apher. Sci. 2014, 50, 154–162. [Google Scholar] [CrossRef] [PubMed]
- Fasano, R.M. Hemolytic disease of the fetus and newborn in the molecular era. Semin. Fetal Neonatal. Med. 2016, 21, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Contro, E.; Bernabini, D.; Farina, A. Cell-Free Fetal DNA for the Prediction of Pre-Eclampsia at the First and Second Trimesters: A Systematic Review and Meta-Analysis. Mol. Diagn. Ther. 2017, 21, 125–135. [Google Scholar] [CrossRef] [PubMed]
- van Boeckel, S.R.; Davidson, D.J.; Norman, J.E.; Stock, S.J. Cell-free Fetal DNA and Spontaneous Preterm Birth. Reproduction 2017, 155, R137–R145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rolnik, D.L.; da Silva Costa, F.; Lee, T.J.; Schmid, M.; McLennan, A.C. Association between fetal fraction on cell-free DNA testing and first trimester markers for pre-eclampsia. Ultrasound Obstet. Gynecol. 2018, 52, 722–727. [Google Scholar] [CrossRef] [Green Version]
- Skrzypek, H.; Hui, L. Noninvasive prenatal testing for fetal aneuploidy and single gene disorders. Best Pract. Res. Clin. Obstet. Gynaecol. 2017, 42, 26–38. [Google Scholar] [CrossRef]
- Mennuti, M.T.; Chandrasekaran, S.; Khalek, N.; Dugoff, L. Cell-free DNA screening and sex chromosome aneuploidies. Prenat. Diagn. 2015, 35, 980–985. [Google Scholar] [CrossRef] [Green Version]
- Wright, C.F.; Burton, H. The use of cell-free fetal nucleic acids in maternal blood for non-invasive prenatal diagnosis. Hum. Reprod. Update 2009, 15, 139–151. [Google Scholar] [CrossRef] [Green Version]
- Bustamante-Aragonés, A.; Rodríguez de Alba, M.; Perlado, S.; Trujillo-Tiebas, M.J.; Arranz, J.P.; Díaz-Recasens, J.; Troyano-Luque, J.; Ramos, C. Non-invasive prenatal diagnosis of single-gene disorders from maternal blood. Gene 2012, 504, 144–149. [Google Scholar] [CrossRef]
- Perlado, S.; Bustamante-Aragonés, A.; Donas, M.; Lorda-Sánchez, I.; Plaza, J.; Rodríguez de Alba, M. Fetal Genotyping in Maternal Blood by Digital PCR: Towards NIPD of Monogenic Disorders Independently of Parental Origin. PLoS ONE 2016, 11, e0153258. [Google Scholar] [CrossRef] [Green Version]
- Hudecova, I. Digital PCR analysis of circulating nucleic acids. Clin. Biochem. 2015, 48, 948–956. [Google Scholar] [CrossRef] [PubMed]
- Hui, W.W.; Jiang, P.; Tong, Y.K.; Lee, W.S.; Cheng, Y.K.; New, M.I.; Kadir, R.A.; Chan, K.C.; Leung, T.Y.; Lo, Y.M.; et al. Universal Haplotype-Based Noninvasive Prenatal Testing for Single Gene Diseases. Clin. Chem. 2017, 63, 513–524. [Google Scholar] [CrossRef] [PubMed]
- Kagan, K.O.; Sonek, J.; Wagner, P.; Hoopmann, M. Principles of first trimester screening in the age of non-invasive prenatal diagnosis: Screening for chromosomal abnormalities. Arch. Gynecol. Obstet. 2017, 296, 645–651. [Google Scholar] [CrossRef] [PubMed]
- Chiu, E.K.L.; Hui, W.W.I.; Chiu, R.W.K. cfDNA screening and diagnosis of monogenic disorders-where are we heading? Prenat. Diagn. 2018, 38, 52–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beulen, L.; Faas, B.H.W.; Feenstra, I.; van Vugt, J.M.G.; Bekker, M.N. Clinical utility of non-invasive prenatal testing in pregnancies with ultrasound anomalies. Ultrasound Obstet. Gynecol. 2017, 49, 721–728. [Google Scholar] [CrossRef]
- Hayward, J.; Chitty, L.S. Beyond screening for chromosomal abnormalities: Advances in non-invasive diagnosis of single gene disorders and fetal exome sequencing. Semin. Fetal Neonatal. Med. 2018, 23, 94–101. [Google Scholar] [CrossRef]
- Lee, S.Y.; Kim, S.J.; Han, S.H.; Park, J.S.; Choi, H.J.; Ahn, J.J.; Oh, M.J.; Shim, S.H.; Cha, D.H.; Hwang, S.Y. A new approach of digital PCR system for non-invasive prenatal screening of trisomy 21. Clin. Chim. Acta 2018, 476, 75–80. [Google Scholar] [CrossRef]
- Sawakwongpra, K.; Tangmansakulchai, K.; Ngonsawan, W.; Promwan, S.; Chanchamroen, S.; Quangkananurug, W.; Sriswasdi, S.; Jantarasaengaram, S.; Ponnikorn, S. Droplet-based digital PCR for non-invasive prenatal genetic diagnosis of α and β-thalassemia. Biomed. Rep. 2021, 15, 82. [Google Scholar] [CrossRef]
- D′Aversa, E.; Breveglieri, G.; Pellegatti, P.; Guerra, G.; Gambari, R.; Borgatti, M. Non-invasive fetal sex diagnosis in plasma of early weeks pregnants using droplet digital PCR. Mol. Med. 2018, 24, 14. [Google Scholar] [CrossRef] [Green Version]
- Pinheiro, L.B.; Coleman, V.A.; Hindson, C.M.; Herrmann, J.; Hindson, B.J.; Bhat, S.; Emslie, K.R. Evaluation of a droplet digital polymerase chain reaction format for DNA copy number quantification. Anal. Chem. 2012, 84, 1003–1011. [Google Scholar] [CrossRef]
- Hindson, B.J.; Ness, K.D.; Masquelier, D.A.; Belgrader, P.; Heredia, N.J.; Makarewicz, A.J.; Bright, I.J.; Lucero, M.Y.; Hiddessen, A.L.; Legler, T.C.; et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal. Chem. 2011, 83, 8604–8610. [Google Scholar] [CrossRef] [PubMed]
- Thein, S.L. The molecular basis of β-thalassemia. Cold Spring Harb. Perspect. Med. 2013, 3, 5. [Google Scholar] [CrossRef] [Green Version]
- Viprakasit, V.; Ekwattanakit, S. Clinical Classification, Screening and Diagnosis for Thalassemia. Hematol. Oncol. Clin. N. Am. 2018, 32, 193–211. [Google Scholar] [CrossRef] [PubMed]
- Sanger, F.; Nicklen, S.; Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 1977, 74, 5463–5467. [Google Scholar] [CrossRef] [Green Version]
- Tan, C.; Chen, X.; Wang, F.; Wang, D.; Cao, Z.; Zhu, X.; Lu, C.; Yang, W.; Gao, N.; Gao, H.; et al. A multiplex droplet digital PCR assay for non-invasive prenatal testing of fetal aneuploidies. Analyst 2019, 144, 2239–2247. [Google Scholar] [CrossRef] [PubMed]
- Bischoff, F.Z.; Lewis, D.E.; Simpson, J.L. Cell-free fetal DNA in maternal blood: Kinetics, source and structure. Hum. Reprod. Update 2005, 11, 59–67. [Google Scholar] [CrossRef] [Green Version]
- Chan, K.C.; Zhang, J.; Hui, A.B.; Wong, N.; Lau, T.K.; Leung, T.N.; Lo, K.W.; Huang, D.W.; Lo, Y.M. Size distributions of maternal and fetal DNA in maternal plasma. Clin. Chem. 2004, 50, 88–92. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Zimmermann, B.; Rusterholz, C.; Kang, A.; Holzgreve, W.; Hahn, S. Size separation of circulatory DNA in maternal plasma permits ready detection of fetal DNA polymorphisms. Clin. Chem. 2004, 50, 1002–1011. [Google Scholar] [CrossRef]
- Liang, B.; Li, H.; He, Q.; Li, H.; Kong, L.; Xuan, L.; Xia, Y.; Shen, J.; Mao, Y.; Li, Y.; et al. Enrichment of the fetal fraction in non-invasive prenatal screening reduces maternal background interference. Sci. Rep. 2018, 8, 17675. [Google Scholar] [CrossRef]
- Breveglieri, G.; D′Aversa, E.; Finotti, A.; Borgatti, M. Non-invasive Prenatal Testing Using Fetal DNA. Mol. Diagn. Ther. 2019, 23, 291–299. [Google Scholar] [CrossRef]
- Gruber, A.; Pacault, M.; El Khattabi, L.A.; Vaucouleur, N.; Orhant, L.; Bienvenu, T.; Girodon, E.; Vidaud, D.; Leturcq, F.; Costa, C.; et al. Non-invasive prenatal diagnosis of paternally inherited disorders from maternal plasma: Detection of NF1 and CFTR mutations using droplet digital PCR. Clin. Chem. Lab. Med. 2018, 56, 728–738. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Zhou, Q.; Zhou, W.; Zhong, M.; Guo, X.; Wang, X.; Fan, X.; Yan, S.; Li, L.; Lai, Y.; et al. Cell-free DNA Barcode-Enabled Single-Molecule Test for Non invasive Prenatal Diagnosis of Monogenic Disorders: Application to β-Thalassemia. Adv. Sci. 2019, 6, 1802332. [Google Scholar] [CrossRef] [PubMed]
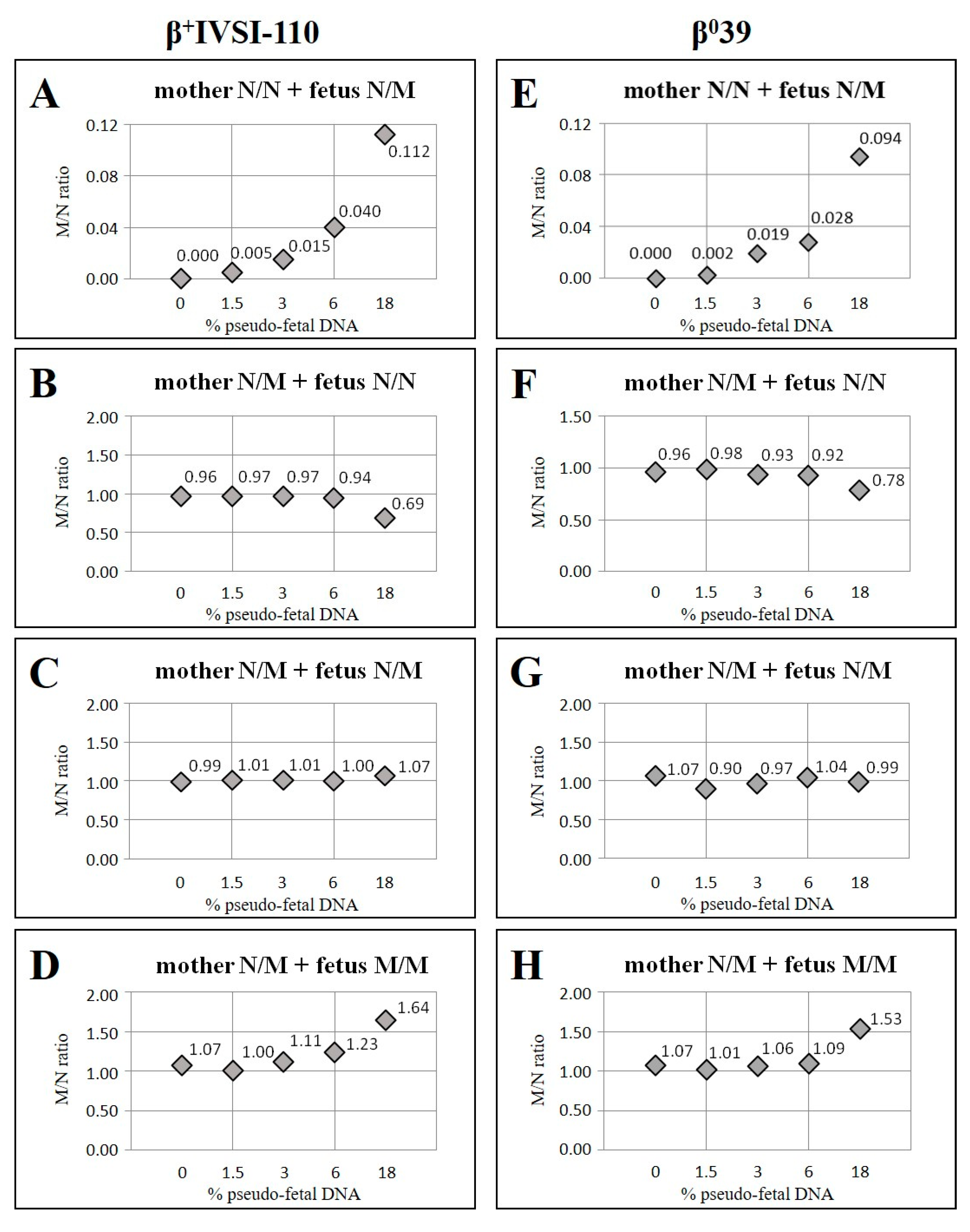
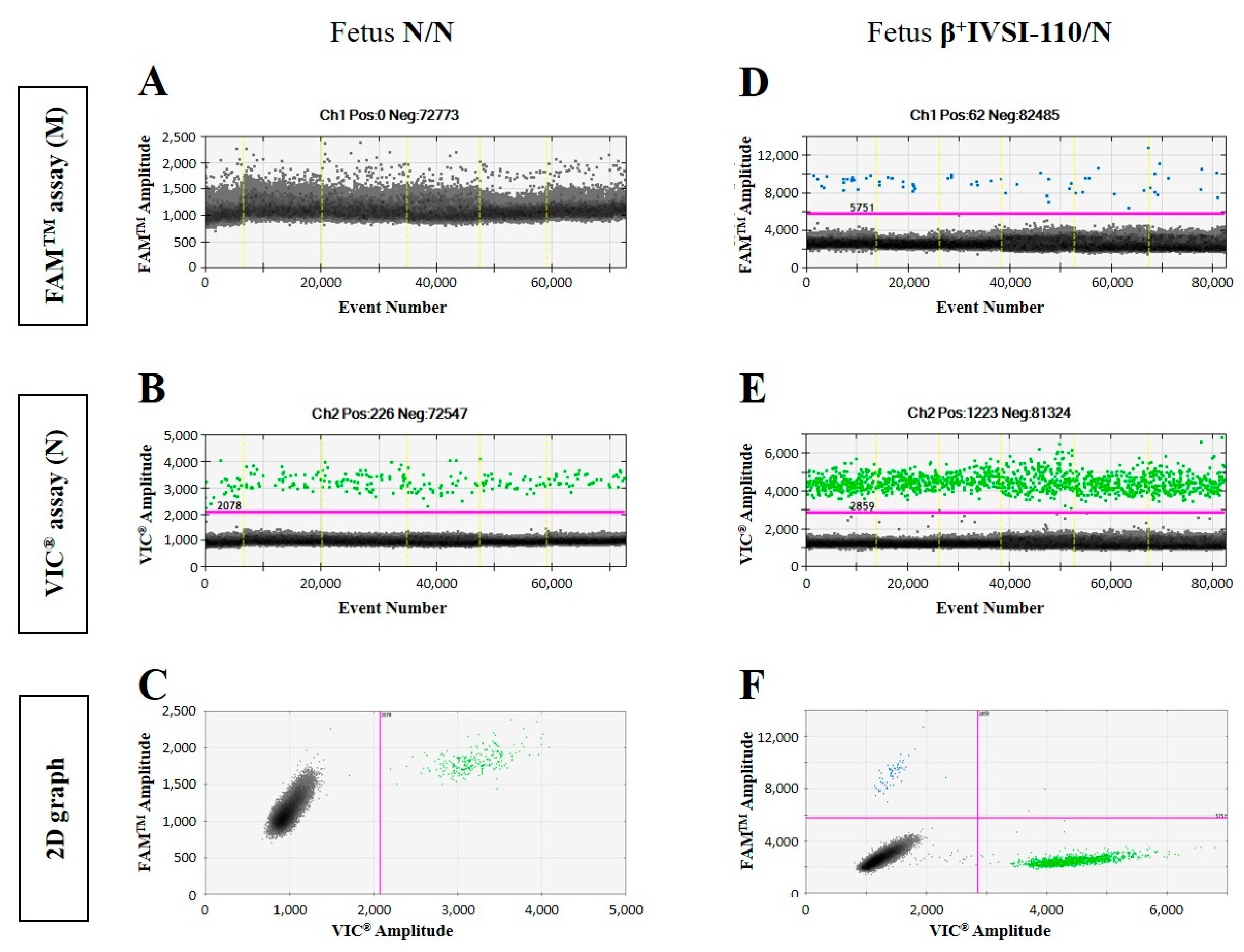
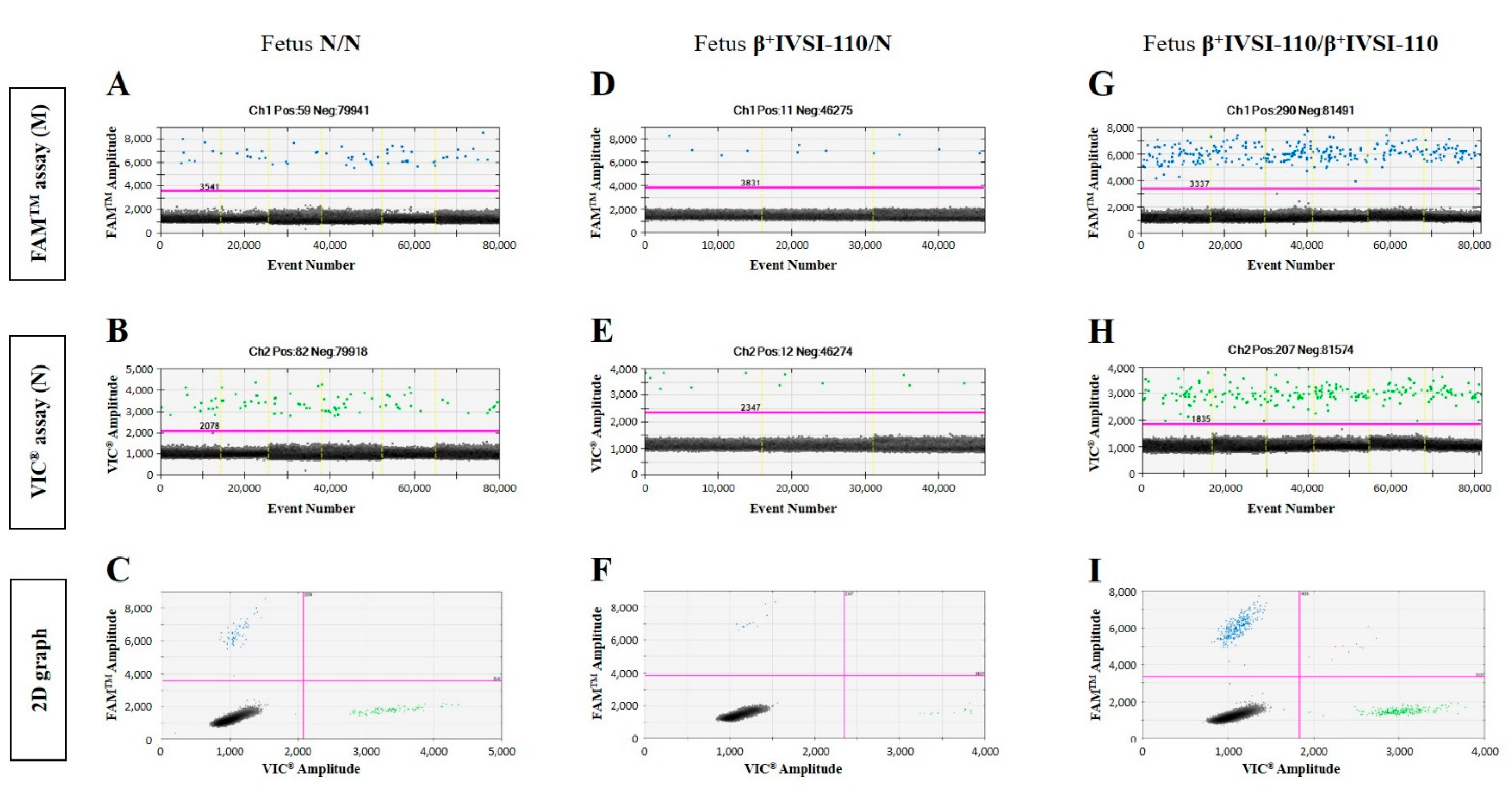
| Mother β+IVSI-110/N with Fetus N/N | Mother β039/N with Fetus N/N | ||||||
|---|---|---|---|---|---|---|---|
| Pseudo-Fetal DNA (%) | Expected M/N Ratio | Obtained M/N Ratio | Obt/Exp × 100 | Pseudo-Fetal DNA (%) | Expected M/N Ratio | Obtained M/N Ratio | Obt/Exp × 100 |
| 0 | 1.00 | 0.96 | 96 | 0 | 1.00 | 0.96 | 96 |
| 1.5 | 0.97 | 0.97 | 100 | 1.5 | 0.97 | 0.98 | 101 |
| 3 | 0.94 | 0.97 | 103 | 3 | 0.94 | 0.93 | 99 |
| 6 | 0.89 | 0.94 | 106 | 6 | 0.89 | 0.92 | 104 |
| 18 | 0.69 | 0.69 | 100 | 18 | 0.69 | 0.78 | 112 |
| Mother β+IVSI-110/N-with Fetus β+IVSI-110/β+IVSI 110 | Mother β039/N with Fetus β039/β039 | ||||||
| Pseudo-Fetal DNA (%) | Expected M/N Ratio | Obtained M/N Ratio | Obt/Exp × 100 | Pseudo-Fetal DNA (%) | Expected M/N Ratio | Obtained M/N Ratio | Obt/Exp × 100 |
| 0 | 1.00 | 1.07 | 107 | 0 | 1.00 | 1.07 | 107 |
| 1.5 | 1.03 | 1.00 | 97 | 1.5 | 1.03 | 1.01 | 98 |
| 3 | 1.06 | 1.11 | 105 | 3 | 1.06 | 1.06 | 100 |
| 6 | 1.13 | 1.23 | 109 | 6 | 1.13 | 1.09 | 97 |
| 18 | 1.44 | 1.64 | 114 | 18 | 1.44 | 1.53 | 106 |
| Mother β+IVSI-110/N with Fetus β+IVSI-110/N | Mother β039/N with Fetus β039/N | ||||||
| Pseudo-Fetal DNA (%) | Expected M/N Ratio | Obtained M/N Ratio | Obt/Exp × 100 | Pseudo-Fetal DNA (%) | Expected M/N Ratio | Obtained M/N Ratio | Obt/Exp × 100 |
| 0 | 1.00 | 0.99 | 99 | 0 | 1.00 | 1.07 | 107 |
| 1.5 | 1.00 | 1.01 | 101 | 1.5 | 1.00 | 0.90 | 90 |
| 3 | 1.00 | 1.01 | 101 | 3 | 1.00 | 0.97 | 97 |
| 6 | 1.00 | 1.00 | 100 | 6 | 1.00 | 1.04 | 104 |
| 18 | 1.00 | 1.07 | 107 | 18 | 1.00 | 0.99 | 99 |
| Pregnant Women N/N with Partner β+IVSI-110/N | Pregnant Women N/N with Partner β039/N | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample | Gestational Weeks | Positive Events for M Allele (no.) | Formulated Diagnosis | Fetal Genotype | Diagnosis Outcome | Sample | Gestational Weeks | Positive Events for M Allele (no.) | Formulated Diagnosis | Fetal Genotype | Diagnosis Outcome |
| 1 | 37 | 0 | N/N | N/N | √ | 25 | 39 | 196 | β039/N | β039/N | √ |
| 2 | 18 | 0 | N/N | N/N | √ | 26 | 35 | 123 | β039/N | β039/N | √ |
| 3 | 16 | 91 | β+IVSI-110/N | β+IVSI-110/N | √ | 27 | 33 | 39 | β039/N | β039/N | √ |
| 4 | 15 | 43 | β+IVSI-110/N | β+IVSI-110/N | √ | 28 | 29 | 19 | β039/N | β039/N | √ |
| 5 | 12 | 1 | N/N | β0IVSII-1/N | √ | 29 | 28 | 78 | β039/N | β039/N | √ |
| 6 | 12 | 12 | β+IVSI-110/N | β+IVSI-110/β039 | √ | 30 | 26 | 33 | β039/N | β039/N | √ |
| 7A | 10 | 178 | β+IVSI-110/N | β+IVSI-110/N | √ | 31 | 25 | 21 | β039/N | β039/N | √ |
| 8 | 9 | 62 | β+IVSI-110/N | β+IVSI-110/N | √ | 32 | 24 | 2 | N/N | N/N | √ |
| 7B | 5 | 3 | β+IVSI-110/N | β+IVSI-110/N | √ | 33 | 24 | 8 | β039/N | β039/N | √ |
| 34 | 21 | 39 | β039/N | β039/N | √ | ||||||
| 35 | 14 | 50 | β039/N | β039/N | √ | ||||||
| 36 | 13 | 4 | β039/N | β039/N | √ | ||||||
| 37 | 7 | 0 | N/N | N/N | √ | ||||||
| 38 | 5 | 26 | β039/N | β039/N | √ | ||||||
| Pregnant Women β+IVSI-110/N with Partner N/N | Pregnant Women β039/N with Partner N/N | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| # Sample | Gestational Weeks | M/N Ratio | z-Score | Formulated Diagnosis | Fetal Genotype (Diagnosis Outcome) | # Sample | Gestational Weeks | M/N Ratio | z-Score | Formulated Diagnosis | Fetal Genotype (Diagnosis Outcome) |
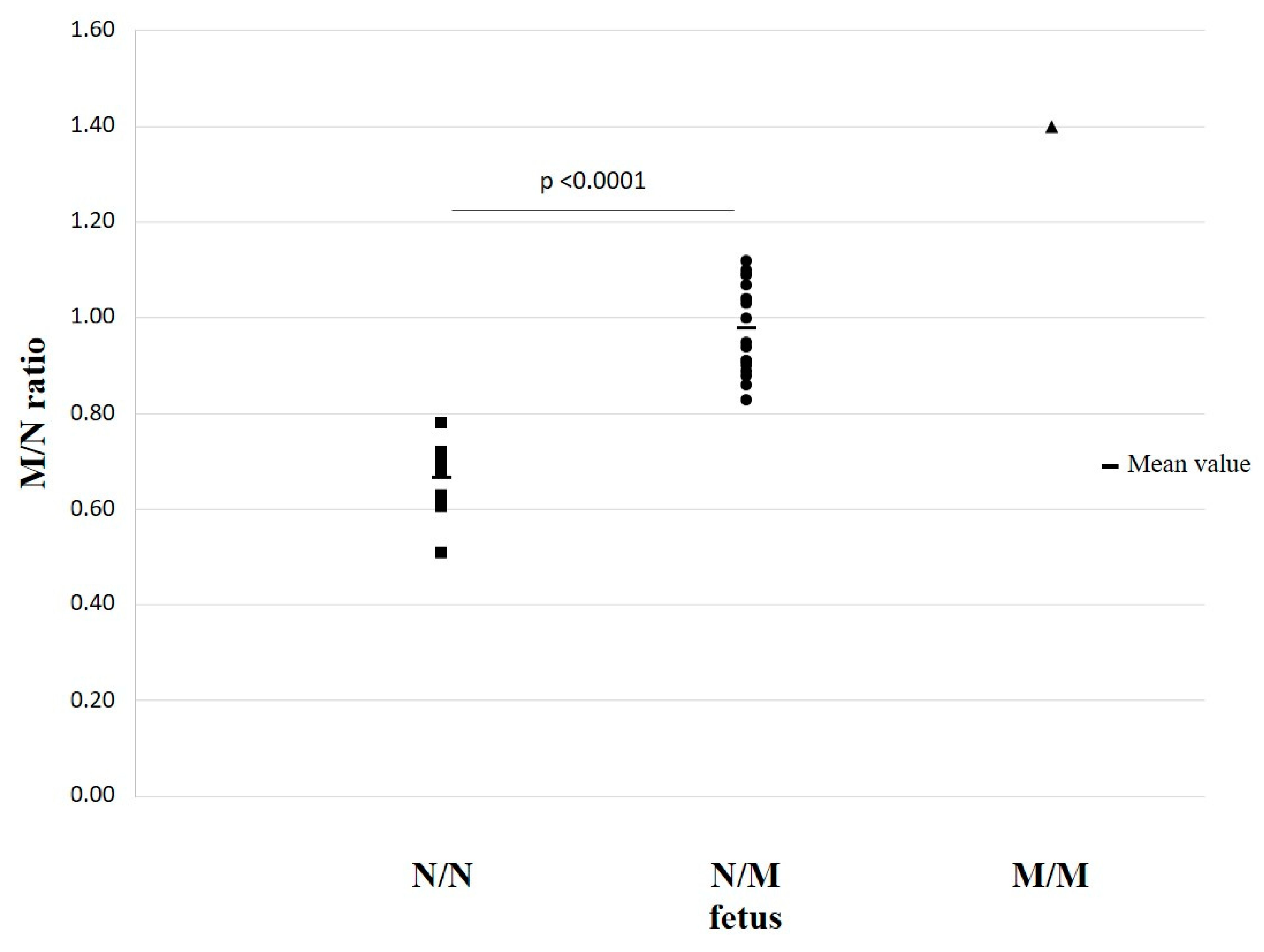
| 9 | 38 | 0.61 | −3.46 | N/N | N/N (√) | 39 | 39 | 1.09 | 0.52 | β039/N | β039/N (√) |
| 10 | 33 | 0.63 | −3.37 | N/N | N/N(√) | 40 | 36 | 0.89 | −1.19 | β039/N | β039/N (√) |
| 11 | 30 | 0.86 | −1.41 | β+IVSI-110/N | β+IVSI-110/N (√) | 41 | 29 | 0.71 | −2.66 | N/N | N/N (√) |
| 12 | 30 | 0.83 | −1.64 | β+IVSI-110/N | β+IVSI-110/N (√) | 42A | 26 | 1.00 | −0.25 | β039/N | β039/N (√) |
| 13 | 14 | 0.91 | −1.02 | β+IVSI-110/N | β+IVSI-110/N (√) | 43 | 26 | 0.51 | −4.36 | N/N | N/N (√) |
| 14 | 12 | 1.03 | 0.00 | β+IVSI-110/N | β+IVSI-110/N (√) | 42B | 22 | 1.09 | 0.49 | β039/N | β039/N (√) |
| 15 | 12 | 0.95 | −0.67 | β+IVSI-110/N | β+IVSI-110/N (√) | 44 | 20 | 0.68 | −2.93 | N/N | N/N (√) |
| 16 | 12 | 0.91 | −1.00 | β+IVSI-110/N | β+IVSI-110/N (√) | 45 | 18 | 0.78 | −2.06 | β039/N | N/N |
| 17 | 11 | 0.91 | −1.00 | β+IVSI-110/N | β+IVSI-110/N (√) | 46 | 14 | 0.63 | −3.31 | N/N | N/N (√) |
| 18 | 10 | 1.04 | 0.09 | β+IVSI-110/N | β+IVSI-110/N (√) | 42C | 13 | 1.12 | 0.75 | β039/N | β039/N (√) |
| 19 | 9 | 0.72 | −2.59 | N/N | N/N (√) | 6 * | 12 | 1.07 | 0.33 | β039/N | β039/β+IVSI-110 (√) |
| Pregnant Women β+IVSI-110/N with Partner β+IVSI-110/N | 47 | 9 | 0.88 | −1.28 | β039/N | β039/N (√) | |||||
| # Sample | Gestational Weeks | M/N Ratio | z-Score | Formulated Diagnosis | Fetal Genotype (Diagnosis Outcome) | 48 | 7 | 1.10 | 0.58 | β039/N | β039/N (√) |
| Pregnant Women β039/N with Partner β039/N | |||||||||||
| # Sample | Gestational Weeks | M/N Ratio | z-Score | Formulated Diagnosis | Fetal Genotype (Diagnosis Outcome) | ||||||
| 20 | 12 | 0.94 | −0.73 | β+IVSI-110/N | β+IVSI-110/N (√) | ||||||
| 21 | 10 | 0.70 | −2.71 | N/N | N/N (√) | ||||||
| 22 | 8 | 1.04 | 0.09 | β+IVSI-110/N | β+IVSI-110/N (√) | 49 | 32 | 0.69 | −2.82 | N/N | N/N (√) |
| 23 | 7 | 1.40 | 3.09 | β+IVSI-110/β+IVSI-110 | β+IVSI-110/β+IVSI-110 (√) | ||||||
| 24 | 7 | 0.90 | −0.94 | β+IVSI-110/N | β+IVSI-110/N (√) | ||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
D’Aversa, E.; Breveglieri, G.; Boutou, E.; Balassopoulou, A.; Voskaridou, E.; Pellegatti, P.; Guerra, G.; Scapoli, C.; Gambari, R.; Borgatti, M. Droplet Digital PCR for Non-Invasive Prenatal Detection of Fetal Single-Gene Point Mutations in Maternal Plasma. Int. J. Mol. Sci. 2022, 23, 2819. https://doi.org/10.3390/ijms23052819
D’Aversa E, Breveglieri G, Boutou E, Balassopoulou A, Voskaridou E, Pellegatti P, Guerra G, Scapoli C, Gambari R, Borgatti M. Droplet Digital PCR for Non-Invasive Prenatal Detection of Fetal Single-Gene Point Mutations in Maternal Plasma. International Journal of Molecular Sciences. 2022; 23(5):2819. https://doi.org/10.3390/ijms23052819
Chicago/Turabian StyleD’Aversa, Elisabetta, Giulia Breveglieri, Effrossyni Boutou, Angeliki Balassopoulou, Ersi Voskaridou, Patrizia Pellegatti, Giovanni Guerra, Chiara Scapoli, Roberto Gambari, and Monica Borgatti. 2022. "Droplet Digital PCR for Non-Invasive Prenatal Detection of Fetal Single-Gene Point Mutations in Maternal Plasma" International Journal of Molecular Sciences 23, no. 5: 2819. https://doi.org/10.3390/ijms23052819









