Application of High Throughput Technologies in the Development of Acute Myeloid Leukemia Therapy: Challenges and Progress
Abstract
:1. Introduction
2. High-Throughput Technologies-Based Targeted Therapies
2.1. Integration of Genomice Sequencing
2.2. Integration of Proteomics
2.3. Integration of Metabolomics
3. High-Throughput Drug Screening-Based Therapies
3.1. Drug Sensitivity Testing (DST)
3.2. Data Analysis
4. Artificial Intelligence (AI) in Cancer Therapy
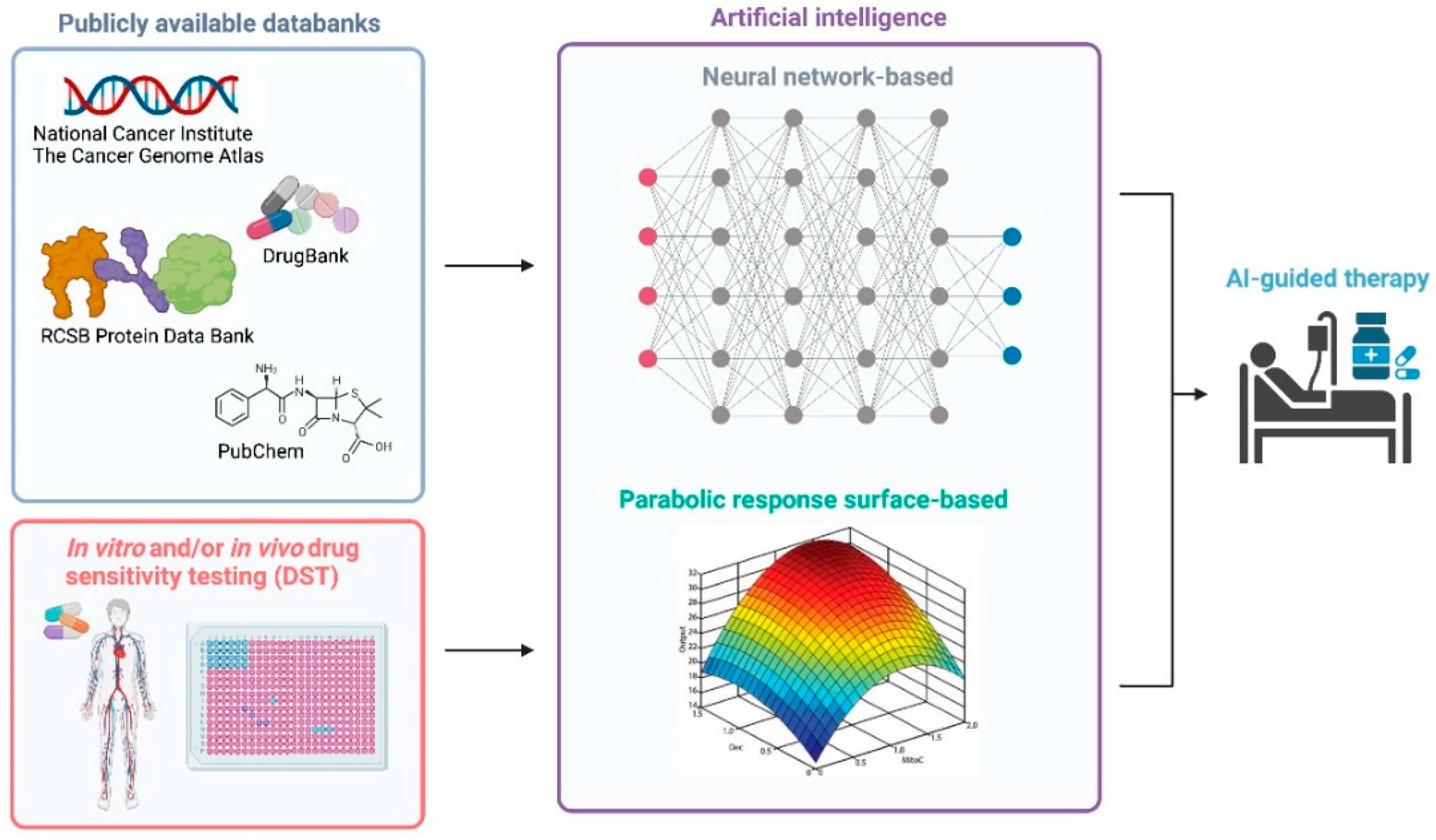
5. A Streamlined Approach of Integrating Genomic Sequencing, HTS-Based DST and AI Technologies for Personalized Treatment in AML
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Roboz, G.J. Novel approaches to the treatment of acute myeloid leukemia. Hematol. Am. Soc. Hematol. Educ. Program 2011, 2011, 43–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papaemmanuil, E.; Gerstung, M.; Bullinger, L.; Gaidzik, V.I.; Paschka, P.; Roberts, N.D.; Potter, N.E.; Heuser, M.; Thol, F.; Bolli, N.; et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. N. Engl. J. Med. 2016, 374, 2209–2221. [Google Scholar] [CrossRef] [PubMed]
- Arber, D.A.; Orazi, A.; Hasserjian, R.; Thiele, J.; Borowitz, M.J.; Le Beau, M.M.; Bloomfield, C.D.; Cazzola, M.; Vardiman, J.W. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 2016, 127, 2391–2405. [Google Scholar] [CrossRef] [PubMed]
- Tyner, J.W.; Tognon, C.E.; Bottomly, D.; Wilmot, B.; Kurtz, S.E.; Savage, S.L.; Long, N.; Schultz, A.R.; Traer, E.; Abel, M.; et al. Functional genomic landscape of acute myeloid leukaemia. Nature 2018, 562, 526–531. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research, N.; Ley, T.J.; Miller, C.; Ding, L.; Raphael, B.J.; Mungall, A.J.; Robertson, A.; Hoadley, K.; Triche, T.J., Jr.; Laird, P.W.; et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 2013, 368, 2059–2074. [Google Scholar] [CrossRef] [Green Version]
- Dohner, H.; Gaidzik, V.I. Impact of genetic features on treatment decisions in AML. Hematol. Am. Soc. Hematol. Educ. Program 2011, 2011, 36–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patel, J.P.; Gonen, M.; Figueroa, M.E.; Fernandez, H.; Sun, Z.; Racevskis, J.; Van Vlierberghe, P.; Dolgalev, I.; Thomas, S.; Aminova, O.; et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N. Engl. J. Med. 2012, 366, 1079–1089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, S.; Cho, B.S.; Kim, H.J. New agents in acute myeloid leukemia (AML). Blood Res 2020, 55 (Suppl. S1), S14–S18. [Google Scholar] [CrossRef]
- DiNardo, C.D.; Cortes, J.E. Mutations in AML: Prognostic and therapeutic implications. Hematol. Am. Soc. Hematol. Educ. Program 2016, 2016, 348–355. [Google Scholar] [CrossRef] [Green Version]
- Lohse, I.; Statz-Geary, K.; Brothers, S.P.; Wahlestedt, C. Precision medicine in the treatment stratification of AML patients: Challenges and progress. Oncotarget 2018, 9, 37790–37797. [Google Scholar] [CrossRef] [Green Version]
- Ho, D. Artificial intelligence in cancer therapy. Science 2020, 367, 982–983. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.W.; Yan, X.J.; Zhou, Z.R.; Yang, F.F.; Wu, Z.Y.; Sun, H.B.; Liang, W.X.; Song, A.X.; Lallemand-Breitenbach, V.; Jeanne, M.; et al. Arsenic trioxide controls the fate of the PML-RARalpha oncoprotein by directly binding PML. Science 2010, 328, 240–243. [Google Scholar] [CrossRef]
- Zhou, G.B.; Zhao, W.L.; Wang, Z.Y.; Chen, S.J.; Chen, Z. Retinoic acid and arsenic for treating acute promyelocytic leukemia. PLoS Med. 2005, 2, e12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, R.; Levis, M.; Piloto, O.; Brown, P.; Baldwin, B.R.; Gorin, N.C.; Beran, M.; Zhu, Z.; Ludwig, D.; Hicklin, D.; et al. FLT3 ligand causes autocrine signaling in acute myeloid leukemia cells. Blood 2004, 103, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Gilliland, D.G.; Griffin, J.D. The roles of FLT3 in hematopoiesis and leukemia. Blood 2002, 100, 1532–1542. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, Y.; Kiyoi, H.; Nakano, Y.; Suzuki, R.; Kodera, Y.; Miyawaki, S.; Asou, N.; Kuriyama, K.; Yagasaki, F.; Shimazaki, C.; et al. Activating mutation of D835 within the activation loop of FLT3 in human hematologic malignancies. Blood 2001, 97, 2434–2439. [Google Scholar] [CrossRef] [Green Version]
- Abbas, H.A.; Alfayez, M.; Kadia, T.; Ravandi-Kashani, F.; Daver, N. Midostaurin In Acute Myeloid Leukemia: An Evidence-Based Review And Patient Selection. Cancer Manag. Res. 2019, 11, 8817–8828. [Google Scholar] [CrossRef] [Green Version]
- Lim, S.H.; Dubielecka, P.M.; Raghunathan, V.M. Molecular targeting in acute myeloid leukemia. J. Transl. Med. 2017, 15, 183. [Google Scholar] [CrossRef]
- Wouters, B.J.; Delwel, R. Epigenetics and approaches to targeted epigenetic therapy in acute myeloid leukemia. Blood 2016, 127, 42–52. [Google Scholar] [CrossRef] [Green Version]
- Plass, C.; Oakes, C.; Blum, W.; Marcucci, G. Epigenetics in acute myeloid leukemia. Semin. Oncol. 2008, 35, 378–387. [Google Scholar] [CrossRef] [Green Version]
- Pleyer, L.; Dohner, H.; Dombret, H.; Seymour, J.F.; Schuh, A.C.; Beach, C.L.; Swern, A.S.; Burgstaller, S.; Stauder, R.; Girschikofsky, M.; et al. Azacitidine for Front-Line Therapy of Patients with AML: Reproducible Efficacy Established by Direct Comparison of International Phase 3 Trial Data with Registry Data from the Austrian Azacitidine Registry of the AGMT Study Group. Int. J. Mol. Sci. 2017, 18, 415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ragon, B.K.; Daver, N.; Garcia-Manero, G.; Ravandi, F.; Cortes, J.; Kadia, T.; Oran, B.; Ohanian, M.; Ferrajoli, A.; Pemmaraju, N.; et al. Minimal residual disease eradication with epigenetic therapy in core binding factor acute myeloid leukemia. Am. J. Hematol. 2017, 92, 845–850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirschbaum, M.; Gojo, I.; Goldberg, S.L.; Bredeson, C.; Kujawski, L.A.; Yang, A.; Marks, P.; Frankel, P.; Sun, X.; Tosolini, A.; et al. A phase 1 clinical trial of vorinostat in combination with decitabine in patients with acute myeloid leukaemia or myelodysplastic syndrome. Br. J. Haematol. 2014, 167, 185–193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DiNardo, C.D.; Stein, E.M.; de Botton, S.; Roboz, G.J.; Altman, J.K.; Mims, A.S.; Swords, R.; Collins, R.H.; Mannis, G.N.; Pollyea, D.A.; et al. Durable Remissions with Ivosidenib in IDH1-Mutated Relapsed or Refractory AML. N. Engl. J. Med. 2018, 378, 2386–2398. [Google Scholar] [CrossRef]
- Stein, E.M.; DiNardo, C.D.; Fathi, A.T.; Pollyea, D.A.; Stone, R.M.; Altman, J.K.; Roboz, G.J.; Patel, M.R.; Collins, R.; Flinn, I.W.; et al. Molecular remission and response patterns in patients with mutant-IDH2 acute myeloid leukemia treated with enasidenib. Blood 2019, 133, 676–687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schaab, C.; Oppermann, F.S.; Klammer, M.; Pfeifer, H.; Tebbe, A.; Oellerich, T.; Krauter, J.; Levis, M.; Perl, A.E.; Daub, H.; et al. Global phosphoproteome analysis of human bone marrow reveals predictive phosphorylation markers for the treatment of acute myeloid leukemia with quizartinib. Leukemia 2014, 28, 716–719. [Google Scholar] [CrossRef] [Green Version]
- Hu, J.; Lin, M.; Liu, T.; Li, J.; Chen, B.; Chen, Y. DIGE-based proteomic analysis identifies nucleophosmin/B23 and nucleolin C23 as over-expressed proteins in relapsed/refractory acute leukemia. Leuk. Res. 2011, 35, 1087–1092. [Google Scholar] [CrossRef] [PubMed]
- Jiang, N.; Kham, S.K.; Koh, G.S.; Suang Lim, J.Y.; Ariffin, H.; Chew, F.T.; Yeoh, A.E. Identification of prognostic protein biomarkers in childhood acute lymphoblastic leukemia (ALL). J Proteomics 2011, 74, 843–857. [Google Scholar] [CrossRef] [PubMed]
- Alcolea, M.P.; Casado, P.; Rodriguez-Prados, J.C.; Vanhaesebroeck, B.; Cutillas, P.R. Phosphoproteomic analysis of leukemia cells under basal and drug-treated conditions identifies markers of kinase pathway activation and mechanisms of resistance. Mol. Cell. Proteom. MCP 2012, 11, 453–466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kornblau, S.M.; Thall, P.F.; Estrov, Z.; Walterscheid, M.; Patel, S.; Theriault, A.; Keating, M.J.; Kantarjian, H.; Estey, E.; Andreeff, M. The prognostic impact of BCL2 protein expression in acute myelogenous leukemia varies with cytogenetics. Clin. Cancer Res. 1999, 5, 1758–1766. [Google Scholar]
- Konopleva, M.; Letai, A. BCL-2 inhibition in AML: An unexpected bonus? Blood 2018, 132, 1007–1012. [Google Scholar] [CrossRef] [Green Version]
- Wei, A.H.; Strickland, S.A., Jr.; Hou, J.Z.; Fiedler, W.; Lin, T.L.; Walter, R.B.; Enjeti, A.; Tiong, I.S.; Savona, M.; Lee, S.; et al. Venetoclax Combined with Low-Dose Cytarabine for Previously Untreated Patients With Acute Myeloid Leukemia: Results From a Phase Ib/II Study. J. Clin. Oncol. 2019, 37, 1277–1284. [Google Scholar] [CrossRef] [PubMed]
- Cortes, J.E.; Dombret, H.; Merchant, A.; Tauchi, T.; DiRienzo, C.G.; Sleight, B.; Zhang, X.; Leip, E.P.; Shaik, N.; Bell, T.; et al. Glasdegib plus intensive/nonintensive chemotherapy in untreated acute myeloid leukemia: BRIGHT AML 1019 Phase III trials. Future Oncol. 2019, 15, 3531–3545. [Google Scholar] [CrossRef] [Green Version]
- Hoff, F.W.; Hu, C.W.; Qutub, A.A.; de Bont, E.; Horton, T.M.; Kornblau, S.M. Shining a light on cell signaling in leukemia through proteomics: Relevance for the clinic. Expert Rev. Proteom. 2018, 15, 613–622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van der Zwet, J.C.G.; Cordo, V.; Cante-Barrett, K.; Meijerink, J.P.P. Multi-omic approaches to improve outcome for T-cell acute lymphoblastic leukemia patients. Adv. Biol. Regul. 2019, 74, 100647. [Google Scholar] [CrossRef]
- Luengo, A.; Gui, D.Y.; Vander Heiden, M.G. Targeting Metabolism for Cancer Therapy. Cell Chem. Biol. 2017, 24, 1161–1180. [Google Scholar] [CrossRef] [Green Version]
- Lagadinou, E.D.; Sach, A.; Callahan, K.; Rossi, R.M.; Neering, S.J.; Minhajuddin, M.; Ashton, J.M.; Pei, S.; Grose, V.; O’Dwyer, K.M.; et al. BCL-2 inhibition targets oxidative phosphorylation and selectively eradicates quiescent human leukemia stem cells. Cell Stem Cell 2013, 12, 329–341. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.L.; Wang, J.H.; Zhao, A.H.; Xu, X.; Wang, Y.H.; Chen, T.L.; Li, J.M.; Mi, J.Q.; Zhu, Y.M.; Liu, Y.F.; et al. A distinct glucose metabolism signature of acute myeloid leukemia with prognostic value. Blood 2014, 124, 1645–1654. [Google Scholar] [CrossRef] [PubMed]
- Sykes, D.B.; Kfoury, Y.S.; Mercier, F.E.; Wawer, M.J.; Law, J.M.; Haynes, M.K.; Lewis, T.A.; Schajnovitz, A.; Jain, E.; Lee, D.; et al. Inhibition of Dihydroorotate Dehydrogenase Overcomes Differentiation Blockade in Acute Myeloid Leukemia. Cell 2016, 167, 171–186.e15. [Google Scholar] [CrossRef] [Green Version]
- Loew, A.; Kohnke, T.; Rehbeil, E.; Pietzner, A.; Weylandt, K.H. A Role for Lipid Mediators in Acute Myeloid Leukemia. Int. J. Mol. Sci. 2019, 20, 2425. [Google Scholar] [CrossRef] [Green Version]
- Wojcicki, A.V.; Kasowski, M.M.; Sakamoto, K.M.; Lacayo, N. Metabolomics in acute myeloid leukemia. Mol. Genet. Metab. 2020, 130, 230–238. [Google Scholar] [CrossRef] [PubMed]
- Stevens, V.L.; Hoover, E.; Wang, Y.; Zanetti, K.A. Pre-Analytical Factors that Affect Metabolite Stability in Human Urine, Plasma, and Serum: A Review. Metabolites 2019, 9, 156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chia, S.; Low, J.L.; Zhang, X.; Kwang, X.L.; Chong, F.T.; Sharma, A.; Bertrand, D.; Toh, S.Y.; Leong, H.S.; Thangavelu, M.T.; et al. Phenotype-driven precision oncology as a guide for clinical decisions one patient at a time. Nat. Commun. 2017, 8, 435. [Google Scholar] [CrossRef] [PubMed]
- Swords, R.T.; Azzam, D.; Al-Ali, H.; Lohse, I.; Volmar, C.H.; Watts, J.M.; Perez, A.; Rodriguez, A.; Vargas, F.; Elias, R.; et al. Ex-vivo sensitivity profiling to guide clinical decision making in acute myeloid leukemia: A pilot study. Leuk. Res. 2018, 64, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Pemovska, T.; Kontro, M.; Yadav, B.; Edgren, H.; Eldfors, S.; Szwajda, A.; Almusa, H.; Bespalov, M.M.; Ellonen, P.; Elonen, E.; et al. Individualized systems medicine strategy to tailor treatments for patients with chemorefractory acute myeloid leukemia. Cancer Discov. 2013, 3, 1416–1429. [Google Scholar] [CrossRef] [Green Version]
- Drenberg, C.D.; Shelat, A.; Dang, J.; Cotton, A.; Orwick, S.J.; Li, M.; Jeon, J.Y.; Fu, Q.; Buelow, D.R.; Pioso, M.; et al. A high-throughput screen indicates gemcitabine and JAK inhibitors may be useful for treating pediatric AML. Nat. Commun. 2019, 10, 2189. [Google Scholar] [CrossRef] [PubMed]
- Sweeney, S.R.; Collins, M.; Pandey, R.; Chiou, J.; Lodi, A.; Tiziani, S. Identification of a synergistic combination of dimethylaminoparthenolide and shikonin alters metabolism and inhibits proliferation of pediatric precursor-B cell acute lymphoblastic leukemia. Mol. Carcinog. 2020, 59, 399–411. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, S.E.; Eide, C.A.; Kaempf, A.; Khanna, V.; Savage, S.L.; Rofelty, A.; English, I.; Ho, H.; Pandya, R.; Bolosky, W.J.; et al. Molecularly targeted drug combinations demonstrate selective effectiveness for myeloid- and lymphoid-derived hematologic malignancies. Proc. Natl. Acad. Sci. USA 2017, 114, E7554–E7563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spinner, M.A.; Aleshin, A.; Santaguida, M.T.; Schaffert, S.A.; Zehnder, J.L.; Patterson, A.S.; Gekas, C.; Heiser, D.; Greenberg, P.L. Ex vivo drug screening defines novel drug sensitivity patterns for informing personalized therapy in myeloid neoplasms. Blood Adv. 2020, 4, 2768–2778. [Google Scholar] [CrossRef]
- Ding, M.; Kaspersson, K.; Murray, D.; Bardelle, C. High-throughput flow cytometry for drug discovery: Principles, applications, and case studies. Drug Discov. Today 2017, 22, 1844–1850. [Google Scholar] [CrossRef] [PubMed]
- Lewis, T.A.; Sykes, D.B.; Law, J.M.; Munoz, B.; Rustiguel, J.K.; Nonato, M.C.; Scadden, D.T.; Schreiber, S.L. Development of ML390: A Human DHODH Inhibitor That Induces Differentiation in Acute Myeloid Leukemia. ACS Med. Chem. Lett. 2016, 7, 1112–1117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yadav, B.; Pemovska, T.; Szwajda, A.; Kulesskiy, E.; Kontro, M.; Karjalainen, R.; Majumder, M.M.; Malani, D.; Murumagi, A.; Knowles, J.; et al. Quantitative scoring of differential drug sensitivity for individually optimized anticancer therapies. Sci. Rep. 2014, 4, 5193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryall, K.A.; Shin, J.; Yoo, M.; Hinz, T.K.; Kim, J.; Kang, J.; Heasley, L.E.; Tan, A.C. Identifying kinase dependency in cancer cells by integrating high-throughput drug screening and kinase inhibition data. Bioinformatics 2015, 31, 3799–3806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, B.; He, H.; Luo, H.; Zhang, T.; Jiang, J. Artificial intelligence and big data facilitated targeted drug discovery. Stroke Vasc. Neurol. 2019, 4, 206–213. [Google Scholar] [CrossRef] [Green Version]
- Menden, M.P.; Iorio, F.; Garnett, M.; McDermott, U.; Benes, C.H.; Ballester, P.J.; Saez-Rodriguez, J. Machine learning prediction of cancer cell sensitivity to drugs based on genomic and chemical properties. PLoS ONE 2013, 8, e61318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rashid, M.; Toh, T.B.; Hooi, L.; Silva, A.; Zhang, Y.; Tan, P.F.; Teh, A.L.; Karnani, N.; Jha, S.; Ho, C.M.; et al. Optimizing drug combinations against multiple myeloma using a quadratic phenotypic optimization platform (QPOP). Sci. Transl. Med. 2018, 10, eaan0941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Mel, S.; Rashid, M.B.M.; Zhang, X.Y.; Goh, J.; Lee, C.T.; Poon, L.M.; Chan, E.H.L.; Liu, X.; Chng, W.J.; Chee, Y.L.; et al. Application of an ex-vivo drug sensitivity platform towards achieving complete remission in a refractory T-cell lymphoma. Blood Cancer J. 2020, 10, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blasiak, A.; Khong, J.; Kee, T. CURATE.AI: Optimizing Personalized Medicine with Artificial Intelligence. SLAS Technol. 2020, 25, 95–105. [Google Scholar] [CrossRef]
- Zarrinpar, A.; Lee, D.K.; Silva, A.; Datta, N.; Kee, T.; Eriksen, C.; Weigle, K.; Agopian, V.; Kaldas, F.; Farmer, D.; et al. Individualizing liver transplant immunosuppression using a phenotypic personalized medicine platform. Sci. Transl. Med. 2016, 8, 333ra49. [Google Scholar] [CrossRef] [Green Version]
- Pantuck, A.J.; Lee, D.K.; Kee, T.; Wang, P.; Lakhotia, S.; Silverman, M.H.; Mathis, C.; Drakaki, A.; Belldegrun, A.S.; Ho, C.M.J.A.T. Modulating BET Bromodomain inhibitor ZEN-3694 and enzalutamide combination dosing in a metastatic prostate cancer patient using CURATE. AI, an artificial intelligence platform. Adv. Ther. 2018, 1, 1800104. [Google Scholar] [CrossRef]
- Johnson, A.; Zeng, J.; Bailey, A.M.; Holla, V.; Litzenburger, B.; Lara-Guerra, H.; Mills, G.B.; Mendelsohn, J.; Shaw, K.R.; Meric-Bernstam, F. The right drugs at the right time for the right patient: The MD Anderson precision oncology decision support platform. Drug Discov. Today 2015, 20, 1433–1438. [Google Scholar] [CrossRef] [PubMed]
- Burd, A.; Levine, R.L.; Ruppert, A.S.; Mims, A.S.; Borate, U.; Stein, E.M.; Patel, P.; Baer, M.R.; Stock, W.; Deininger, M.; et al. Precision medicine treatment in acute myeloid leukemia using prospective genomic profiling: Feasibility and preliminary efficacy of the Beat AML Master Trial. Nat. Med. 2020, 26, 1852–1858. [Google Scholar] [CrossRef] [PubMed]


| Study Name | Approaches | Cancer Type | Outcome | Year | Reference |
|---|---|---|---|---|---|
| Ex vivo drug screening defines novel drug sensitivity patterns for informing personalized therapy in myeloid neoplasms | DST-based HTS | MDS | The platform had a positive predictive value of 0.92, negative predictive value of 0.82, and overall accuracy of 0.85. | 2020 | [49] |
| Application of an ex-vivo drug sensitivity platform towards achieving complete remission in a refractory T-cell lymphoma | QPOP Co-clinical trial | T-cell lymphoma | Patient achieved CR with an actionable drug combination identified within one week of sample collection | 2020 | [57] |
| Ex Vivo Drug Sensitivity Testing and Mutation Profiling | DST-based HTS Genome sequencing | Solid Tumors and Leukemias | Ongoing clinical trial | 2019 | ClinicalTrials.gov Identifier: NCT03860376 |
| Precision medicine treatment in acute myeloid leukemia using prospective genomic profiling: feasibility and preliminary efficacy of the Beat AML Master Trial | Genome sequencing | AML | Thirty-day mortality was less frequent and overall survival was significantly longer for patients enrolled on the Beat AML sub-studies versus those who elected SOC | 2017 | [62] |
| Phenotype-driven precision oncology as a guide for clinical decisions one patient at a time | DST-based HTS Co-clinical trial | head and neck squamous cell carcinomas | Can guide real-time therapeutic decisions | 2017 | [43] |
| Beat AML Core Study | genome sequencing | AML | Not available | 2016–2020 | ClinicalTrials.gov Identifier: NCT02927106 |
| High Throughput Drug Sensitivity Assay and Genomics- Guided Treatment of Patients With Relapsed or Refractory Acute Leukemia | DST-based HTS genome sequencing | AML | Ongoing clinical trial | 2015 | ClinicalTrials.gov Identifier: NCT02551718 |
| A distinct glucose metabolism signature of acute myeloid leukemia with prognostic value | Metabolomic profiling with GC-TOFMS. | AML | Suggests the use of serum metabolites and metabolic pathways as prognostic markers and potential therapeutic targets for AML | 2014 | [38] |
| Global phosphoproteome analysis of human bone marrow reveals predictive phosphorylation markers for the treatment of acute myeloid leukemia with quizartinib. | MS based- phosphoproteome analysis | AML | A signature consisting of five phosphorylation sites predicted the response to quizartinib in AML patients | 2014 | [26] |
| Individualized systems medicine strategy to tailor treatments for patients with chemorefractory acute myeloid leukemia | DST-based HTS genome sequencing Co-clinical trial | AML | Can predict clinical responses | 2013 | [45] |
| Treatment for Relapsed/Refractory AML Based on a High Throughput Drug Sensitivity Assay | DST-based HTS | AML | Total 9 treated patients 1 CR with MRD 2 CRi | 2013 | ClinicalTrials.gov Identifier: NCT01872819 |
| Phosphoproteomic analysis of leukemia cells under basal and drug-treated conditions identifies markers of kinase pathway activation and mechanisms of resistance | LC-MS/MS-based phosphoproteomic analysis | AML | Provides valuable information to personalize therapies based on kinase inhibitors | 2012 | [29] |
| DIGE-based proteomic analysis identifies nucleophosmin/B23 and nucleolin C23 as over-expressed proteins in relapsed/refractory acute leukemia | DIGE-based proteomic analysis | AML | Upregulation of B23 and C23 could be related to resistance of leukemia | 2011 | [27] |
| Identification of prognostic protein biomarkers in childhood acute lymphoblastic leukemia | Proteomic analysis | AML | PCNA as highly predictive of prednisolone response in patients | 2011 | [28] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiang, W.; Lam, Y.H.; Periyasamy, G.; Chuah, C. Application of High Throughput Technologies in the Development of Acute Myeloid Leukemia Therapy: Challenges and Progress. Int. J. Mol. Sci. 2022, 23, 2863. https://doi.org/10.3390/ijms23052863
Xiang W, Lam YH, Periyasamy G, Chuah C. Application of High Throughput Technologies in the Development of Acute Myeloid Leukemia Therapy: Challenges and Progress. International Journal of Molecular Sciences. 2022; 23(5):2863. https://doi.org/10.3390/ijms23052863
Chicago/Turabian StyleXiang, Wei, Yi Hui Lam, Giridharan Periyasamy, and Charles Chuah. 2022. "Application of High Throughput Technologies in the Development of Acute Myeloid Leukemia Therapy: Challenges and Progress" International Journal of Molecular Sciences 23, no. 5: 2863. https://doi.org/10.3390/ijms23052863
APA StyleXiang, W., Lam, Y. H., Periyasamy, G., & Chuah, C. (2022). Application of High Throughput Technologies in the Development of Acute Myeloid Leukemia Therapy: Challenges and Progress. International Journal of Molecular Sciences, 23(5), 2863. https://doi.org/10.3390/ijms23052863





