A Deeper Insight in Metal Binding to the hCtr1 N-terminus Fragment: Affinity, Speciation and Binding Mode of Binuclear Cu2+ and Mononuclear Ag+ Complex Species
Abstract
:1. Introduction
2. Results and Discussion
2.1. Ctr1(1-14) Can Bind Two Cu2+ Equivalents
2.2. Cu2+ Complexes with Ctr1(1-14) Oxidize Ascorbic Acid When Binuclear Species Form
2.3. Ag+ Forms Mononuclear Complex with Ctr1(1-14)
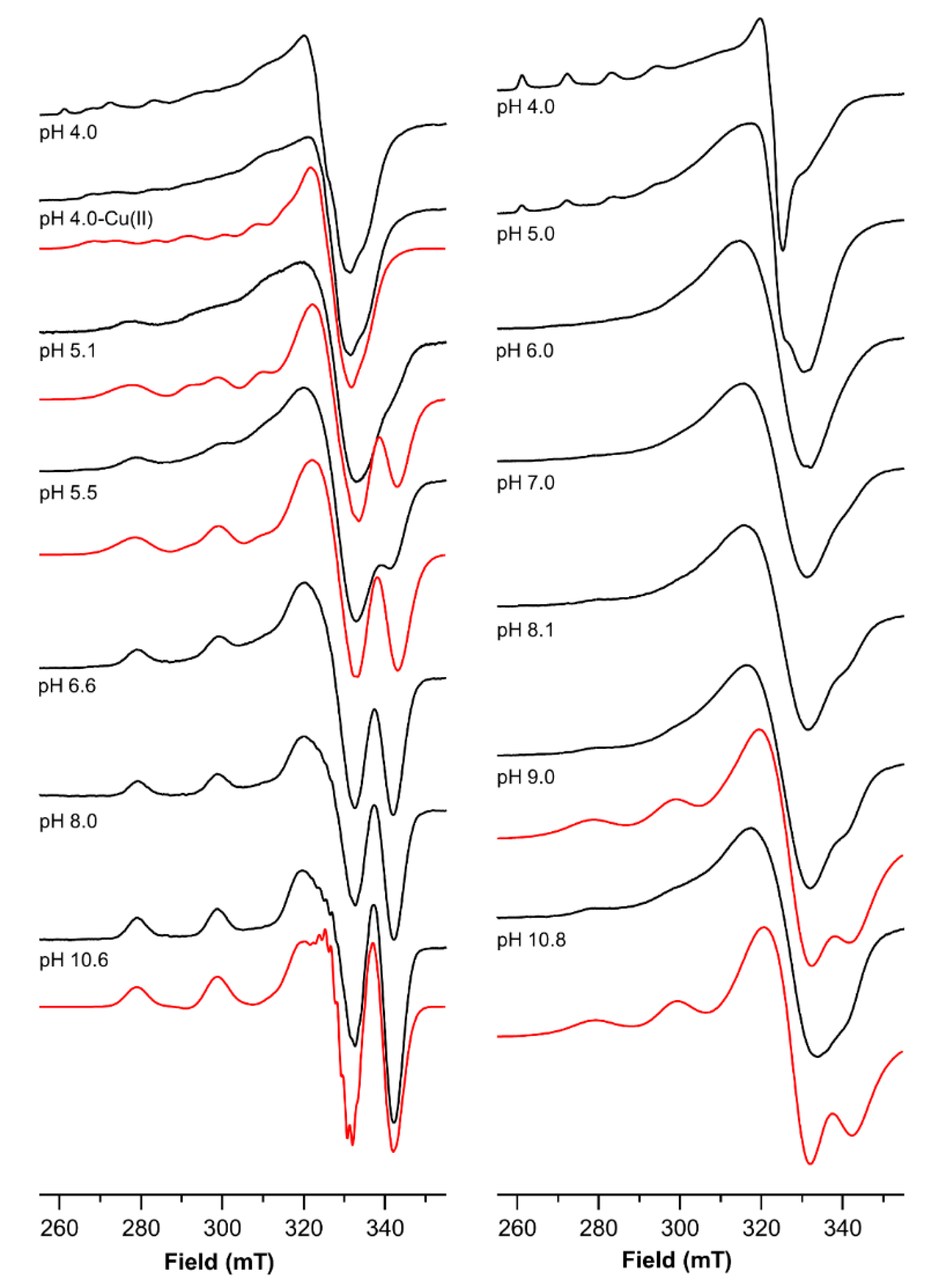
2.4. The Magnetic Parameters Values Obtained by Electro Spin Resonance (ESR) Measurements Indicate That the Two Binding Sites of Ctr1(1-14) Experience a Different Coordination Environment
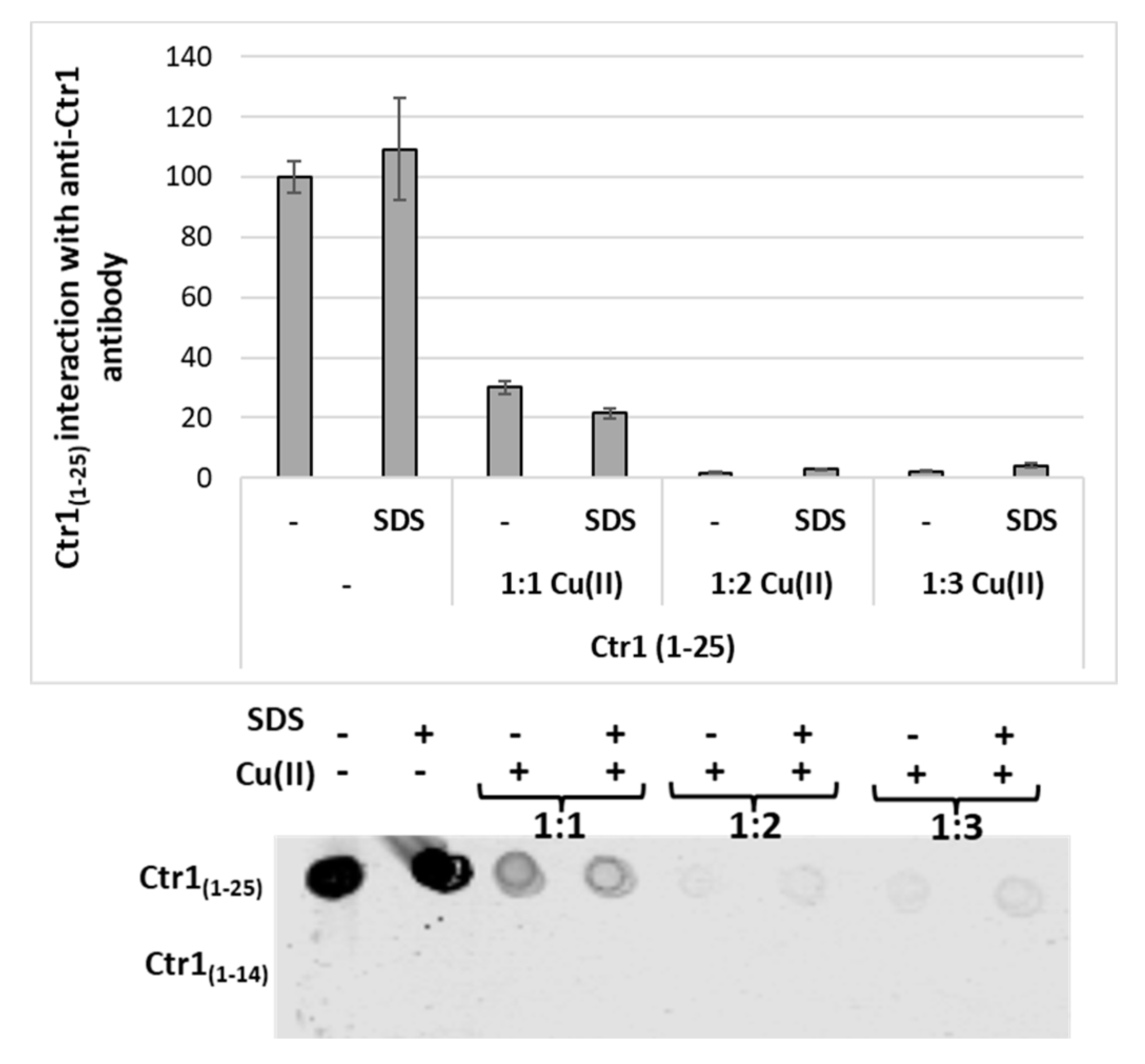
2.5. Immunoreactivity of Ctr1 Peptides of Different Length with Anti-Ctr1 Extracellular Domain Antibody
2.6. Concluding Remarks
3. Materials and Methods
3.1. Chemicals
3.2. Electron Spin Resonance Spectroscopy (ESR)
3.3. Potentiometric Titrations
3.4. Ultraviolet-visible (UV-vis) Measurements
3.5. Circular Dichroism (CD) Experiments
3.6. Ultraviolet (UV) Determination of Ascorbate Consumption
3.7. Dot Blot Assay
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Linder, M.C. Introduction and Overview of Copper as an Element Essential for Life. In Biochemistry of Copper; Springer: Boston, MA, USA, 1991. [Google Scholar]
- Uriu-Adams, J.Y.; Keen, C.L. Copper, oxidative stress, and human health. Mol. Asp. Med. 2005, 26, 268–298. [Google Scholar] [CrossRef]
- Valko, M.; Jomova, K.; Rhodes, C.J.; Kuča, K.; Musílek, K. Redox- and non-redox-metal-induced formation of free radicals and their role in human disease. Arch. Toxicol. 2016, 90, 1–37. [Google Scholar] [CrossRef]
- Maung, M.T.; Carlson, A.; Olea-Flores, M.; Elkhadragy, L.; Schachtschneider, K.M.; Navarro-Tito, N.; Padilla-Benavides, T. The molecular and cellular basis of copper dysregulation and its relationship with human pathologies. FASEB J. 2021, 35, e21810. [Google Scholar] [CrossRef]
- Yang, N.; Cao, D.F.; Yin, X.X.; Zhou, H.H.; Mao, X.Y. Lysyl oxidases: Emerging biomarkers and therapeutic targets for various diseases. Biomed. Pharmacother. 2020, 131, 110791. [Google Scholar] [CrossRef]
- Kim, H.; Wu, X.; Lee, J. SLC31 (CTR) family of copper transporters in health and disease. Mol. Aspects Med. 2013, 34, 561–570. [Google Scholar] [CrossRef] [Green Version]
- Nose, Y.; Kim, B.E.; Thiele, D.J. Ctr1 drives intestinal copper absorption and is essential for growth, iron metabolism, and neonatal cardiac function. Cell Metab. 2006, 4, 235–244. [Google Scholar] [CrossRef] [Green Version]
- La Fontaine, S.; Mercer, J.F. Trafficking of the copper-ATPases, ATP7A and ATP7B: Role in copper homeostasis. Arch. Biochem. Biophys. 2007, 463, 149–167. [Google Scholar] [CrossRef]
- Lutsenko, S. Human copper homeostasis: A network of interconnected pathways. Curr. Opin. Chem. Biol. 2010, 14, 211–217. [Google Scholar] [CrossRef]
- Puig, S.; Lee, J.; Lau, M.; Thiele, D.J. Biochemical and genetic analyses of yeast and human high affinity copper transporters suggest a conserved mechanism for copper uptake. J. Biol. Chem. 2002, 277, 26021–26030. [Google Scholar] [CrossRef] [Green Version]
- Mandal, T.; Kar, S.; Maji, S.; Sen, S.; Gupta, A. Structural and functional diversity among the members of CTR, the membrane copper transporter family. J. Membr. Biol. 2020, 253, 459–468. [Google Scholar] [CrossRef]
- De Feo, C.J.; Aller, S.G.; Siluvai, G.S.; Blackburn, N.J.; Unger, V.-M. Three-dimensional structure of the human copper transporter hCTR1. Proc. Natl. Acad. Sci. USA 2009, 106, 4237–4242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, F.; Logeman, B.L.; Zhang, X.; Liu, Y.; Thiele, D.J.; Yuan, P. X-ray structures of the high-affinity copper transporter Ctr1. Nat. Commun. 2019, 10, 1386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klomp, A.E.; Juijn, J.A.; van der Gun, L.T.; van den Berg, I.E.; Berger, R.; Klomp, L.W. The N-terminus of the human copper transporter 1 (hCTR1) is localized extracellularly, and interacts with itself. Biochem. J. 2003, 370, 881–889. [Google Scholar] [CrossRef] [Green Version]
- Kim, B.E.; Nevitt, T.; Thiele, D.J. Mechanisms for copper acquisition, distribution and regulation. Nat. Chem. Biol. 2008, 4, 176–185. [Google Scholar] [CrossRef] [PubMed]
- Ohgami, R.S.; Campagna, D.R.; McDonald, A.; Fleming, M.D. The Steap proteins are metalloreductases. Blood 2006, 108, 1388–1394. [Google Scholar] [CrossRef] [PubMed]
- Stefaniak, E.; Płonka, D.; Drew, S.C.; Bossak-Ahmad, K.; Haas, K.L.; Pushie, M.J.; Faller, P.; Wezynfeld, N.E.; Bal, W. The N-terminal 14-mer model peptide of human Ctr1 can collect Cu(II) from albumin. Implications for copper uptake by Ctr1. Metallomics 2018, 10, 1723–1727. [Google Scholar] [CrossRef] [PubMed]
- Hassett, R.; Kosman, D.J. Evidence for Cu(II) reduction as a component of copper uptake by Saccharomyces cerevisiae. J. Biol. Chem. 1995, 270, 128–134. [Google Scholar] [CrossRef] [Green Version]
- Georgatsou, E.; Mavrogiannis, L.A.; Fragiadakis, G.S.; Alexandraki, D. The yeast Fre1p/Fre2p cupric reductases facilitate copper uptake and are regulated by the copper-modulated Mac1p activator. J. Biol. Chem. 1997, 272, 13786–13792. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Peña, M.M.; Nose, Y.; Thiele, D.J. Biochemical characterization of the human copper transporter Ctr1. J. Biol. Chem. 2002, 277, 4380–4387. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Petris, M.J.; Thiele, D.J. Characterization of mouse embryonic cells deficient in the Ctr1 high affinity copper transporter. Identification of a Ctr1-independent copper transport system. J. Biol. Chem. 2002, 277, 40253–40259. [Google Scholar] [CrossRef] [Green Version]
- Scheiber, I.F.; Mercer, J.F.; Dringen, R. Copper accumulation by cultured astrocytes. Neurochem. Int. 2010, 56, 451–460. [Google Scholar] [CrossRef] [PubMed]
- Haas, K.L.; Putterman, A.B.; White, D.R.; Thiele, D.J.; Franz, K.J. Model peptides provide new insights into the role of histidine residues as potential ligands in human cellular copper acquisition via Ctr1. J. Am. Chem. Soc. 2011, 133, 4427–4437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harford, C.; Sarkar, B. Amino Terminal Cu(II)- and Ni(II)-Binding (ATCUN) motif of proteins and peptides: metal binding, DNA cleavage, and other properties. Acc. Chem. Res. 1997, 30, 123–130. [Google Scholar] [CrossRef]
- Rubino, J.T.; Riggs-Gelasco, P.; Franz, K.J. Methionine motifs of copper transport proteins provide general and flexible thioether-only binding sites for Cu(I) and Ag(I). J. Biol. Inorg. Chem. 2010, 15, 1033–1049. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Nadas, I.A.; Kim, M.A.; Franz, K.J. A Mets motif peptide found in copper transport proteins selectively binds Cu(I) with methionine-only coordination. Inorg. Chem. 2005, 44, 9787–9794. [Google Scholar] [CrossRef]
- Pushie, M.J.; Shaw, K.; Franz, K.J.; Shearer, J.; Haas, K.L. Model peptide studies reveal a mixed histidine-methionine Cu(I) binding site at the N-terminus of human copper transporter 1. Inorg. Chem. 2015, 54, 8544–8551. [Google Scholar] [CrossRef]
- Schwab, S.; Shearer, J.; Conklin, S.E.; Alies, B.; Haas, K.L. Sequence proximity between Cu(II) and Cu(I) binding sites of human copper transporter 1 model peptides defines reactivity with ascorbate and O2. J. Inorg. Biochem. 2016, 158, 70–76. [Google Scholar] [CrossRef] [Green Version]
- Shenberger, Y.; Marciano, O.; Gottlieb, H.E.; Ruthstein, S. Insights into the N-terminal Cu(II) and Cu(I) binding sites of the human copper transporter CTR1. J. Coord. Chem. 2018, 71, 1985–2002. [Google Scholar] [CrossRef] [Green Version]
- Du, X.; Li, H.; Wang, X.; Liu, Q.; Ni, J.; Sun, H. Kinetics and thermodynamics of metal binding to the N-terminus of a human copper transporter, hCTR1. Chem. Commun. 2013, 49, 9134–9136. [Google Scholar] [CrossRef]
- Bossak-Ahmad, K.; Frączyk, T.; Bal, W.; Drew, S.C. The sub-picomolar Cu2+ dissociation constant of human serum albumin. Chembiochem 2020, 21, 331–334. [Google Scholar] [CrossRef]
- Bossak, K.; Drew, S.C.; Stefaniak, E.; Płonka, D.; Bonna, A.; Bal, W. The Cu(II) affinity of the N-terminus of human copper transporter CTR1: Comparison of human and mouse sequences. J. Inorg. Biochem. 2018, 182, 230–237. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhu, Y.; Hu, H.; Cheng, L.; Liu, M.; Ma, G.; Yuan, S.; Cui, P.; Liu, Y. Cuprous binding promotes interaction of copper transport protein hCTR1 with cell membranes. Chem. Commun. 2019, 55, 11107–11110. [Google Scholar] [CrossRef] [PubMed]
- Galler, T.; Lebrun, V.; Raibaut, L.; Faller, P.; Wezynfeld, N.E. How trimerization of CTR1 N-terminal model peptides tunes Cu-binding and redox-chemistry. Chem. Commun. 2020, 56, 12194–12197. [Google Scholar] [CrossRef] [PubMed]
- Santoro, A.; Wezynfeld, N.E.; Stefaniak, E.; Pomorski, A.; Płonka, D.; Krężel, A.; Bal, W.; Faller, P. Cu transfer from amyloid-β4-16 to metallothionein-3: The role of the neurotransmitter glutamate and metallothionein-3 Zn(II)-load states. Chem. Commun. 2018, 54, 12634–12637. [Google Scholar] [CrossRef] [PubMed]
- Kotuniak, R.; Strampraad, M.J.F.; Bossak-Ahmad, K.; Wawrzyniak, U.E.; Ufnalska, I.; Hagedoorn, P.L.; Bal, W. Key intermediate species reveal the copper(II)-exchange pathway in biorelevant ATCUN/NTS complexes. Angew. Chem. Int. Ed. Engl. 2020, 59, 11234–11239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wezynfeld, N.E.; Vileno, B.; Faller, P. Cu(II) binding to the N-terminal model peptide of the human Ctr2 transporter at lysosomal and extracellular pH. Inorg. Chem. 2019, 58, 7488–7498. [Google Scholar] [CrossRef]
- Mindell, J.A. Lysosomal acidification mechanisms. Annu. Rev. Physiol. 2012, 74, 69–86. [Google Scholar] [CrossRef] [Green Version]
- La Mendola, D.; Magrì, A.; Vagliasindi, L.I.; Hansson, Ö.; Bonomo, R.P.; Rizzarelli, E. Copper(II) complex formation with a linear peptide encompassing the putative cell binding site of angiogenin. Dalton Trans. 2010, 39, 10678–10684. [Google Scholar] [CrossRef]
- Gonzalez, P.; Bossak, K.; Stefaniak, E.; Hureau, C.; Raibaut, L.; Bal, W.; Faller, P. N-Terminal Cu-binding motifs (Xxx-Zzz-His, Xxx-His) and their derivatives: Chemistry, biology and medicinal applications. Chemistry 2018, 24, 8029–8041. [Google Scholar] [CrossRef]
- La Mendola, D.; Magrì, A.; Santoro, A.M.; Nicoletti, V.G.; Rizzarelli, E. Copper(II) interaction with peptide fragments of histidine-proline-rich glycoprotein: Speciation, stability and binding details. J. Inorg. Biochem. 2012, 111, 59–69. [Google Scholar] [CrossRef]
- Kowalik-Jankowska, T.; Ruta-Dolejsz, M.; Wiśniewska, K.; Łankiewicz, L.; Kozłowski, H. Copper(II) complexation by human and mouse fragments (11–16) of β-amyloid peptide. J. Chem. Soc. Dalton Trans. 2000, 4511–4519. [Google Scholar] [CrossRef]
- Scarpa, M.; Vianello, F.; Signor, L.; Zennaro, L.; Rigo, A. Ascorbate oxidation catalyzed by bis(histidine)copper(II). Inorg. Chem. 1996, 35, 5201–5206. [Google Scholar] [CrossRef]
- Maiti, B.K.; Govil, N.; Kundu, T.; Moura, J.J.G. Designed metal-ATCUN derivatives: Redox- and non-redox-based applications relevant for chemistry, biology, and medicine. IScience 2020, 23, 101792. [Google Scholar] [CrossRef] [PubMed]
- Wezynfeld, N.E.; Stefaniak, E.; Stachucy, K.; Drozd, A.; Płonka, D.; Drew, S.C.; Krężel, A.; Bal, W. Resistance of Cu(Aβ4-16) to copper capture by metallothionein-3 supports a function for the Aβ4-42 peptide as a synaptic Cu(II) scavenger. Angew. Chem. Int. Ed. Engl. 2016, 55, 8235–8238. [Google Scholar] [CrossRef] [PubMed]
- Valensin, D.; Padula, E.M.; Hecel, A.; Luczkowski, M.; Kozlowski, H. Specific binding modes of Cu(I) and Ag(I) with neurotoxic domain of the human prion protein. J. Inorg. Biochem. 2016, 155, 26–35. [Google Scholar] [CrossRef]
- Arnesano, F.; (Department of Chemistry, University of Bari Aldo Moro, Bari, Italy). Personal communication, 2022.
- Gutowska, I.; Machoy, Z.; Machaliński, B. The role of bivalent metals in hydroxyapatite structures as revealed by molecular modeling with the HyperChem software. J. Biomed. Mater. Res. A 2005, 75, 788–793. [Google Scholar] [CrossRef]
- Kanazhevskii, V.V.; Kotsarenko, N.S.; Kolomiichuk, V.N.; Kochubey, D.I. A hierarchic structure in zirconium butoxide complexes in n-butanol solutions. J. Struct. Chem. 2011, 52, 75–82. [Google Scholar] [CrossRef]
- Valentine, J.S.; Silverstein, A.J.; Soos, Z.G. Interdimer exchange in linear chain copper acetate-pyrazine. J. Am. Chem. Soc. 1974, 96, 97–103. [Google Scholar] [CrossRef]
- Larson, C.A.; Adams, P.L.; Jandial, D.D.; Blair, B.G.; Safaei, R.; Howell, S.B. The role of the N-terminus of mammalian copper transporter 1 in the cellular accumulation of cisplatin. Biochem. Pharmacol. 2010, 80, 448–454. [Google Scholar] [CrossRef] [Green Version]
- Bonaccorso, C.; Brancatelli, G.; Forte, G.; Arena, G.; Geremia, S.; Sciotto, D.; Sgarlata, C. Factors driving the self-assembly of water-soluble calix[4]arene and gemini guests: A combined solution, computational and solid-state study. RSC Adv. 2014, 4, 53575–53587. [Google Scholar] [CrossRef]
- Sgarlata, C.; Bonaccorso, C.; Gulino, F.; Zito, V.; Arena, G.; Sciotto, D. Stereochemistry and thermodynamics of the inclusion of aliphatic and aromatic anionic guests in a tetracationic calix[4]arene in acidic and neutral aqueous solutions. New J. Chem. 2009, 33, 991–997. [Google Scholar] [CrossRef]
- Sgarlata, C.; Bonaccorso, C.; Gulino, F.; Zito, V.; Arena, G.; Sciotto, D. Inclusion of aromatic and aliphatic anions into a cationic water-soluble calix[4]arene at different pH values. Tetrahedron Lett. 2009, 50, 1610–1613. [Google Scholar] [CrossRef]
- Lund, A.; Vanngard, T. Note on the determination of the principal fine and hyperfine coupling constants in ESR. J. Chem. Phys. 1965, 42, 2979–2980. [Google Scholar] [CrossRef]
- Gersmann, H.R.; Swalen, J.D. Electron paramagnetic resonance spectra of copper complexes. J. Chem. Phys. 1962, 36, 3221–3233. [Google Scholar] [CrossRef]
- Pilbrow, J.R.; Winfield, M.E. Computer simulation of low symmetry E.S.R. spectra due to vitamin B12r and model systems. Mol. Phys. 1973, 25, 1073–1092. [Google Scholar] [CrossRef]
- Corrigan, M.F.; Murray, K.S.; West, B.O.; Pilbrow, J.R. Synthesis and electron spin resonance study of a tetradentate copper(II) mercaptobenzaldimine complex. Aust. J. Chem. 1977, 30, 2455–2463. [Google Scholar] [CrossRef]
- Gans, P.; Sabatini, A.; Vacca, A. Investigation of equilibria in solution. Determination of equilibrium constants with the HYPERQUAD suite of programs. Talanta 1996, 43, 1739–1753. [Google Scholar] [CrossRef]
- Alderighi, L.; Gans, P.; Ienco, A.; Peters, D.; Sabatini, A.; Vacca, A. Hyperquad simulation and speciation (HySS): A utility program for the investigation of equilibria involving soluble and partially soluble species. Coord. Chem. Rev. 1999, 184, 311–318. [Google Scholar] [CrossRef]
- Flashka, H.A. EDTA Titrations, 2nd ed.; Elsevier: London, UK, 1964. [Google Scholar]
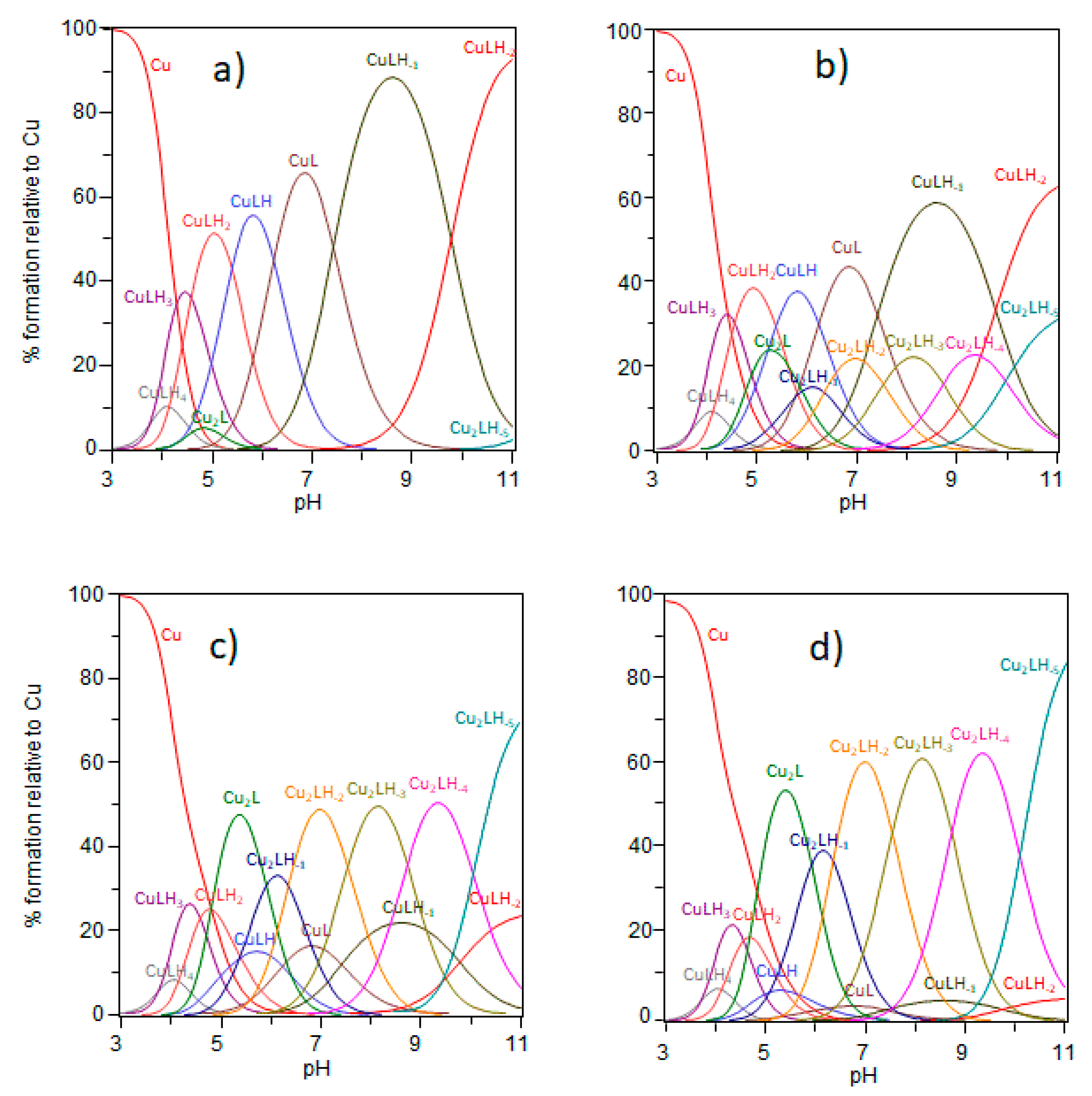
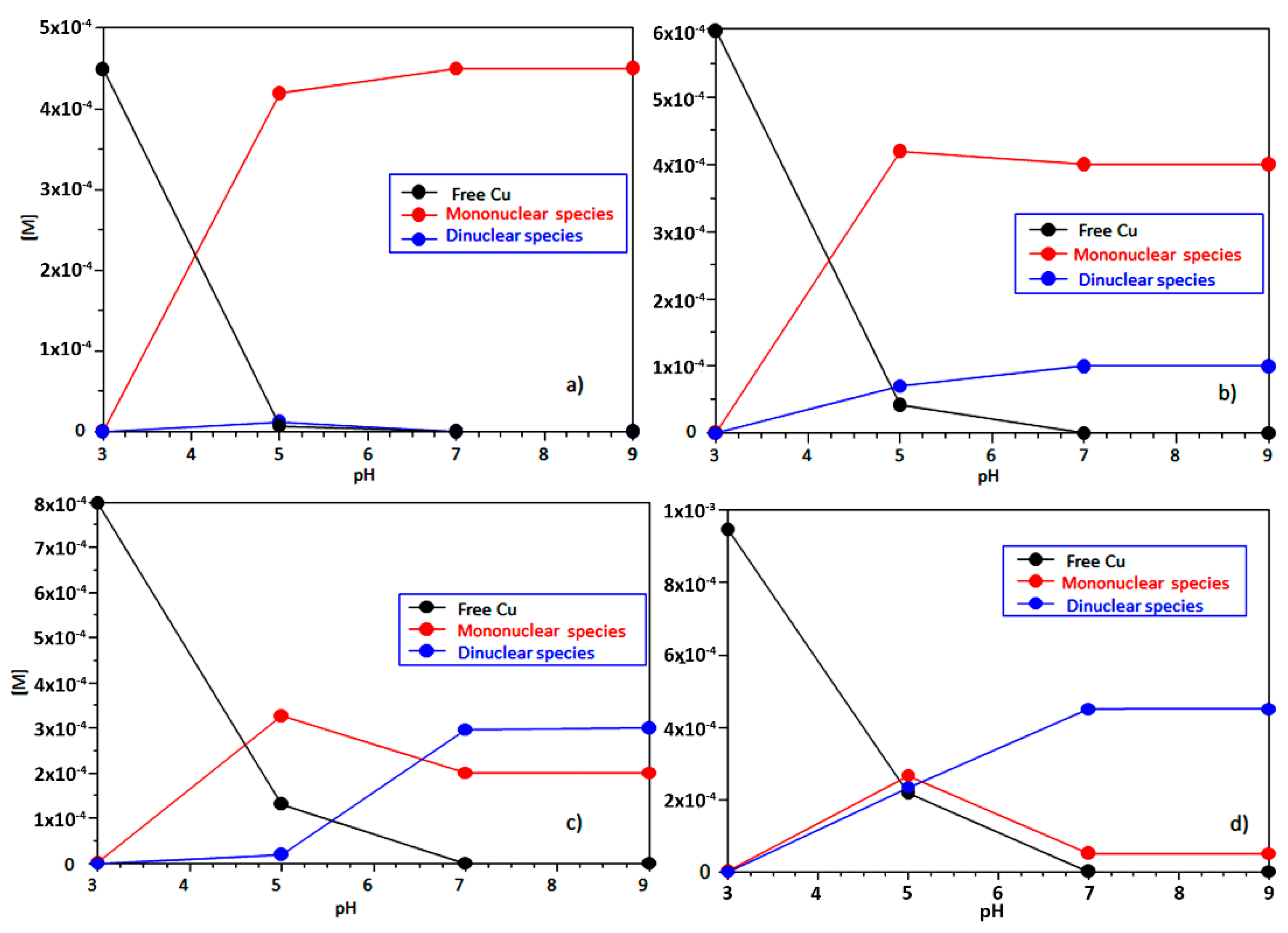
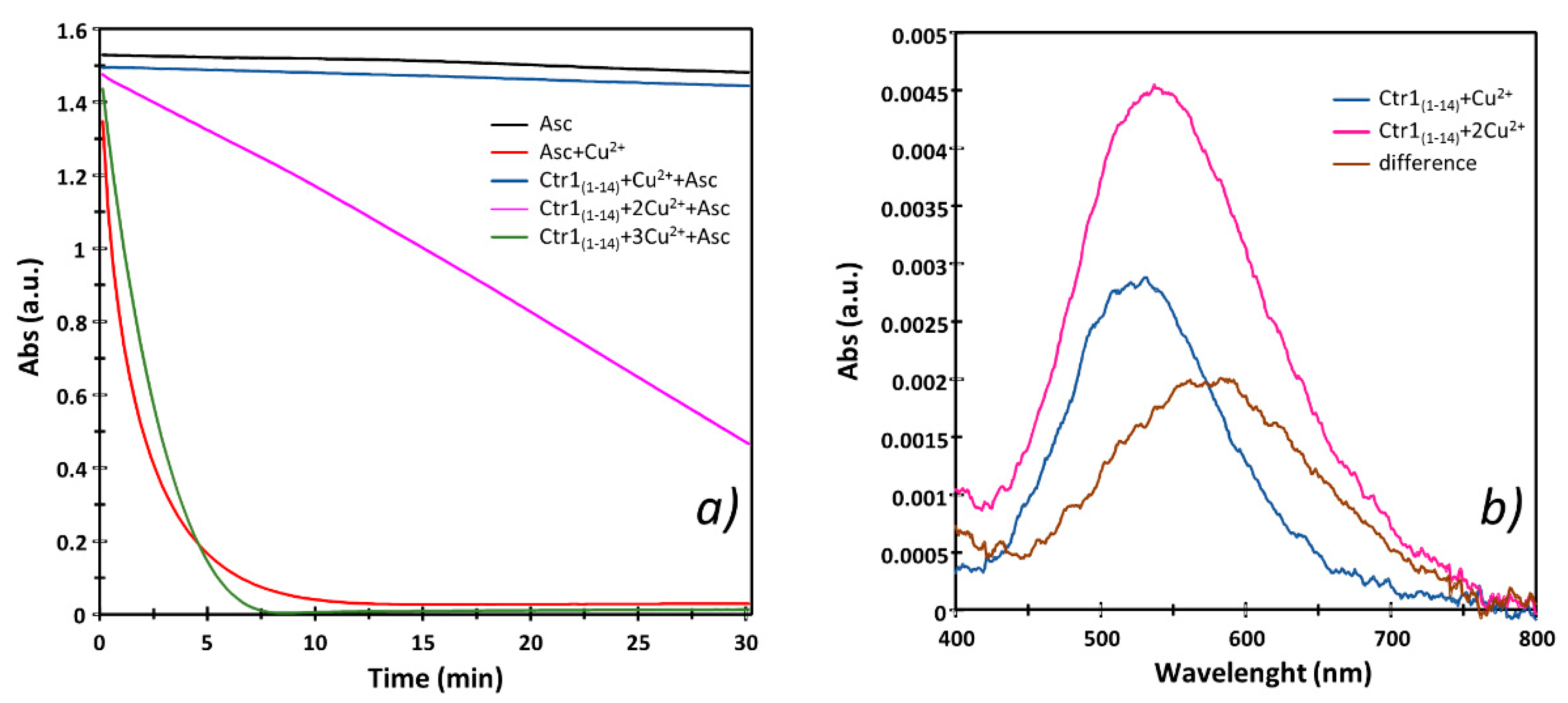
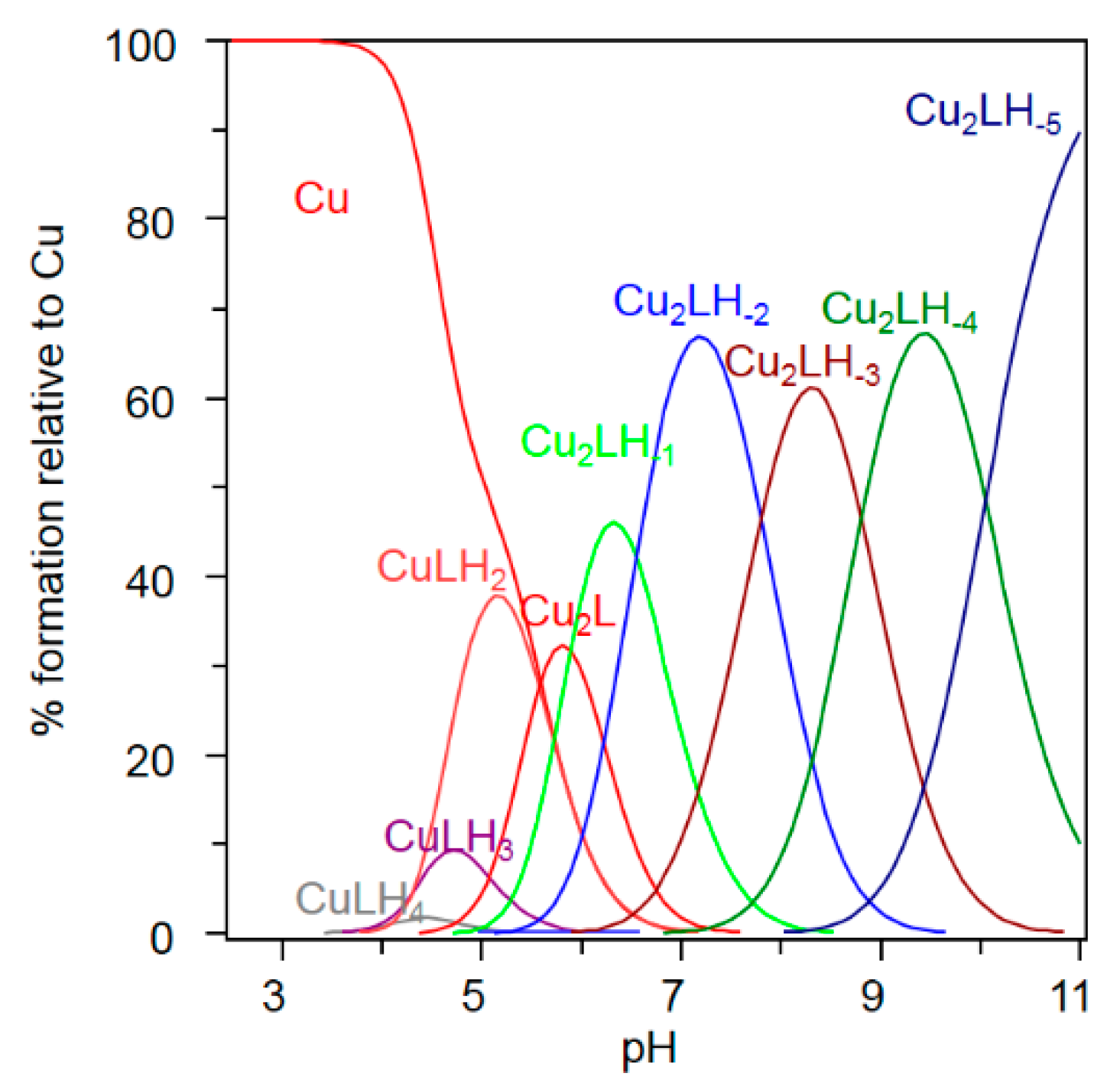

| Species [CupLqHr] | log β a,b | pK b | log β c | pK c | Coordination Mode d | |
|---|---|---|---|---|---|---|
| pqr | ||||||
| 114 | 35.07 (7) | - | 35.37 | - | — | |
| 113 | 31.38 (3) | 3.69 | 31.62 | 3.75 | — | |
| 112 | 26.82 (3) | 4.56 | 27.04 | 4.58 | — | |
| 111 | 21.44 (4) | 5.39 | 21.64 | 5.40 | — | |
| 110 | 15.21 (4) | 6.23 | 15.37 | 6.27 | — | |
| 11-1 | 7.80 (5) | 7.41 | 8.25 | 7.12 | — | |
| 11-2 | −1.96 (6) | 9.77 | −1.74 | 9.99 | — | |
| 210 | 20.63 (3) | — | — | — | 4N {NH2, 2N-, NIm} | 1N1O {NIm, OCOO-} |
| 21-1 | 14.72 (3) | 5.91 | — | — | 4N {NH2, 2N-, NIm} | 2N1O {2NIm, OCOO-} |
| 21-2 | 8.37 (5) | 6.34 | — | — | 4N {NH2, 2N-, NIm} | 3N1O {2NIm, N-, OCOO-} |
| 21-3 | 0.85 (4) | 7.53 | — | — | 4N {NH2, 2N-, NIm} | 4N {2NIm, 2N-} |
| 21-4 | −7.87 (4) | 8.71 | — | — | 4N {NH2, 3N-} | 4N {NIm, 3N-} |
| 21-5 | −18.59 (4) | 9.96 | — | — | 4N {NH2, 3N-} | 4N {NIm, 3N-} |
| Species [CupLqHr] | UV-vis λ, nm (ε, M−1 cm−1) | CD λ, nm (∆ε, M−1 cm−1) | g∥ | A∥ × 104 cm−1 | |
|---|---|---|---|---|---|
| pqr | |||||
| 210 | 523 (130) | 704 (35) | 272 (−1.36), 314 (+0.39), 490 (+0.34), 580 (−0.56) | — | — |
| 21-1 | — | — | — | — | — |
| 21-2 | 522 (130) | 590 (90) | 260 (+0.94), 281 (−0.75), 314 (+0.76), 492 (+0.97), 580 (−0.90) | — | — |
| 21-3 | 522 (130) | 560 (110) | 260 (+0.91), 281 (−0.91), 323 (+1.03), 492 (+1.18), 580 (−0.94) | 2.177 2.230 | 205 185 |
| 21-4 | 522 (130) | 540 (125) | 260 (+0.71), 284 (−1.33), 323 (+1.16), 492 (+1.24), 580 (−1.01) | 2.181 | 203 |
| 21-5 | 522 (130) | 540 (130) | 260 (+0.69), 284 (−1.30), 320 (+1.12), 492 (+1.10), 580 (−1.01) | 2.181 | 203 |
| Motif | UV-vis | CD | EPR | ||
|---|---|---|---|---|---|
| λmax | d-d Transitions | CT | g|| | A|| (104 × cm−1) | |
| XZH | ~520 nm | ~480 nm ~560 nm | ~310 nm | ~2.190 | ~200 |
| Species [AgpLqHr] | log β a | pK | log K* | Coordination Mode |
|---|---|---|---|---|
| pqr | ||||
| 114 | 33.45 (3) | — | 2.98 | 1N {NIm/1N1O1S} |
| 113 | 27.40 (4) | 6.05 | 3.22 | 2N {2 NIm/2N1O1S} |
| 112 | 21.05 (3) | 6.35 | 3.78 | 2N {2 NIm/2N1OS} |
| 111 | 13.72 (2) | 7.33 | 4.28 | 2N {2 NIm/2N1OS} |
| 10-1 | −8.56 b | — | — | — |
| L:Cu 1:1 | L:Cu 1:2 | ||||
|---|---|---|---|---|---|
| pH | g∥ | A∥ × 104 cm−1 | pH | g∥ | A∥ × 104 cm−1 |
| 4.0 | 2.245(3) | 178(3) | 4.0 | 2.424(3) | 124(3) |
| 2.307(3) | 172(3) | lbr | lbr | ||
| 2.424(2) | 124(2) | ||||
| 5.1 | 2.177(2) | 205(3) | 5.0 | 2.424(3) | 124(3) |
| 2.230(3) | 185(3) | lbr | lbr | ||
| 5.5 | 2.177(2) | 205(3) | |||
| 2.230(3) | 185(3) | ||||
| 6.1 | lbr | lbr | |||
| 6.6 | 2.177(2) | 205(3) | 7.0 | 2.181(5) | 203(5) |
| lbr | lbr | ||||
| 8.0 | 2.181(3) | 203(3) | 8.1 | 2.181(5) | 203(5) |
| lbr | lbr | ||||
| 9.0 | 2.181(5) | 205(5) | |||
| 2.210(5) | 185(5) | ||||
| 10.6 | 2.181(3) | 203(3) | 10.8 | 2.181(5) | 205(5) |
| 2.210(5) | 185(5) | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Magrì, A.; Tabbì, G.; Naletova, I.; Attanasio, F.; Arena, G.; Rizzarelli, E. A Deeper Insight in Metal Binding to the hCtr1 N-terminus Fragment: Affinity, Speciation and Binding Mode of Binuclear Cu2+ and Mononuclear Ag+ Complex Species. Int. J. Mol. Sci. 2022, 23, 2929. https://doi.org/10.3390/ijms23062929
Magrì A, Tabbì G, Naletova I, Attanasio F, Arena G, Rizzarelli E. A Deeper Insight in Metal Binding to the hCtr1 N-terminus Fragment: Affinity, Speciation and Binding Mode of Binuclear Cu2+ and Mononuclear Ag+ Complex Species. International Journal of Molecular Sciences. 2022; 23(6):2929. https://doi.org/10.3390/ijms23062929
Chicago/Turabian StyleMagrì, Antonio, Giovanni Tabbì, Irina Naletova, Francesco Attanasio, Giuseppe Arena, and Enrico Rizzarelli. 2022. "A Deeper Insight in Metal Binding to the hCtr1 N-terminus Fragment: Affinity, Speciation and Binding Mode of Binuclear Cu2+ and Mononuclear Ag+ Complex Species" International Journal of Molecular Sciences 23, no. 6: 2929. https://doi.org/10.3390/ijms23062929
APA StyleMagrì, A., Tabbì, G., Naletova, I., Attanasio, F., Arena, G., & Rizzarelli, E. (2022). A Deeper Insight in Metal Binding to the hCtr1 N-terminus Fragment: Affinity, Speciation and Binding Mode of Binuclear Cu2+ and Mononuclear Ag+ Complex Species. International Journal of Molecular Sciences, 23(6), 2929. https://doi.org/10.3390/ijms23062929









