BoPEP4, a C-Terminally Encoded Plant Elicitor Peptide from Broccoli, Plays a Role in Salinity Stress Tolerance
Abstract
:1. Introduction
2. Results
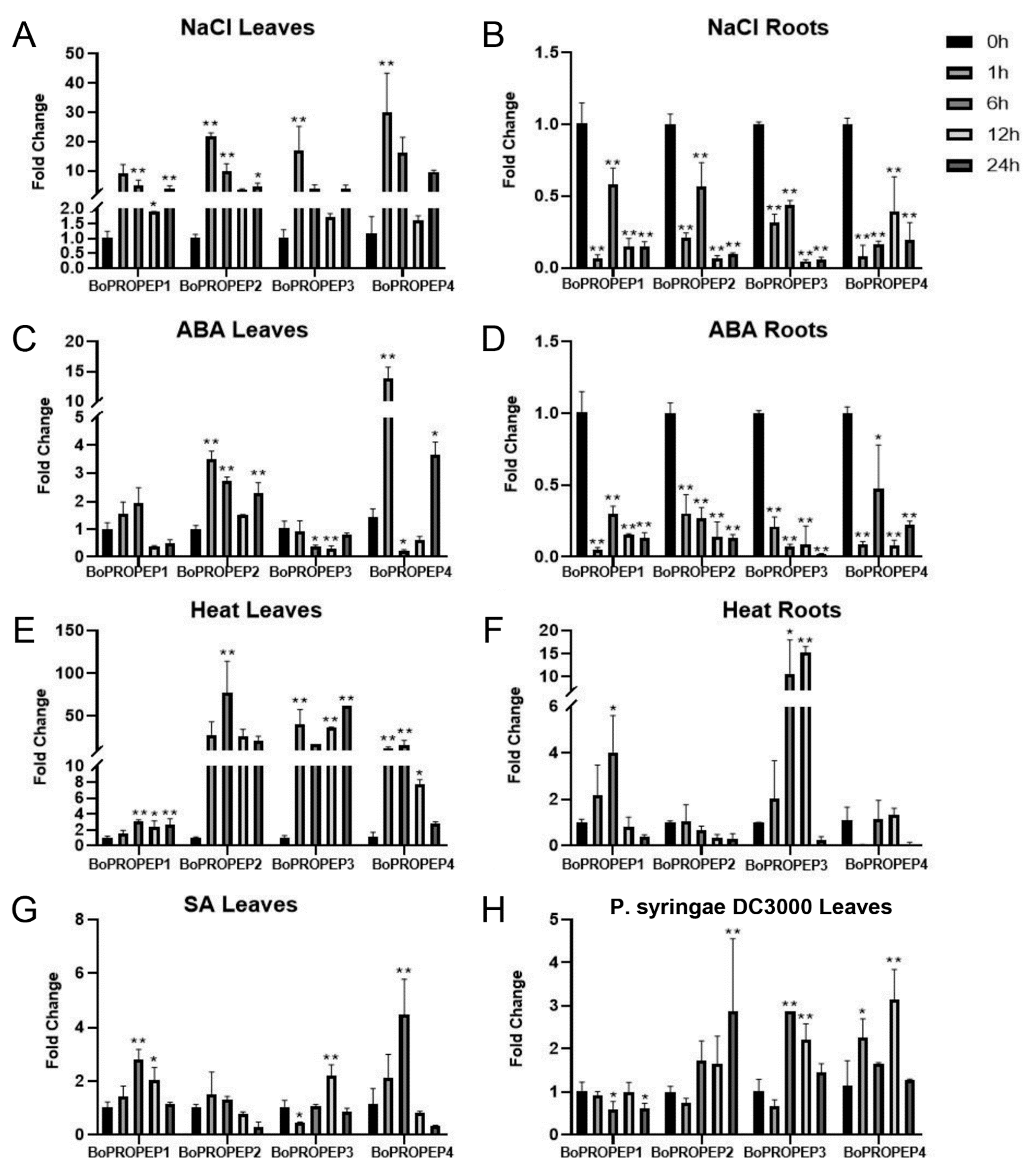
2.1. Identification and Expression Patterns of BoPROPEP Genes in Broccoli
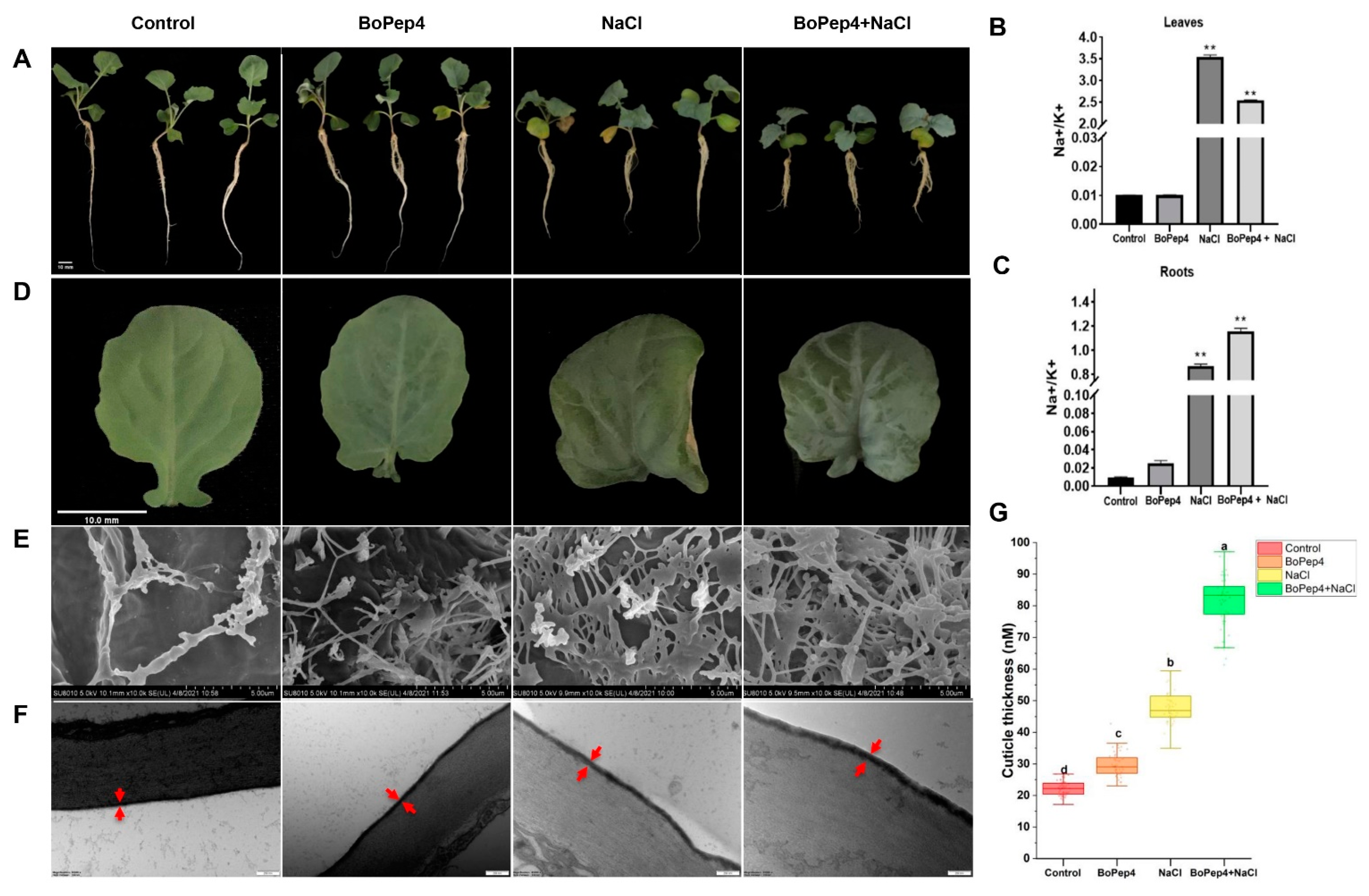
2.2. Exogenous Application of BoPep4 Alleviated Salinity-Induced Stress in Broccoli Seedlings
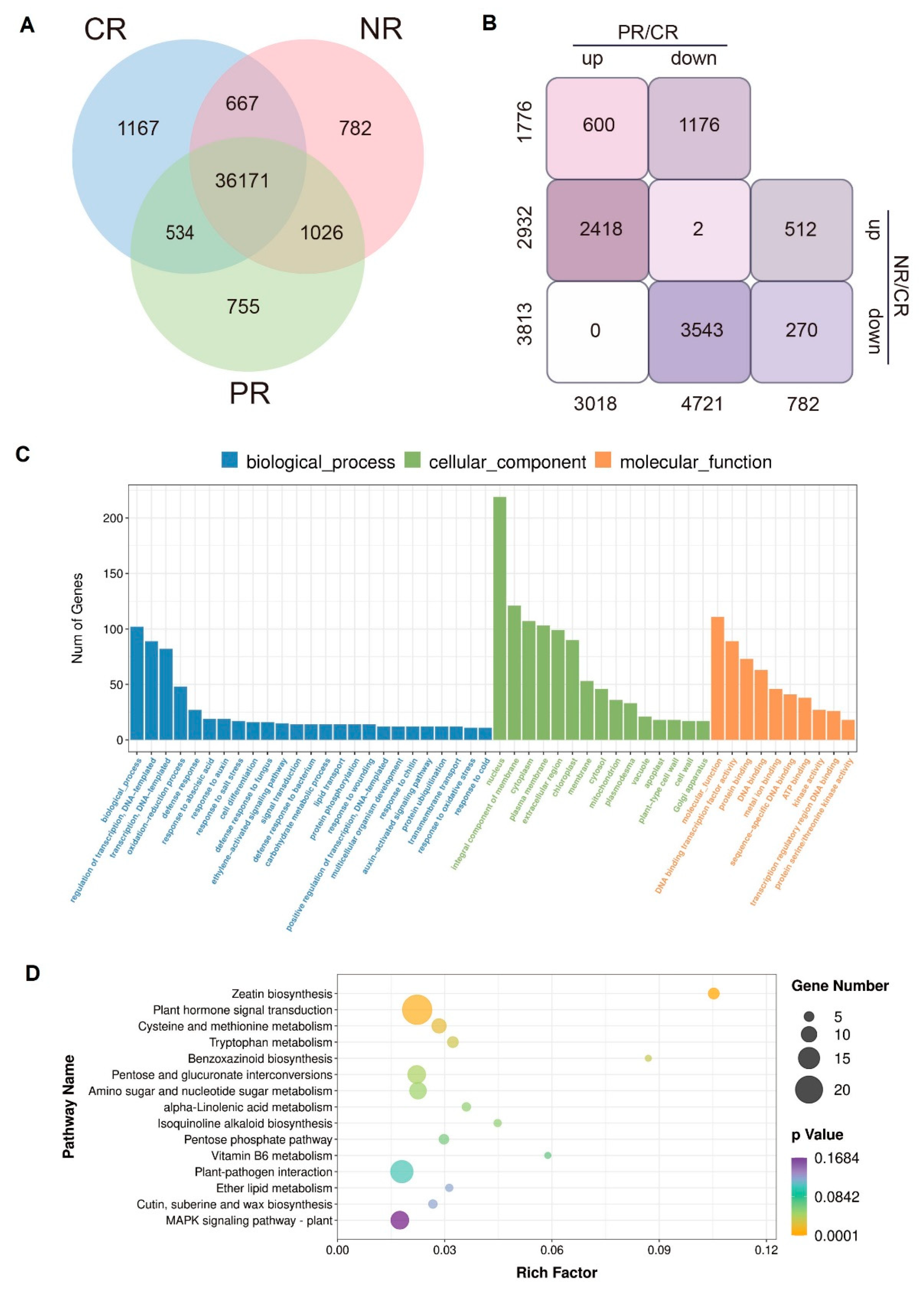
2.3. Identification of Differentially Expressed Genes Regulated by BoPep4 in Response to NaCl Stress
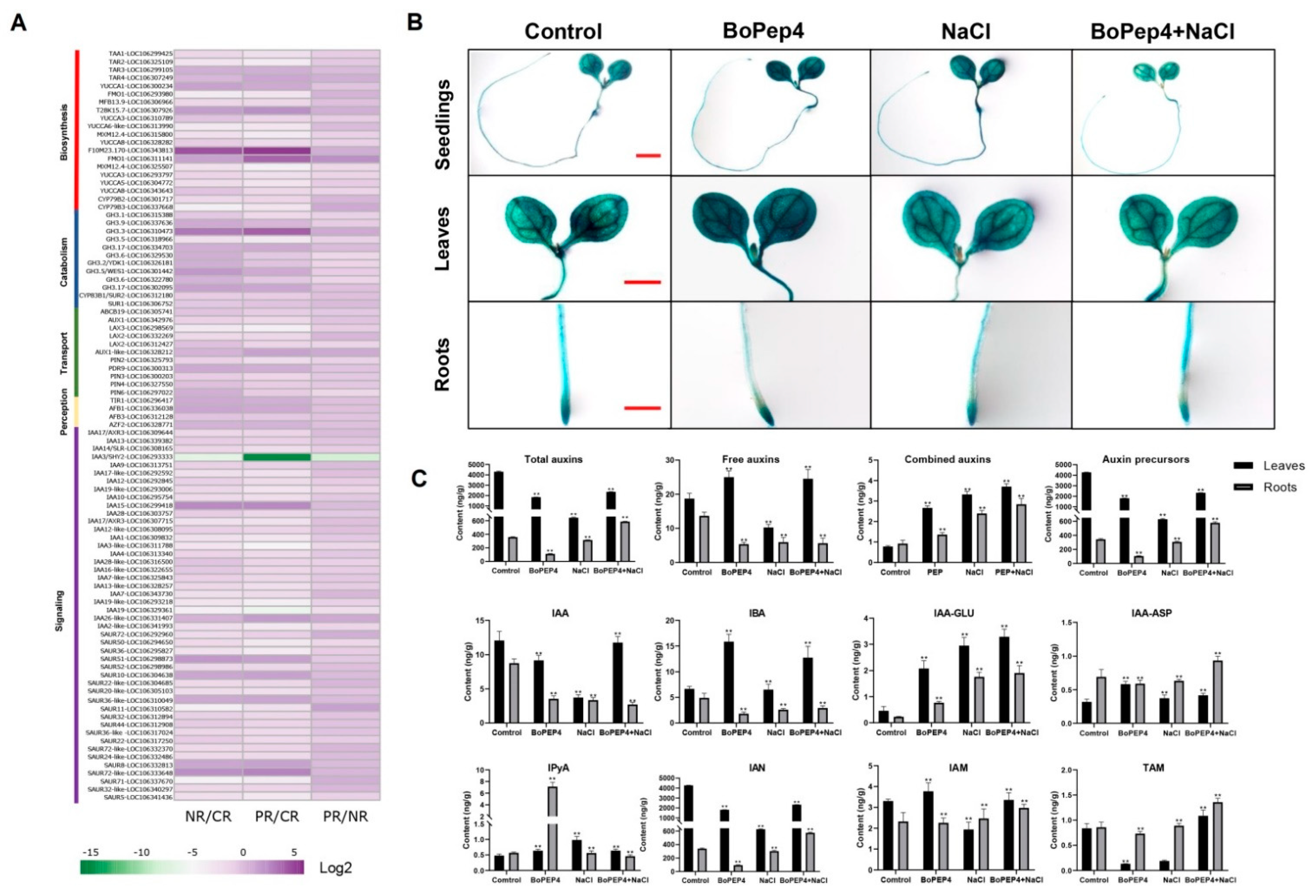
2.4. BoPep4 Influences Auxin Homeostasis during Salinity Stress
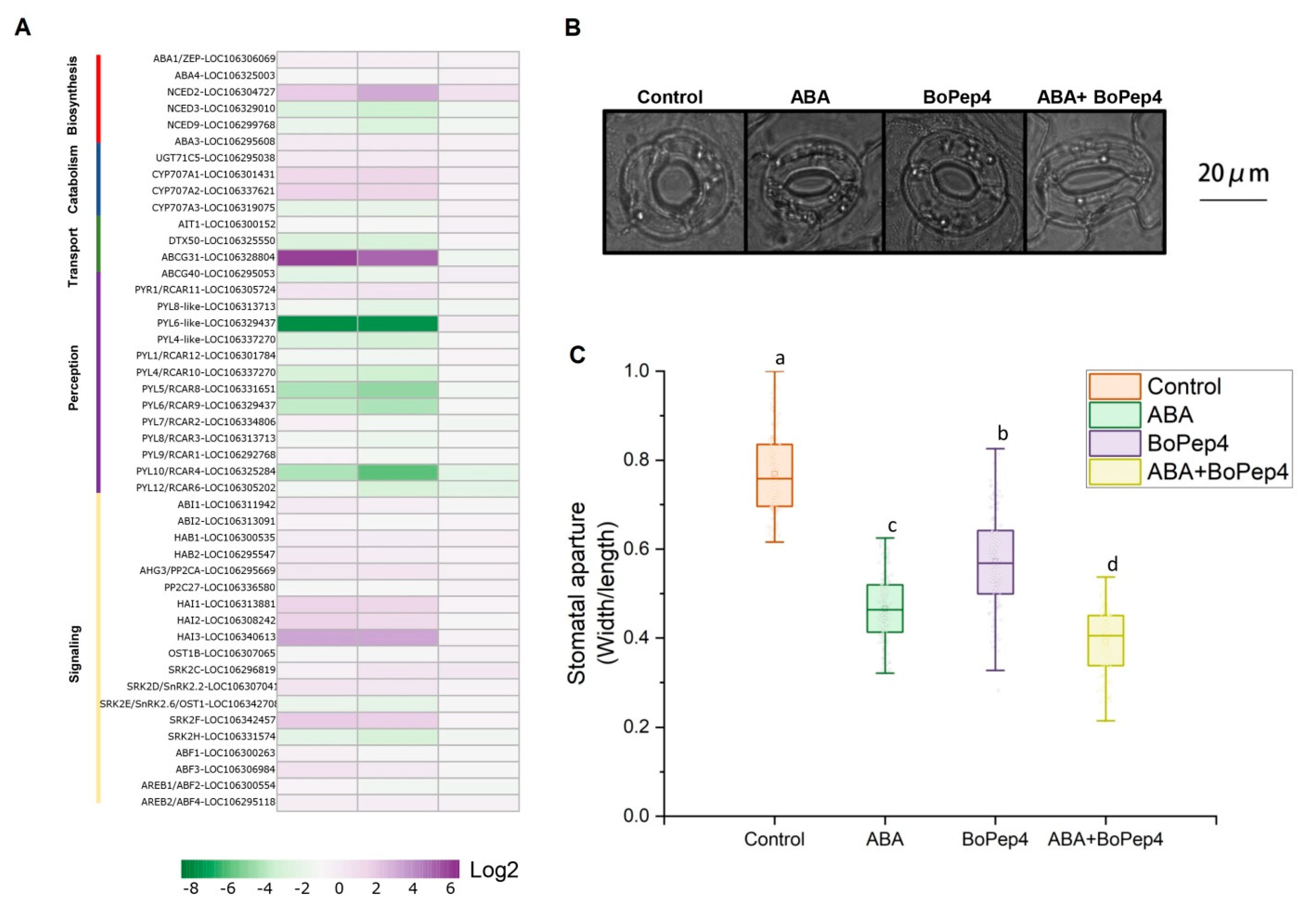
2.5. BoPep4 Accentuated ABA Response under Salinity Stress
2.6. BoPep4 Enhances Wax and Cutin Accumulation under Salinity Stress
3. Discussion
4. Materials and Methods
4.1. Identification and Characterization of BoPROPEP Genes
4.2. Plant Growth and Treatments
4.3. RNA Extraction and qRT-PCR Analysis
4.4. Peptide Synthesis and Treatment Assay
4.5. Measurement of Na+ and K+ Contents
4.6. Scanning and Transmission Electron Microscopy Analyses
4.7. Transcriptome Sequencing and Data Analysis
4.8. GUS Staining of DR5 Reporter Arabidopsis
4.9. Detection of Auxin Contents
4.10. Measurement of Stomatal Aperture
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhu, J.-K. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Zhao, Y.; Zhu, J.-K. Thriving under Stress: How Plants Balance Growth and the Stress Response. Dev. Cell 2020, 55, 529–543. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, F.; Hanada, K.; Kondo, T.; Shinozaki, K. Hormone-like peptides and small coding genes in plant stress signaling and development. Curr. Opin. Plant Biol. 2019, 51, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.-L.; Lu, H.; Hassan, M.; Zhang, J.; Yuan, G.; Abraham, P.E.; Shrestha, H.K.; Solis, M.I.V.; Chen, J.-G.; Tschaplinski, T.J.; et al. Advances and perspectives in discovery and functional analysis of small secreted proteins in plants. Hortic. Res. 2021, 8, 130. [Google Scholar] [CrossRef] [PubMed]
- Huffaker, A.; Pearce, G.; Ryan, C.A. An endogenous peptide signal in Arabidopsis activates components of the innate immune response. Proc. Natl. Acad. Sci. USA 2006, 103, 10098–10103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hander, T.; Fernández-Fernández, D.; Kumpf, R.P.; Willems, P.; Schatowitz, H.; Rombaut, D.; Staes, A.; Nolf, J.; Pottie, R.; Yao, P.; et al. Damage on plants activates Ca 2+ -dependent metacaspases for release of immunomodulatory peptides. Science 2019, 363, eaar7486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, W.; Liu, J.; Li, J.-F. Type-II Metacaspases Mediate the Processing of Plant Elicitor Peptides in Arabidopsis. Mol. Plant 2019, 12, 1524–1533. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, Y.; Huffaker, A.; Bryan, A.C.; Tax, F.; Ryan, C.A. PEPR2 Is a Second Receptor for the Pep1 and Pep2 Peptides and Contributes to Defense Responses in Arabidopsis. Plant Cell 2010, 22, 508–522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakaminami, K.; Okamoto, M.; Higuchi-Takeuchi, M.; Yoshizumi, T.; Yamaguchi, Y.; Fukao, Y.; Shimizu, M.; Ohashi, C.; Tanaka, M.; Matsui, M.; et al. AtPep3 is a hormone-like peptide that plays a role in the salinity stress tolerance of plants. Proc. Natl. Acad. Sci. USA 2018, 115, 5810–5815. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, X.; Kang, S.; Jing, Y.; Ren, Z.; Li, L.; Zhou, J.-M.; Berkowitz, G.; Shi, J.; Fu, A.; Lan, W.; et al. Danger-Associated Peptides Close Stomata by OST1-Independent Activation of Anion Channels in Guard Cells. Plant Cell 2018, 30, 1132–1146. [Google Scholar] [CrossRef] [Green Version]
- Jing, Y.; Zheng, X.; Zhang, D.; Shen, N.; Wang, Y.; Yang, L.; Fu, A.; Shi, J.; Zhao, F.; Lan, W.; et al. Danger-Associated Peptides Interact with PIN-Dependent Local Auxin Distribution to Inhibit Root Growth in Arabidopsis. Plant Cell 2019, 31, 1767–1787. [Google Scholar] [CrossRef] [PubMed]
- Jing, Y.P.; Shen, N.; Zheng, X.J.; Fu, A.G.; Zhao, F.G.; Lan, W.Z.; Luan, S. Danger-Associated Peptide Regulates Root Immune Responses and Root Growth by Affecting ROS Formation in Arabidopsis. Int. J. Mol. Sci. 2020, 21, 4590. [Google Scholar] [CrossRef] [PubMed]
- Shen, N.; Jing, Y.; Tu, G.; Fu, A.; Lan, W. Danger-Associated Peptide Regulates Root Growth by Promoting Protons Extrusion in an AHA2-Dependent Manner in Arabidopsis. Int. J. Mol. Sci. 2020, 21, 7963. [Google Scholar] [CrossRef] [PubMed]
- Huffaker, A.; Dafoe, N.J.; Schmelz, E. ZmPep1, an Ortholog of Arabidopsis Elicitor Peptide 1, Regulates Maize Innate Immunity and Enhances Disease Resistance. Plant Physiol. 2011, 155, 1325–1338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shinya, T.; Yasuda, S.; Hyodo, K.; Tani, R.; Hojo, Y.; Fujiwara, Y.; Hiruma, K.; Ishizaki, T.; Fujita, Y.; Saijo, Y.; et al. Integration of danger peptide signals with herbivore-associated molecular pattern signaling amplifies anti-herbivore defense responses in rice. Plant J. 2018, 94, 626–637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, W.; Zhang, X.; Liu, J.; Tao, K.; Li, C.; Xiao, S.; Zhang, W.; Li, J. Plant elicitor peptide signaling confers rice resistance to piercing-sucking insect herbivores and pathogens. Plant Biotechnol. J. 2022. [Google Scholar] [CrossRef]
- Ruiz, C.; Nadal, A.; Foix, L.; Montesinos, L.; Montesinos, E.; Pla, M. Diversity of plant defense elicitor peptides within the Rosaceae. BMC Genet. 2018, 19, 11. [Google Scholar] [CrossRef] [Green Version]
- Foix, L.; Nadal, A.; Zagorščak, M.; Ramšak, Z.; Esteve-Codina, A.; Gruden, K.; Pla, M. Prunus persica plant endogenous peptides PpPep1 and PpPep2 cause PTI-like transcriptome reprogramming in peach and enhance resistance to Xanthomonas arboricola pv. pruni. BMC Genom. 2021, 22, 360. [Google Scholar] [CrossRef]
- Yamaguchi, Y.; Barona, G.; Ryan, C.A.; Pearce, G. GmPep914, an Eight-Amino Acid Peptide Isolated from Soybean Leaves, Activates Defense-Related Genes. Plant Physiol. 2011, 156, 932–942. [Google Scholar] [CrossRef]
- Gao, J.; Yu, X.; Ma, F.; Li, J. RNA-Seq Analysis of Transcriptome and Glucosinolate Metabolism in Seeds and Sprouts of Broccoli (Brassica oleracea var. italic). PLoS ONE 2014, 9, e88804. [Google Scholar] [CrossRef]
- Yu, Z.; Duan, X.; Luo, L.; Dai, S.; Ding, Z.; Xia, G. How Plant Hormones Mediate Salt Stress Responses. Trends Plant Sci. 2020, 25, 1117–1130. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Liu, X.; Gai, X.; Ren, J.; Liu, X.; Cai, Y.; Wang, Q.; Ren, H. Cucumis sativusL.WAX2Plays a Pivotal Role in Wax Biosynthesis, Influencing Pollen Fertility and Plant Biotic and Abiotic Stress Responses. Plant Cell Physiol. 2015, 56, 1339–1354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huffaker, A.; Pearce, G.; Veyrat, N.; Erb, M.; Turlings, T.C.J.; Sartor, R.; Shen, Z.; Briggs, S.P.; Vaughan, M.M.; Alborn, H.T.; et al. Plant elicitor peptides are conserved signals regulating direct and indirect antiherbivore defense. Proc. Natl. Acad. Sci. USA 2013, 110, 5707–5712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lori, M.; Van Verk, M.C.; Hander, T.; Schatowitz, H.; Klauser, D.; Flury, P.; Gehring, C.A.; Boller, T.; Bartels, S. Evolutionary divergence of the plant elicitor peptides (Peps) and their receptors: Interfamily incompatibility of perception but compatibility of downstream signalling. J. Exp. Bot. 2015, 66, 5315–5325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olsson, V.; Joos, L.; Zhu, S.; Gevaert, K.; Butenko, M.A.; De Smet, I. Look Closely, the Beautiful May Be Small: Precursor-Derived Peptides in Plants. Annu. Rev. Plant Biol. 2019, 70, 153–186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bartels, S.; Lori, M.; Mbengue, M.; Van Verk, M.; Klauser, D.; Hander, T.; Böni, R.; Robatzek, S.; Boller, T. The family of Peps and their precursors in Arabidopsis: Differential expression and localization but similar induction of pattern-triggered immune responses. J. Exp. Bot. 2013, 64, 5309–5321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klauser, D.; Desurmont, G.A.; Glauser, G.; Vallat, A.; Flury, P.; Boller, T.; Turlings, T.C.J.; Bartels, S. TheArabidopsisPep-PEPR system is induced by herbivore feeding and contributes to JA-mediated plant defence against herbivory. J. Exp. Bot. 2015, 66, 5327–5336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saijo, Y.; Loo, E.P. Plant immunity in signal integration between biotic and abiotic stress responses. New Phytol. 2020, 225, 87–104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Safaeizadeh, M.; Boller, T. Differential and tissue-specific activation pattern of the AtPROPEP and AtPEPR genes in response to biotic and abiotic stress in Arabidopsis thaliana. Plant Signal. Behav. 2019, 14, e1590094. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, L.; Yang, R.; Wang, Z.; Guo, Q.; Gu, Z. Effect of NaCl stress on health-promoting compounds and antioxidant activity in the sprouts of three broccoli cultivars. Int. J. Food Sci. Nutr. 2014, 65, 476–481. [Google Scholar] [CrossRef]
- Ross, A.; Yamada, K.; Hiruma, K.; Yamashita-Yamada, M.; Lu, X.; Takano, Y.; Tsuda, K.; Saijo, Y. The Arabidopsis PEPR pathway couples local and systemic plant immunity. EMBO J. 2014, 33, 62–75. [Google Scholar] [CrossRef] [PubMed]
- Saijo, Y.; Loo, E.; Tajima, Y.; Yamada, K.; Kido, S.; Hirase, T.; Ariga, H.; Fujiwara, T.; Tanaka, K.; Taji, T.; et al. Recognition of microbe/damage-associated molecular patterns by leucine-rich repeat pattern recognition receptor kinases confers salt tolerance in plants. Mol. Plant-Microbe Interact. 2021. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Li, M.; Yu, L.; Zhou, Z.; Liang, X.; Liu, Z.; Cai, G.; Gao, L.; Zhang, X.; Wang, Y.; et al. The FLS2-Associated Kinase BIK1 Directly Phosphorylates the NADPH Oxidase RbohD to Control Plant Immunity. Cell Host Microbe 2014, 15, 329–338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Z.; Wu, Y.; Yang, F.; Zhang, Y.; Chen, S.; Xie, Q.; Tian, X.; Zhou, J.-M. BIK1 interacts with PEPRs to mediate ethylene-induced immunity. Proc. Natl. Acad. Sci. USA 2013, 110, 6205–6210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Veronese, P.; Nakagami, H.; Bluhm, B.; AbuQamar, S.; Chen, X.; Salmeron, J.; Dietrich, R.A.; Hirt, H.; Mengiste, T. The Membrane-Anchored BOTRYTIS-INDUCED KINASE1 Plays Distinct Roles in Arabidopsis Resistance to Necrotrophic and Biotrophic Pathogens. Plant Cell 2006, 18, 257–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeFalco, T.A.; Zipfel, C. Molecular mechanisms of early plant pattern-triggered immune signaling. Mol. Cell 2021, 81, 3449–3467. [Google Scholar] [CrossRef] [PubMed]
- Bartels, S.; Boller, T. Quo vadis, Pep? Plant elicitor peptides at the crossroads of immunity, stress, and development. J. Exp. Bot. 2015, 66, 5183–5193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Li, Y.; Bao, Q.; Wang, H.; Hou, S. Plant elicitor peptide 1 fortifies root cell walls and triggers a systemic root-to-shoot immune signaling in Arabidopsis. Plant Signal. Behav. 2022, 2034270. [Google Scholar] [CrossRef] [PubMed]
- Jiang, K.; Moe-Lange, J.; Hennet, L.; Feldman, L.J. Salt Stress Affects the Redox Status of Arabidopsis Root Meristems. Front. Plant Sci. 2016, 7, 81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y. The role of local biosynthesis of auxin and cytokinin in plant development. Curr. Opin. Plant Biol. 2008, 11, 16–22. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.W.; Li, C.H.; Cao, J.; Zhang, Y.C.; Zhang, S.Q.; Xia, Y.F.; Sun, D.Y.; Sun, Y. Altered architecture and enhanced drought tolerance in rice via the down-regulation of indole-3-acetic acid by TLD1/OsGH3.13 activation. Plant Physiol. 2009, 151, 1889–1901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koochak, H.; Ludwig-Müller, J. Physcomitrium patens Mutants in Auxin Conjugating GH3 Proteins Show Salt Stress Tolerance but Auxin Homeostasis Is Not Involved in Regulation of Oxidative Stress Factors. Plants 2021, 10, 1398. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Li, G.J.; Bressan, R.A.; Song, C.P.; Zhu, J.K.; Zhao, Y. Abscisic acid dynamics, signaling, and functions in plants. J. Integr. Plant Biol. 2020, 62, 25–54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Li, X.; He, Z.; Zhao, X.; Wang, Q.; Zhou, B.; Yu, D.; Huang, X.; Tang, D.; Guo, X.; et al. Molecular character of a phosphatase 2C (PP2C) gene relation to stress tolerance in Arabidopsis thaliana. Mol. Biol. Rep. 2013, 40, 2633–2644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhaskara, G.B.; Nguyen, T.T.; Verslues, P.E. Unique Drought Resistance Functions of theHighly ABA-InducedClade A Protein Phosphatase 2Cs. Plant Physiol. 2012, 160, 379–395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, S.; Zhang, S.; Liu, Y.; Jiang, Y.; Wang, Y.; Li, J.; Ni, Y. A combination of genome-wide association study and transcriptome analysis in leaf epidermis identifies candidate genes involved in cuticular wax biosynthesis in Brassica napus. BMC Plant Biol. 2020, 20, 458. [Google Scholar] [CrossRef] [PubMed]
- Kunst, L.; Samuels, A.L. Biosynthesis and secretion of plant cuticular wax. Prog. Lipid. Res. 2003, 42, 51–80. [Google Scholar] [CrossRef]
- Guo, W.; Wu, Q.; Yang, L.; Hu, W.; Liu, D.; Liu, Y. Ectopic Expression of CsKCS6 From Navel Orange Promotes the Production of Very-Long-Chain Fatty Acids (VLCFAs) and Increases the Abiotic Stress Tolerance of Arabidopsis thaliana. Front. Plant Sci. 2020, 11, 564656. [Google Scholar] [CrossRef]
- Wang, Y.; Jin, S.; Xu, Y.; Li, S.; Zhang, S.; Yuan, Z.; Li, J.; Ni, Y. Overexpression of BnKCS1-1, BnKCS1-2, and BnCER1-2 promotes cuticular wax production and increases drought tolerance in Brassica napus. Crop J. 2020, 8, 26–37. [Google Scholar] [CrossRef]
- Wang, M.; Wu, H.; Xu, J.; Li, C.; Wang, Y.; Wang, Z. Five Fatty Acyl-Coenzyme A Reductases Are Involved in the Biosynthesis of Primary Alcohols in Aegilops tauschii Leaves. Front. Plant Sci. 2017, 8, 1012. [Google Scholar] [CrossRef] [Green Version]
- Domergue, F.; Vishwanath, S.J.; Joubès, J.; Ono, J.; Lee, J.A.; Bourdon, M.; Alhattab, R.; Lowe, C.; Pascal, S.; Lessire, R.; et al. Three Arabidopsis Fatty Acyl-Coenzyme A Reductases, FAR1, FAR4, and FAR5, Generate Primary Fatty Alcohols Associated with Suberin Deposition. Plant Physiol. 2010, 153, 1539–1554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Wang, G.; Xu, Z.; Li, J.; Sun, M.; Guo, J.; Ji, W. Organization and Regulation of Soybean SUMOylation System under Abiotic Stress Conditions. Front. Plant Sci. 2017, 8, 1458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ji, W.; Cong, R.; Li, S.; Li, R.; Qin, Z.; Li, Y.; Zhou, X.; Chen, S.; Li, J. Comparative Proteomic Analysis of Soybean Leaves and Roots by iTRAQ Provides Insights into Response Mechanisms to Short-Term Salt Stress. Front. Plant Sci. 2016, 7, 573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Tian, X.; Zhao, Q.; Liu, Z.; Li, X.; Ren, Y.; Tang, J.; Fang, J.; Xu, Q.; Bu, Q. The E3 Ligase DROUGHT HYPERSENSITIVE Negatively Regulates Cuticular Wax Biosynthesis by Promoting the Degradation of Transcription Factor ROC4 in Rice. Plant Cell 2018, 30, 228–244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kong, W.; Li, J.; Yu, Q.; Cang, W.; Xu, R.; Wang, Y.; Ji, W. Two Novel Flavin-Containing Monooxygenases Involved in Biosynthesis of Aliphatic Glucosinolates. Front. Plant Sci. 2016, 7, 1292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 Years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, A.; Guo, J.; Wang, S.; Zhang, Y.; Lu, F.; Duan, J.; Liu, Z.; Ji, W. BoPEP4, a C-Terminally Encoded Plant Elicitor Peptide from Broccoli, Plays a Role in Salinity Stress Tolerance. Int. J. Mol. Sci. 2022, 23, 3090. https://doi.org/10.3390/ijms23063090
Wang A, Guo J, Wang S, Zhang Y, Lu F, Duan J, Liu Z, Ji W. BoPEP4, a C-Terminally Encoded Plant Elicitor Peptide from Broccoli, Plays a Role in Salinity Stress Tolerance. International Journal of Molecular Sciences. 2022; 23(6):3090. https://doi.org/10.3390/ijms23063090
Chicago/Turabian StyleWang, Anyi, Jingsong Guo, Sibo Wang, Ying Zhang, Fangfang Lu, Jingbin Duan, Zhao Liu, and Wei Ji. 2022. "BoPEP4, a C-Terminally Encoded Plant Elicitor Peptide from Broccoli, Plays a Role in Salinity Stress Tolerance" International Journal of Molecular Sciences 23, no. 6: 3090. https://doi.org/10.3390/ijms23063090
APA StyleWang, A., Guo, J., Wang, S., Zhang, Y., Lu, F., Duan, J., Liu, Z., & Ji, W. (2022). BoPEP4, a C-Terminally Encoded Plant Elicitor Peptide from Broccoli, Plays a Role in Salinity Stress Tolerance. International Journal of Molecular Sciences, 23(6), 3090. https://doi.org/10.3390/ijms23063090





