A Homozygous Deep Intronic Variant Causes Von Willebrand Factor Deficiency and Lack of Endothelial-Specific Secretory Organelles, Weibel–Palade Bodies
Abstract
1. Introduction
2. Results
2.1. Patient Characterization
2.1.1. Patient Phenotype Description
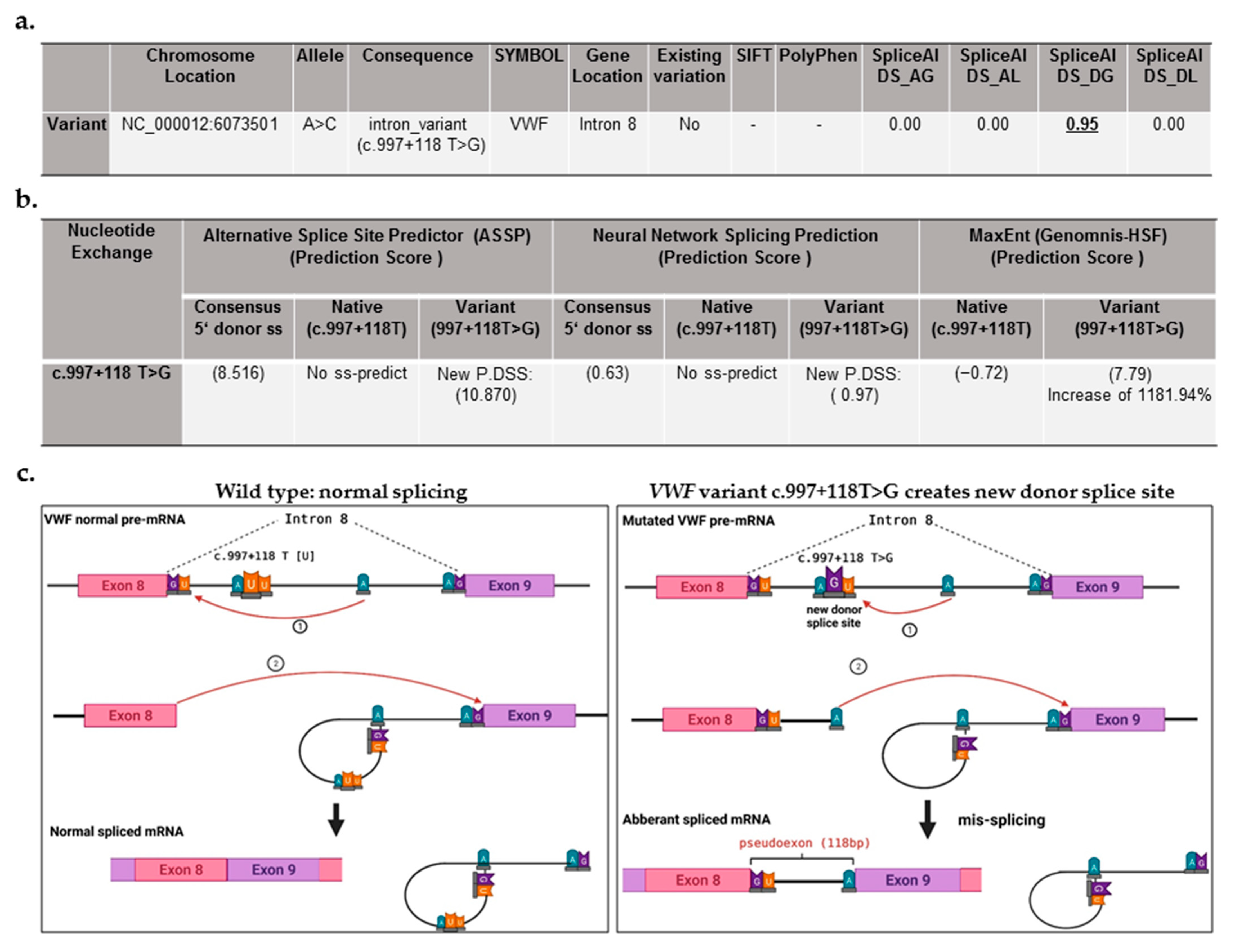
2.1.2. Patient Genotype: Detection of a Deep Intronic Variant, Creating New Donor Splice Site
| Mutation Routine DNA Analysis (Sanger Seq. and MLPA) | Mutation NGS of Whole VWF | Age (Years) | VWF:Ag IU/dL | VWF:GPIbM IU/dL | FVIII:C IU/dL | Bleeding Manifestations | |
|---|---|---|---|---|---|---|---|
| Index Patient | No mutation | c.997 + 118 T > G (g.6073501 A > C) | 26 | <4 | <4 | 2.0 | Frequent nose bleeding, easy bruising, menorrhagia, bleeding after injury |
| Normal Range | ― | ― | ― | 65–165 | 64–150 | 70–157 | ― |
2.2. ECFC Characterization: Confirming Endothelial Cells’ Phenotype
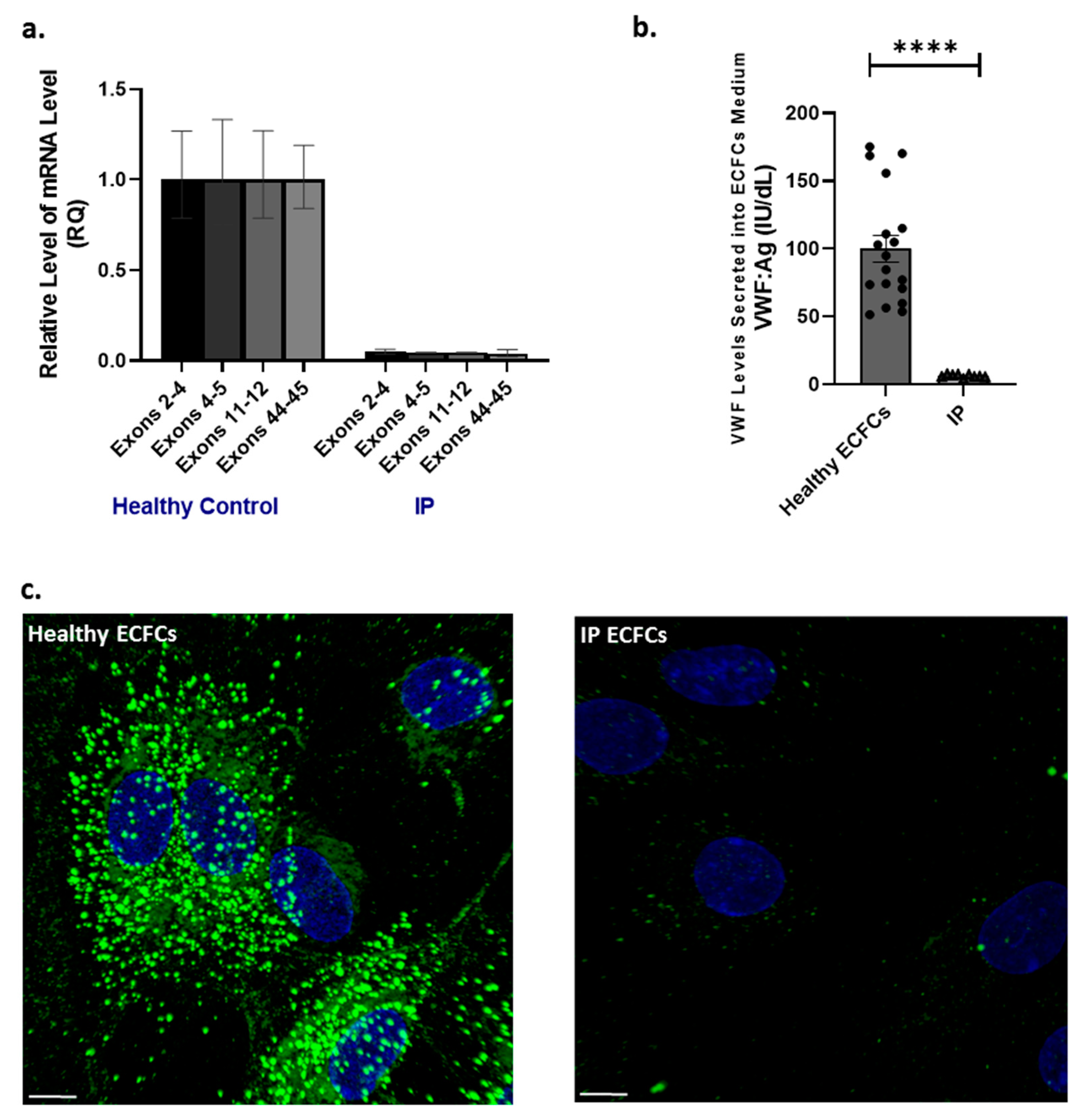
2.3. VWF RNA Transcripts, Endogenous VWF Biosynthesis, and Storage in ECFCs
2.4. Alternative Trafficking of WPBs’ Proinflammatory Ang2 Cargo in IP Cells
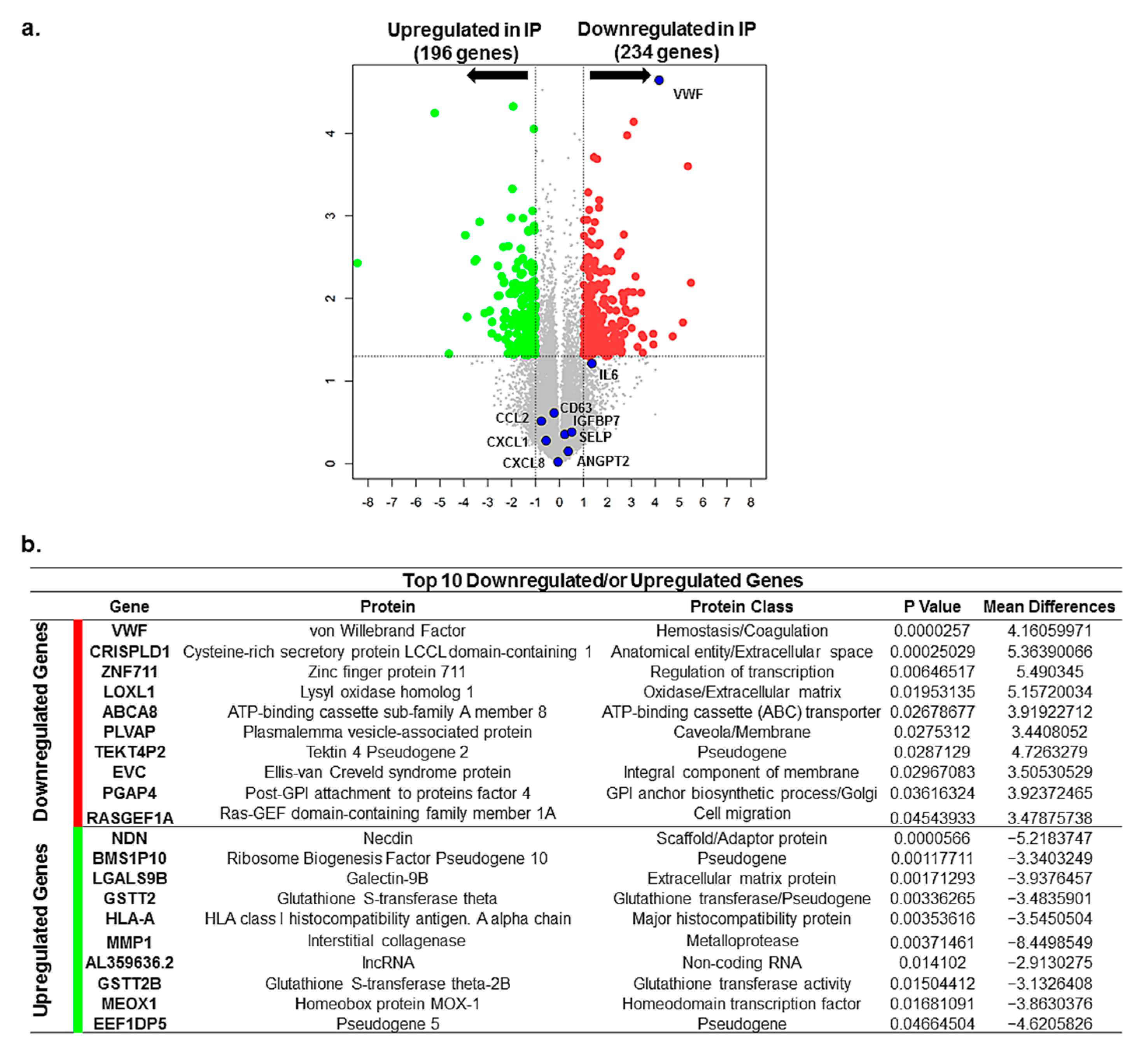
2.5. Differential Gene Expression and Altered Signaling Pathways in IP ECFCs
3. Discussion
4. Materials and Methods
4.1. Patient: Phenotype and Genetic Analyses
4.2. Isolation and Ex Vivo Expansion of ECFCS
4.3. VWF Transcript Analysis and Quantitative Real-Time PCR
4.4. Whole-Transcriptome RNA-Seq Analysis
4.5. VWF Production Evaluation in ECFCs
4.6. Immunofluorescence Microscopy
4.7. Data Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lenting, P.J.; Christophe, O.D.; Denis, C.V. Von Willebrand factor biosynthesis, secretion, and clearance: Connecting the far ends. Blood 2015, 125, 2019–2028. [Google Scholar] [CrossRef] [PubMed]
- Leebeek, F.W.G.; Eikenboom, J.C.J. Von Willebrand’s Disease. N. Engl. J. Med. 2017, 376, 701–702. [Google Scholar] [CrossRef]
- Lillicrap, D. Von Willebrand disease: Advances in pathogenetic understanding, diagnosis, and therapy. Blood 2013, 122, 3735–3740. [Google Scholar] [CrossRef]
- Castaman, G. How I treat von Willebrand disease. Thromb. Res. 2020, 196, 618–625. [Google Scholar] [CrossRef]
- James, P.D.; Connell, N.T.; Ameer, B.; Di Paola, J.; Eikenboom, J.; Giraud, N.; Haberichter, S.; Jacobs-Pratt, V.; Konkle, B.; McLintock, C.; et al. ASH ISTH NHF WFH 2021 guidelines on the diagnosis of von Willebrand disease. Blood Adv. 2021, 5, 280–300. [Google Scholar] [CrossRef] [PubMed]
- Sadler, J.E.; Budde, U.; Eikenboom, J.C.J.; Favaloro, E.J.; Hill, F.G.H.; Holmberg, L.; INGERSLEV, J.; Lee, C.A.; Lillicrap, D.; Mannucci, P.M.; et al. Update on the pathophysiology and classification of von Willebrand disease: A report of the Subcommittee on von Willebrand Factor. J. Thromb. Haemost. 2006, 4, 2103–2114. [Google Scholar] [CrossRef] [PubMed]
- Pagliari, M.T.; Rosendaal, F.R.; Ahmadinejad, M.; Badiee, Z.; Baghaipour, M.-R.; Baronciani, L.; Benítez Hidalgo, O.; Bodó, I.; Budde, U.; Castaman, G.; et al. Von Willebrand factor propeptide and pathophysiological mechanisms in European and Iranian patients with type 3 von Willebrand disease enrolled in the 3WINTERS-IPS study. J. Thromb. Haemost. 2022. [Google Scholar] [CrossRef]
- de Jong, A.; Eikenboom, J. Von Willebrand disease mutation spectrum and associated mutation mechanisms. Thromb. Res. 2017, 159, 65–75. [Google Scholar] [CrossRef]
- Mancuso, D.J.; Tuley, E.A.; Westfield, L.A.; Worrall, N.K.; Shelton-Inloes, B.B.; Sorace, J.M.; Alevy, Y.G.; Sadler, J.E. Structure of the gene for human von Willebrand factor. J. Biol. Chem. 1989, 264, 19514–19527. [Google Scholar] [CrossRef]
- Zhou, Y.-F.; Eng, E.T.; Zhu, J.; Lu, C.; Walz, T.; Springer, T.A. Sequence and structure relationships within von Willebrand factor. Blood 2012, 120, 449–458. [Google Scholar] [CrossRef]
- Yadegari, H.; Biswas, A.; Ahmed, S.; Naz, A.; Oldenburg, J. Von Willebrand factor propeptide missense variants affect anterograde transport to Golgi resulting in ER retention. Hum. Mutat. 2021, 42, 731–744. [Google Scholar] [CrossRef] [PubMed]
- Purvis, A.R.; Sadler, J.E. A covalent oxidoreductase intermediate in propeptide-dependent von Willebrand factor multimerization. J. Biol. Chem. 2004, 279, 49982–49988. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.-F.; Springer, T.A. Highly reinforced structure of a C-terminal dimerization domain in von Willebrand factor. Blood 2014, 123, 1785–1793. [Google Scholar] [CrossRef] [PubMed]
- Vischer, U.M.; Wagner, D.D. Von Willebrand factor proteolytic processing and multimerization precede the formation of Weibel-Palade bodies. Blood 1994, 83, 3536–3544. [Google Scholar] [CrossRef] [PubMed]
- Lippok, S.; Kolsek, K.; Lof, A.; Eggert, D.; Vanderlinden, W.; Muller, J.P.; Konig, G.; Obser, T.; Rohrs, K.; Schneppenheim, S.; et al. Von Willebrand factor is dimerized by protein disulfide isomerase. Blood 2016, 127, 1183–1191. [Google Scholar] [CrossRef]
- Valentijn, K.M.; Sadler, J.E.; Valentijn, J.A.; Voorberg, J.; Eikenboom, J. Functional architecture of Weibel-Palade bodies. Blood 2011, 117, 5033–5043. [Google Scholar] [CrossRef]
- Schillemans, M.; Kat, M.; Westeneng, J.; Gangaev, A.; Hofman, M.; Nota, B.; van Alphen Floris, P.; de Boer, M.; van den Biggelaar, M.; Margadant, C.; et al. Alternative trafficking of Weibel-Palade body proteins in CRISPR/Cas9-engineered von Willebrand factor-deficient blood outgrowth endothelial cells. Res. Pract. Thromb. Haemost. 2019, 3, 718–732. [Google Scholar] [CrossRef]
- Nightingale, T.D.; McCormack, J.J.; Grimes, W.; Robinson, C.; Lopes da Silva, M.; White, I.J.; Vaughan, A.; Cramer, L.P.; Cutler, D.F. Tuning the endothelial response: Differential release of exocytic cargos from Weibel-Palade bodies. J. Thromb. Haemost. 2018, 16, 1873–1886. [Google Scholar] [CrossRef]
- McCormack, J.J.; Lopes da Silva, M.; Ferraro, F.; Patella, F.; Cutler, D.F. Weibel-Palade bodies at a glance. J. Cell Sci. 2017, 130, 3611–3617. [Google Scholar] [CrossRef]
- Yadegari, H.; Driesen, J.; Pavlova, A.; Biswas, A.; Hertfelder, H.-J.; Oldenburg, J. Mutation distribution in the von Willebrand factor gene related to the different von Willebrand disease (VWD) types in a cohort of VWD patients. Thromb. Haemost. 2012, 108, 662–671. [Google Scholar] [CrossRef]
- Ahmad, F.; Budde, U.; Jan, R.; Oyen, F.; Kannan, M.; Saxena, R.; Schneppenheim, R. Phenotypic and molecular characterisation of type 3 von Willebrand disease in a cohort of Indian patients. Thromb. Haemost. 2013, 109, 652–660. [Google Scholar] [CrossRef] [PubMed]
- Baronciani, L.; Cozzi, G.; Canciani, M.T.; Peyvandi, F.; Srivastava, A.; Federici, A.B.; Mannucci, P.M. Molecular defects in type 3 von Willebrand disease: Updated results from 40 multiethnic patients. Blood Cells Mol. Dis. 2003, 30, 264–270. [Google Scholar] [CrossRef]
- Bowman, M.; Tuttle, A.; Notley, C.; Brown, C.; Tinlin, S.; Deforest, M.; Leggo, J.; Blanchette, V.S.; Lillicrap, D.; James, P. The genetics of Canadian type 3 von Willebrand disease: Further evidence for co-dominant inheritance of mutant alleles. J. Thromb. Haemost. 2013, 11, 512–520. [Google Scholar] [CrossRef] [PubMed]
- Gupta, P.K.; Saxena, R.; Adamtziki, E.; Budde, U.; Oyen, F.; Obser, T.; Schneppenheim, R. Genetic defects in von Willebrand disease type 3 in Indian and Greek patients. Blood Cells Mol. Dis. 2008, 41, 219–222. [Google Scholar] [CrossRef]
- Sutherland, M.S.; Keeney, S.; Bolton-Maggs, P.H.B.; Hay, C.R.M.; Will, A.; Cumming, A.M. The mutation spectrum associated with type 3 von Willebrand disease in a cohort of patients from the North West of England. Haemophilia 2009, 15, 1048–1057. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.; Yadegari, H.; Naz, A.; Biswas, A.; Budde, U.; Saqlain, N.; Amanat, S.; Tariq, S.; Raziq, F.; Masood, S.; et al. Characterization of the mutation spectrum in a Pakistani cohort of type 3 von Willebrand disease. Haemophilia 2019, 25, 1035–1044. [Google Scholar] [CrossRef] [PubMed]
- Yadegari, H.; Driesen, J.; Hass, M.; Budde, U.; Pavlova, A.; Oldenburg, J. Large deletions identified in patients with von Willebrand disease using multiple ligation-dependent probe amplification. J. Thromb. Haemost. 2011, 9, 1083–1086. [Google Scholar] [CrossRef] [PubMed]
- Yadegari, H.; Oldenburg, J. The Current Understanding of Molecular Pathogenesis of Quantitative von Willebrand Disease, Types 1 and 3. Hamostaseologie 2020, 40, 105–118. [Google Scholar] [CrossRef]
- de Boer, S.; Eikenboom, J. Von Willebrand Disease: From In Vivo to In Vitro Disease Models. Hemasphere 2019, 3, e297. [Google Scholar] [CrossRef]
- Liang, Q.; Lin, X.; Wu, X.; Shao, Y.; Chen, C.; Dai, J.; Lu, Y.; Wu, W.; Ding, Q.; Wang, X. Unraveling the molecular basis underlying nine putative splice site variants of von Willebrand factor. Hum. Mutat. 2021, 43, 215–227. [Google Scholar] [CrossRef]
- Hawke, L.; Bowman, M.L.; Poon, M.-C.; Scully, M.-F.; Rivard, G.-E.; James, P.D. Characterization of aberrant splicing of von Willebrand factor in von Willebrand disease: An underrecognized mechanism. Blood 2016, 128, 584–593. [Google Scholar] [CrossRef]
- Gallinaro, L.; Sartorello, F.; Pontara, E.; Cattini, M.G.; Bertomoro, A.; Bartoloni, L.; Pagnan, A.; Casonato, A. Combined partial exon skipping and cryptic splice site activation as a new molecular mechanism for recessive type 1 von Willebrand disease. Thromb. Haemost. 2006, 96, 711–716. [Google Scholar] [CrossRef] [PubMed]
- Yadegari, H.; Biswas, A.; Akhter, M.S.; Driesen, J.; Ivaskevicius, V.; Marquardt, N.; Oldenburg, J. Intron retention resulting from a silent mutation in the VWF gene that structurally influences the 5’ splice site. Blood 2016, 128, 2144–2152. [Google Scholar] [CrossRef] [PubMed]
- Conboy, J.G. A Deep Exon Cryptic Splice Site Promotes Aberrant Intron Retention in a Von Willebrand Disease Patient. Int. J. Mol. Sci. 2021, 22, 13248. [Google Scholar] [CrossRef] [PubMed]
- Vaz-Drago, R.; Custódio, N.; Carmo-Fonseca, M. Deep intronic mutations and human disease. Hum. Genet. 2017, 136, 1093–1111. [Google Scholar] [CrossRef]
- Khan, A.O.; Becirovic, E.; Betz, C.; Neuhaus, C.; Altmüller, J.; Maria Riedmayr, L.; Motameny, S.; Nürnberg, G.; Nürnberg, P.; Bolz, H.J. A deep intronic CLRN1 (USH3A) founder mutation generates an aberrant exon and underlies severe Usher syndrome on the Arabian Peninsula. Sci. Rep. 2017, 7, 1411. [Google Scholar] [CrossRef]
- Laflamme, N.; Lace, B.; Thonta Setty, S.; Rioux, N.; Labrie, Y.; Droit, A.; Chrestian, N.; Rivest, S. A Homozygous Deep Intronic Mutation Alters the Splicing of Nebulin Gene in a Patient With Nemaline Myopathy. Front. Neurol. 2021, 12, 660113. [Google Scholar] [CrossRef]
- Malekkou, A.; Sevastou, I.; Mavrikiou, G.; Georgiou, T.; Vilageliu, L.; Moraitou, M.; Michelakakis, H.; Prokopiou, C.; Drousiotou, A. A novel mutation deep within intron 7 of the GBA gene causes Gaucher disease. Mol. Genet. Genom. Med. 2020, 8, e1090. [Google Scholar] [CrossRef]
- Hujová, P.; Souček, P.; Grodecká, L.; Grombiříková, H.; Ravčuková, B.; Kuklínek, P.; Hakl, R.; Litzman, J.; Freiberger, T. Deep Intronic Mutation in SERPING1 Caused Hereditary Angioedema Through Pseudoexon Activation. J. Clin. Immunol. 2020, 40, 435–446. [Google Scholar] [CrossRef]
- Qian, X.; Wang, J.; Wang, M.; Igelman, A.D.; Jones, K.D.; Li, Y.; Wang, K.; Goetz, K.E.; Birch, D.G.; Yang, P.; et al. Identification of Deep-Intronic Splice Mutations in a Large Cohort of Patients With Inherited Retinal Diseases. Front. Genet. 2021, 12, 276. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, Y.; Ding, J.; Wang, F. mRNA analysis identifies deep intronic variants causing Alport syndrome and overcomes the problem of negative results of exome sequencing. Sci. Rep. 2021, 11, 18097. [Google Scholar] [CrossRef]
- Pezeshkpoor, B.; Zimmer, N.; Marquardt, N.; Nanda, I.; Haaf, T.; Budde, U.; Oldenburg, J.; El-Maarri, O. Deep intronic ‘mutations’ cause hemophilia A: Application of next generation sequencing in patients without detectable mutation in F8 cDNA. J. Thromb. Haemost. 2013, 11, 1679–1687. [Google Scholar] [CrossRef] [PubMed]
- Groeneveld, D.J.; van Bekkum, T.; Dirven, R.J.; Wang, J.-W.; Voorberg, J.; Reitsma, P.H.; Eikenboom, J. Angiogenic characteristics of blood outgrowth endothelial cells from patients with von Willebrand disease. J. Thromb. Haemost. 2015, 13, 1854–1866. [Google Scholar] [CrossRef] [PubMed]
- Starke, R.D.; Ferraro, F.; Paschalaki, K.E.; Dryden, N.H.; McKinnon, T.A.J.; Sutton, R.E.; Payne, E.M.; Haskard, D.O.; Hughes, A.D.; Cutler, D.F.; et al. Endothelial von Willebrand factor regulates angiogenesis. Blood 2011, 117, 1071–1080. [Google Scholar] [CrossRef] [PubMed]
- Denis, C.V.; André, P.; Saffaripour, S.; Wagner, D.D. Defect in regulated secretion of P-selectin affects leukocyte recruitment in von Willebrand factor-deficient mice. Proc. Natl. Acad. Sci. USA 2001, 98, 4072–4077. [Google Scholar] [CrossRef] [PubMed]
- Haberichter, S.L.; Merricks, E.P.; Fahs, S.A.; Christopherson, P.A.; Nichols, T.C.; Montgomery, R.R. Re-establishment of VWF-dependent Weibel-Palade bodies in VWD endothelial cells. Blood 2005, 105, 145–152. [Google Scholar] [CrossRef][Green Version]
- Gebrane-Younès, J.; Drouet, L.; Caen, J.P.; Orcel, L. Heterogeneous distribution of Weibel-Palade bodies and von Willebrand factor along the porcine vascular tree. Am. J. Pathol. 1991, 139, 1471–1484. [Google Scholar]
- Yadegari, H.; Jamil, M.A.; Müller, J.; Marquardt, N.; Rawley, O.; Budde, U.; El-Maarri, O.; Lillicrap, D.; Oldenburg, J. Multifaceted Pathomolecular Mechanism of a VWF Large Deletion Involved in the Pathogenesis of Severe VWD. Blood Adv. 2022, 6, 1038–1053. [Google Scholar] [CrossRef]
- Thurston, G.; Daly, C. The complex role of angiopoietin-2 in the angiopoietin-tie signaling pathway. Cold Spring Harb. Perspect. Med. 2012, 2, a006550. [Google Scholar] [CrossRef]
- Korhonen, E.A.; Lampinen, A.; Giri, H.; Anisimov, A.; Kim, M.; Allen, B.; Fang, S.; D’Amico, G.; Sipilä, T.J.; Lohela, M.; et al. Tie1 controls angiopoietin function in vascular remodeling and inflammation. J. Clin. Investig. 2016, 126, 3495–3510. [Google Scholar] [CrossRef]
- Fiedler, U.; Scharpfenecker, M.; Koidl, S.; Hegen, A.; Grunow, V.; Schmidt, J.M.; Kriz, W.; Thurston, G.; Augustin, H.G. The Tie-2 ligand angiopoietin-2 is stored in and rapidly released upon stimulation from endothelial cell Weibel-Palade bodies. Blood 2004, 103, 4150–4156. [Google Scholar] [CrossRef] [PubMed]
- Jang, C.; Koh, Y.J.; Lim, N.K.; Kang, H.J.; Kim, D.H.; Park, S.K.; Lee, G.M.; Jeon, C.J.; Koh, G.Y. Angiopoietin-2 exocytosis is stimulated by sphingosine-1-phosphate in human blood and lymphatic endothelial cells. Arterioscler. Thromb. Vasc. Biol. 2009, 29, 401–407. [Google Scholar] [CrossRef] [PubMed]
- Rodeghiero, F.; Tosetto, A.; Abshire, T.; Arnold, D.M.; Coller, B.; James, P.; Neunert, C.; Lillicrap, D. ISTH/SSC bleeding assessment tool: A standardized questionnaire and a proposal for a new bleeding score for inherited bleeding disorders. J. Thromb. Haemost. 2010, 8, 2063–2065. [Google Scholar] [CrossRef] [PubMed]
- McLaren, W.; Gil, L.; Hunt, S.E.; Riat, H.S.; Ritchie, G.R.S.; Thormann, A.; Flicek, P.; Cunningham, F. The Ensembl Variant Effect Predictor. Genome Biol. 2016, 17, 122. [Google Scholar] [CrossRef]
- Jaganathan, K.; Panagiotopoulou, S.K.; McRae, J.F.; Darbandi, S.F.; Knowles, D.; Li, Y.I.; Kosmicki, J.A.; Arbelaez, J.; Cui, W.; Schwartz, G.B.; et al. Predicting Splicing from Primary Sequence with Deep Learning. Cell 2019, 176, 535–548.e24. [Google Scholar] [CrossRef]
- Reese, M.G.; Eeckman, F.H.; Kulp, D.; Haussler, D. Improved splice site detection in Genie. J. Comput. Biol. 1997, 4, 311–323. [Google Scholar] [CrossRef]
- Wang, M.; Marín, A. Characterization and prediction of alternative splice sites. Gene 2006, 366, 219–227. [Google Scholar] [CrossRef]
- Martin-Ramirez, J.; Hofman, M.; van den Biggelaar, M.; Hebbel, R.P.; Voorberg, J. Establishment of outgrowth endothelial cells from peripheral blood. Nat. Protoc. 2012, 7, 1709–1715. [Google Scholar] [CrossRef]
- Ormiston, M.L.; Toshner, M.R.; Kiskin, F.N.; Huang, C.J.Z.; Groves, E.; Morrell, N.W.; Rana, A.A. Generation and Culture of Blood Outgrowth Endothelial Cells from Human Peripheral Blood. J. Vis. Exp. 2015, e53384. [Google Scholar] [CrossRef]
- Andrés-León, E.; Núñez-Torres, R.; Rojas, A.M. miARma-Seq: A comprehensive tool for miRNA, mRNA and circRNA analysis. Sci. Rep. 2016, 6, 25749. [Google Scholar] [CrossRef]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Yadegari, H.; Driesen, J.; Pavlova, A.; Biswas, A.; Ivaskevicius, V.; Klamroth, R.; Oldenburg, J. Insights into pathological mechanisms of missense mutations in C-terminal domains of von Willebrand factor causing qualitative or quantitative von Willebrand disease. Haematologica 2013, 98, 1315–1323. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yadegari, H.; Jamil, M.A.; Marquardt, N.; Oldenburg, J. A Homozygous Deep Intronic Variant Causes Von Willebrand Factor Deficiency and Lack of Endothelial-Specific Secretory Organelles, Weibel–Palade Bodies. Int. J. Mol. Sci. 2022, 23, 3095. https://doi.org/10.3390/ijms23063095
Yadegari H, Jamil MA, Marquardt N, Oldenburg J. A Homozygous Deep Intronic Variant Causes Von Willebrand Factor Deficiency and Lack of Endothelial-Specific Secretory Organelles, Weibel–Palade Bodies. International Journal of Molecular Sciences. 2022; 23(6):3095. https://doi.org/10.3390/ijms23063095
Chicago/Turabian StyleYadegari, Hamideh, Muhammad Ahmer Jamil, Natascha Marquardt, and Johannes Oldenburg. 2022. "A Homozygous Deep Intronic Variant Causes Von Willebrand Factor Deficiency and Lack of Endothelial-Specific Secretory Organelles, Weibel–Palade Bodies" International Journal of Molecular Sciences 23, no. 6: 3095. https://doi.org/10.3390/ijms23063095
APA StyleYadegari, H., Jamil, M. A., Marquardt, N., & Oldenburg, J. (2022). A Homozygous Deep Intronic Variant Causes Von Willebrand Factor Deficiency and Lack of Endothelial-Specific Secretory Organelles, Weibel–Palade Bodies. International Journal of Molecular Sciences, 23(6), 3095. https://doi.org/10.3390/ijms23063095






