Abstract
Staphylococcus epidermidis is a common cause of device related infections on which pathogens form biofilms (i.e., multilayered cell populations embedded in an extracellular matrix). Here, we report that the transcription factor SpoVG is essential for the capacity of S. epidermidis to form such biofilms on artificial surfaces under in vitro conditions. Inactivation of spoVG in the polysaccharide intercellular adhesin (PIA) producing S. epidermidis strain 1457 yielded a mutant that, unlike its parental strain, failed to produce a clear biofilm in a microtiter plate-based static biofilm assay. A decreased biofilm formation capacity was also observed when 1457 ΔspoVG cells were co-cultured with polyurethane-based peripheral venous catheter fragments under dynamic conditions, while the cis-complemented 1457 ΔspoVG::spoVG derivative formed biofilms comparable to the levels seen with the wild-type. Transcriptional studies demonstrated that the deletion of spoVG significantly altered the expression of the intercellular adhesion (ica) locus by upregulating the transcription of the ica operon repressor icaR and down-regulating the transcription of icaADBC. Electrophoretic mobility shift assays (EMSA) revealed an interaction between SpoVG and the icaA-icaR intergenic region, suggesting SpoVG to promote biofilm formation of S. epidermidis by modulating ica expression. However, when mice were challenged with the 1457 ΔspoVG mutant in a foreign body infection model, only marginal differences in biomasses produced on the infected catheter fragments between the mutant and the parental strain were observed. These findings suggest that SpoVG is critical for the PIA-dependent biofilm formation of S. epidermis under in vitro conditions, but is largely dispensable for biofilm formation of this skin commensal under in vivo conditions.
1. Introduction
Implanted medical devices are becoming more and more important in modern medicine. However, at the same time, these implants pose a serious risk for the patient to develop an implantable device related infection, as they may serve as seeding ground for microorganisms to form a multicellular biofilm on these artificial surfaces. Of the nearly 2 million healthcare-associated infections reported by the Centers for Disease Control per year since the beginning of the 21st century, more than 50% can be attributed to indwelling medical devices [1]. Staphylococcus aureus and S. epidermidis are the most frequently reported bacterial species causing implantable device related infections [2]. Identification of the latter bacterial species—a highly abundant commensal of the human skin flora [3]—in positive blood cultures was considered for a long time primarily as contamination, however, in recent years, S. epidermidis has been also recognized as a relevant source for implantable device-related bloodstream infections [4] and late-onset neonatal sepsis [5]. In fact, S. epidermidis is nowadays identified as the most important cause of nosocomial catheter related blood stream infections (CRBSI) in numerous countries including Argentina [6], Germany [7], Portugal [8], South Africa [9], Switzerland [10], and the United States [11].
One of the reasons for the success of S. epidermidis as a nosocomial pathogen lies in its ability to attach to various materials including metal and plastic, to survive on these materials for several days, and to form biofilms on these materials if the environment allows for this (e.g., after the insertion of the implant material into the host). After primary attachment to a surface, the opportunistic pathogen starts to multiply and accumulate within an extracellular matrix (ECM), which is followed by the maturation of the biofilm and finally the dispersal of single cells (reviewed in [12,13]). Biofilm-released cells may spread to other organ systems, and are reported to induce a prompt and stronger inflammatory response than planktonic or biofilm-grown cells of this species [14].
The majority of S. epidermidis strains isolated from explanted medical devices produce an extracellular matrix that includes the polysaccharide intercellular adhesin (PIA) as main exopolysaccharide component, a partially deacetylated, positively charged poly-β(1-6)-N-acetylglucosamine, whose synthesis is mediated by the ica locus (reviewed in [15]). Beside various environmental stimuli such as temperature, iron- and oxygen limitation, and the amount of available nutrient sources [16,17], PIA-dependent biofilm formation of S. epidermidis underlies several internal regulation mechanisms (reviewed in [13]). The alternative transcription factor σB and the staphylococcal accessory regulator SarA positively affect PIA production by enhancing the transcription of the polycistronic transcript icaADBC [18,19], while IcaR, a transcriptional regulator which is also part of the ica locus, reduces PIA production by repressing icaADBC transcription [20]. Expression of the ica locus in S. epidermidis is further affected by serine/threonine kinases Stk1 [21], the catabolite control protein A [22], SarX [23], TcaR [24,25], VraSR [26], non-coding RNAs [27,28], and by phase variation via the insertion/excision of the insertion sequence IS256 into different regions of the ica locus [29].
The lack of an apparent σB binding consensus within the promoter region of the ica locus suggests that the stimulating effect of this alternative sigma factor on icaADBC transcription is probably of indirect nature. One putative mediator of σB activity could be SpoVG (aka BarB in S. epidermidis), a DNA/RNA-binding protein that is highly conserved in eubacteria [30]. SpoVG is a known regulator of virulence in a number of pathogenic firmicutes such as Bacillus cereus [31], Listeria monocytogenes [32], and S. aureus [33,34,35]. The regulatory protein is also reported to affect biofilm formation in B. cereus [31]. Given that σB controls the transcription of SpoVG in S. aureus [33] and that activity of SpoVG is enhanced by Stk1-mediated phosphorylation in the latter species [36], we wondered whether and how SpoVG might affect biofilm formation and ica transcription in S. epidermidis. Here, we report on the deletion of spoVG in the PIA-biofilm producing S. epidermidis central venous catheter isolate 1457 [37] and its impact on growth, ica transcription and biofilm formation under in vitro and in vivo conditions.
2. Results
2.1. Genetic Organization of the yabJ-spoVG Locus in S. epidermidis 1457
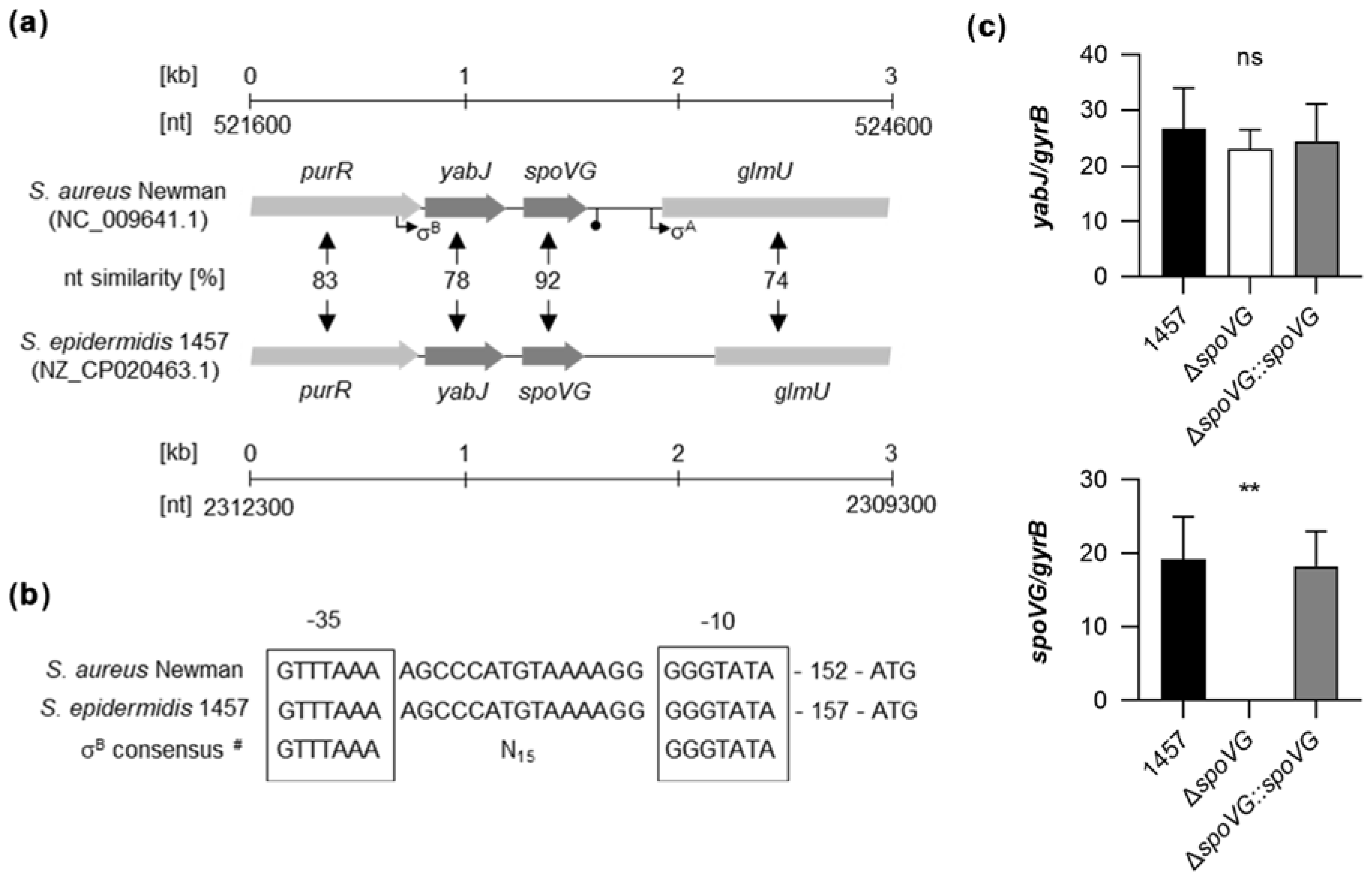
Genome comparison indicates that the genetic organization of the purR-yabJ-spoVG-glmU region is highly conserved among the staphylococcal species S. aureus and S. epidermidis (Figure 1a). Given that yabJ-spoVG is co-transcribed in S. aureus under the control of a σB-dependent promoter [33], and that the nucleotide sequences of the σB promoter in front of yabJ are identical between S. aureus Newman and S. epidermidis 1457 (Figure 1b), one can assume that yabJ and spoVG are also co-transcribed in S. epidermidis, asking for a careful deletion strategy that avoids polar effects on yabJ.
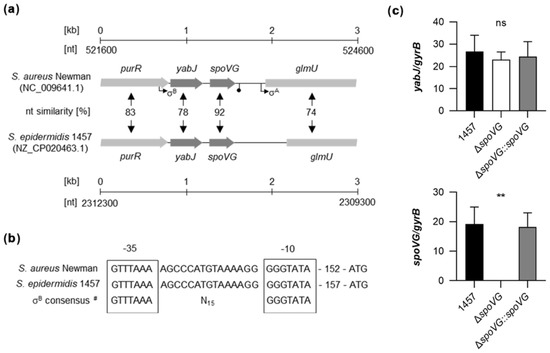
Figure 1.
Genetic organization of the yabJ-spoVG locus in staphylococci. (a) Schematic representation of the yabJ-spoVG regions in S. aureus Newman and S. epidermidis 1457. Open reading frames (ORFs), promoters, terminators, and nucleotide similarities between homologous ORFs are indicated. (b) Nucleotide sequences of the σB-promoter regions in front of yabJ. σ-factor binding sites are framed. #, σB-promoter consensus motif of S. aureus derived from reference [38]. (c) Quantitative transcript analyses of yabJ and spoVG by qRT-PCR in S. epidermidis 1457, 1457 ΔspoVG, and 1457 ΔspoVG::spoVG cells grown in TSB at 37 °C and 225 rpm for 3 h. Transcripts were quantified in reference to the transcription of gyrB (in copies per copy of gyrB). Data are presented as mean + SD of five biological replicates. **, p < 0.01; ns, not significant (Kruskal-Wallis test and Dunn’s multiple comparisons test. Only differences between 1457 and mutants are shown).
To confirm that yabJ was not affected by our spoVG deletion in S. epidermidis 1457, we first tested the transcription rates of yabJ and spoVG by quantitative Real-Time Reverse-Transcription PCR (qRT-PCR) in the strain triplet 1457, 1457 ΔspoVG, and 1457 ΔspoVG::spoVG (Figure 1c). After 3 h of growth, comparable yabJ transcript rates were seen in all three derivatives, while spoVG transcripts were missing in the 1457 ΔspoVG derivative, but detected at a comparable level in the wild-type and the cis-complemented strain. These findings confirmed that spoVG was successfully deleted in strain 1457 ΔspoVG without altering yabJ transcription, and that cis-complementation of the spoVG deletion with a functional spoVG allowed for a complete restoration of spoVG transcription in 1457 ΔspoVG::spoVG.
2.2. Impact of the spoVG Deletion on Growth of S. epidermidis 1457
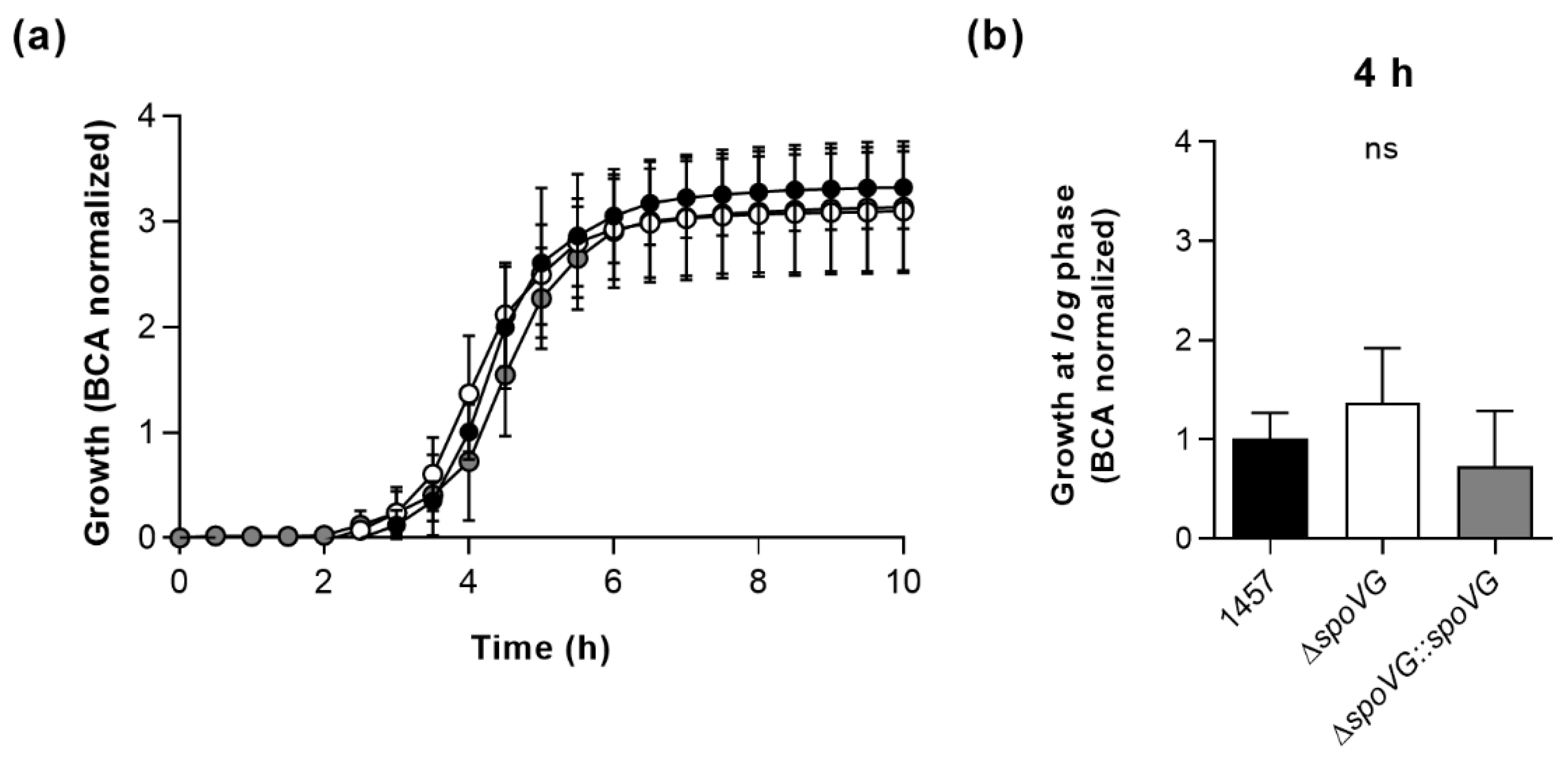
In order to determine the impact of the spoVG deletion on in vitro growth, the strain triplet was first cultured under static conditions in 96-well microtiter plates on an oCelloScope system [39] in Luria-Bertani (LB) broth at 37 °C and 5% CO2 (Figure 2). Under these microaerophilic growth conditions, all three strains exhibited comparable growth kinetics (Figure 2a), although 1457 ΔspoVG cultures tended to produce slightly higher cell densities after 4 h of growth than wild-type and 1457 ΔspoVG::spoVG cultures (Figure 2b), however, this effect was not statistically significant (p ≥ 0.275, Kruskal Wallis test and Dunn’s multiple comparisons test).
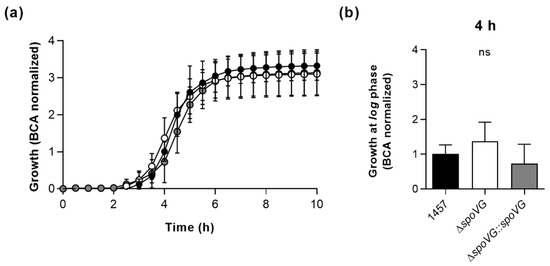
Figure 2.
Effect of the spoVG deletion on growth of S. epidermidis strain 1457 under static conditions in Luria Bertani (LB) broth. Bacterial cells were cultured in LB broth under static conditions at 37 °C and 5% CO2 for 10 h. Growth was measured by optical density using the oCelloScope BCA algorithm. (a) Growth kinetics of 1457 (black symbols), 1457 ΔspoVG (white symbols) and 1457 ΔspoVG::spoVG (grey symbols) cell suspensions. The graphs represent the average growth values ± SD of three biological experiments carried out in triplicate. (b) BCA normalized optical densities of the cultures at 4 h of growth. ns, not significant (Kruskal-Wallis test and Dunn’s multiple comparisons test).
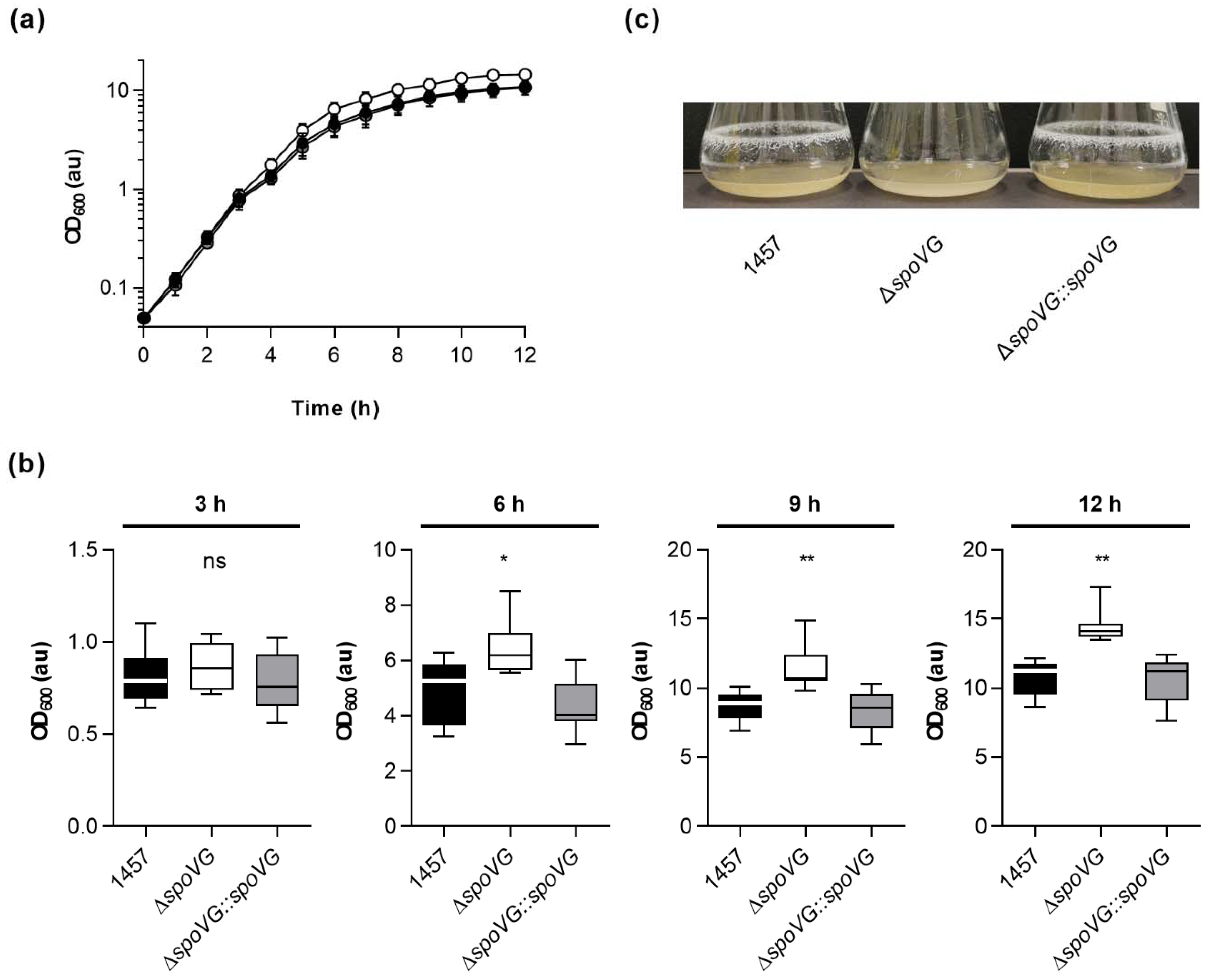
To test whether SpoVG might render the growth behavior of strain 1457 under more aerobic conditions, growth kinetics of the strain triplet were next analyzed in glass flasks in the bacterial cell culture medium tryptic soy broth (TSB) at 37 °C and 225 rpm (culture to flask volume 1:10). During the early growth stages (i.e., lag phase and early exponential growth phase), again no clear differences in growth rates were observed between all three strains (Figure 3a and Table 1).
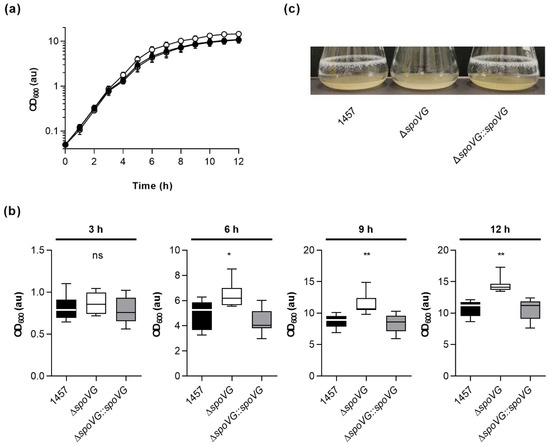
Figure 3.
Effect of the spoVG deletion on growth of S. epidermidis strain 1457 under dynamic conditions in tryptic soy broth (TSB). Bacteria were inoculated to an optical density at 600 nm (OD600) of 0.05 in TSB and cultured aerobically at 37 °C and 225 rpm in a culture to flask volume of 1:10. OD600 measurements of the cultures were determined hourly. Samples were diluted in PBS when an OD600 value of 0.8 was reached. The dilution factor was used to multiply with the measured result for the OD600 values displayed in the graphs. Calculated values are given as arbitrary units (au). (a) Growth kinetics of 1457 (black symbols), 1457 ΔspoVG (white symbols), and 1457 ΔspoVG::spoVG (grey symbols) cell suspensions. The results are the mean ± SD of eight biological replicates. (b) OD600 values (au) of the cell cultures at 3, 6, 9, and 12 h of growth, respectively. Data are presented as box and whisker plot showing the interquartile range (25–75%, box), the median (horizontal line) and the standard deviation (bars) of eight biological replicates. * p < 0.05; ** p < 0.01; ns, not significant (Kruskal-Wallis test and Dunn’s multiple comparisons test. Only differences between 1457 and mutants are shown). (c) Representative images of biofilms formed on glass flasks by cell suspensions cultured in TSB at 37 °C and 225 rpm for 12 h.

Table 1.
Generation time of S. epidermidis exponential growth phase cells cultivated in TSB under aerobic conditions.
However, at about 4 h of cultivation, first cell aggregates on the glass surface at the air-liquid interface became visible with the wild-type and 1457 ΔspoVG::spoVG cultures but not with the 1457 ΔspoVG cultures, suggesting that parts of the wild-type and 1457 ΔspoVG::spoVG cell populations started to switch from planktonic growth to a biofilm-mode of growth.
In line with this partial change in growth mode, optical densities of the wild-type and 1457 ΔspoVG::spoVG cultures at 600 nm (OD600) fell significantly back behind the OD600 values seen with 1457 ΔspoVG cultures (Figure 3b). This growth behavior was maintained by all three strains until the end of the observation period of 12 h, at which strong biofilms on the glass surface at the air-liquid interface were observed for the wild-type and 1457 ΔspoVG::spoVG cultures, which were completely missing with the 1457 ΔspoVG cultures (Figure 3c). These findings suggest that SpoVG affects the growth behavior and biofilm formation capacity of S. epidermidis strain 1457, if cultured in TSB.
2.3. Impact of the spoVG Deletion on Biofilm Formation of S. epidermidis 1457
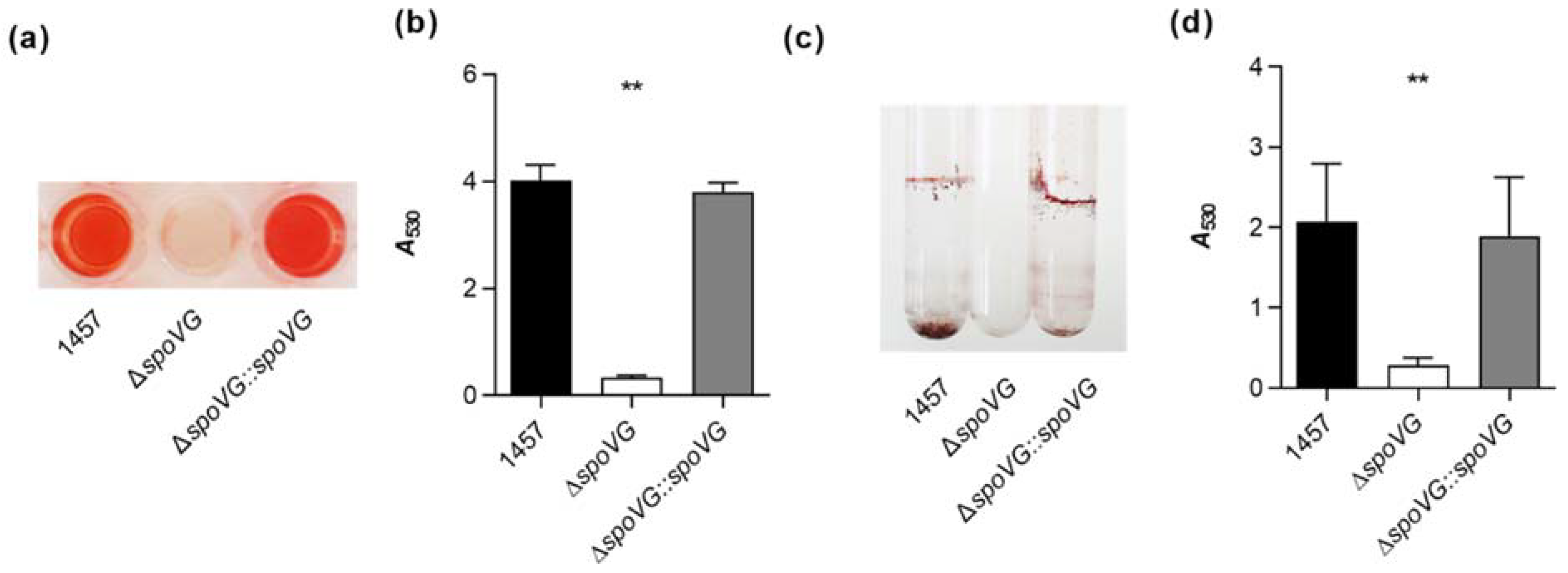
To substantiate our hypothesis that the spoVG deletion in S. epidermidis 1457 might affect biofilm formation, three different in vitro biofilm assays were carried out. We first studied the biofilm formation capacity of the strain triplet under static conditions in cell culture-treated 96-well flat-bottom microplates as described in [40]. After 18 h of growth in TSB, cultures of the wild-type and the cis-complemented strain produced strong biofilms on the bottom of the microtiter plate wells (Figure 4a). Wells inoculated with the ΔspoVG mutant, however, produced after 18 h only weak biofilms that were about one log lower than the ones observed with strains 1457 and 1457 ΔspoVG::spoVG (Figure 4b). Next, we assayed the biofilm formation of the strain triplet under dynamic conditions in a glass tube assay. Here, after cultivation for 18 h at 37 °C and 150 rpm, cultures of the wild-type and the cis-complemented strain produced clearly visible biofilms at the air-liquid interface and the bottom of the glass tubes, while the glass tubes of cultures inoculated with the ΔspoVG mutant remained virtually free from cell populations sticking to the glass wall (Figure 4c).
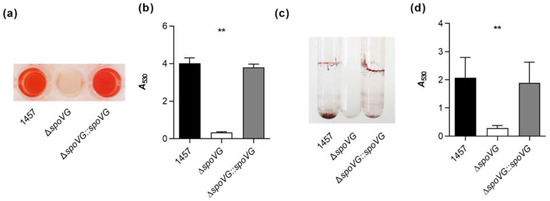
Figure 4.
Effect of the spoVG deletion on in vitro biofilm formation of S. epidermidis strain 1457. (a,b) Biofilm growth of S. epidermidis strains 1457 (black symbols), 1457 ΔspoVG (white symbols), and 1457 ΔspoVG::spoVG (grey symbols) in a static 96-well microplate assay. Cells were incubated for 18 h at 37 °C in TSB. (a) Representative images of safranin-stained biofilms formed at the bottom of the wells. (b) Absorption rates of safranin-stained biofilms at 530 nm. The data show the mean + SD of six biological replicates carried out in triplicate. (c,d) Biofilm growth of the strain triplet under dynamic conditions in glass tubes. (c) Representative image of safranin-stained biofilms formed on the glass surfaces after 18 h of growth in TSB at 37 °C and 150 rpm. (d) Absorption rates of safranin-stained biofilms at 530 nm. The data show the mean + SD of six biological replicates. ** p < 0.01 (Kruskal-Wallis test and Dunn’s multiple comparisons test, only differences between 1457 and mutants are shown).
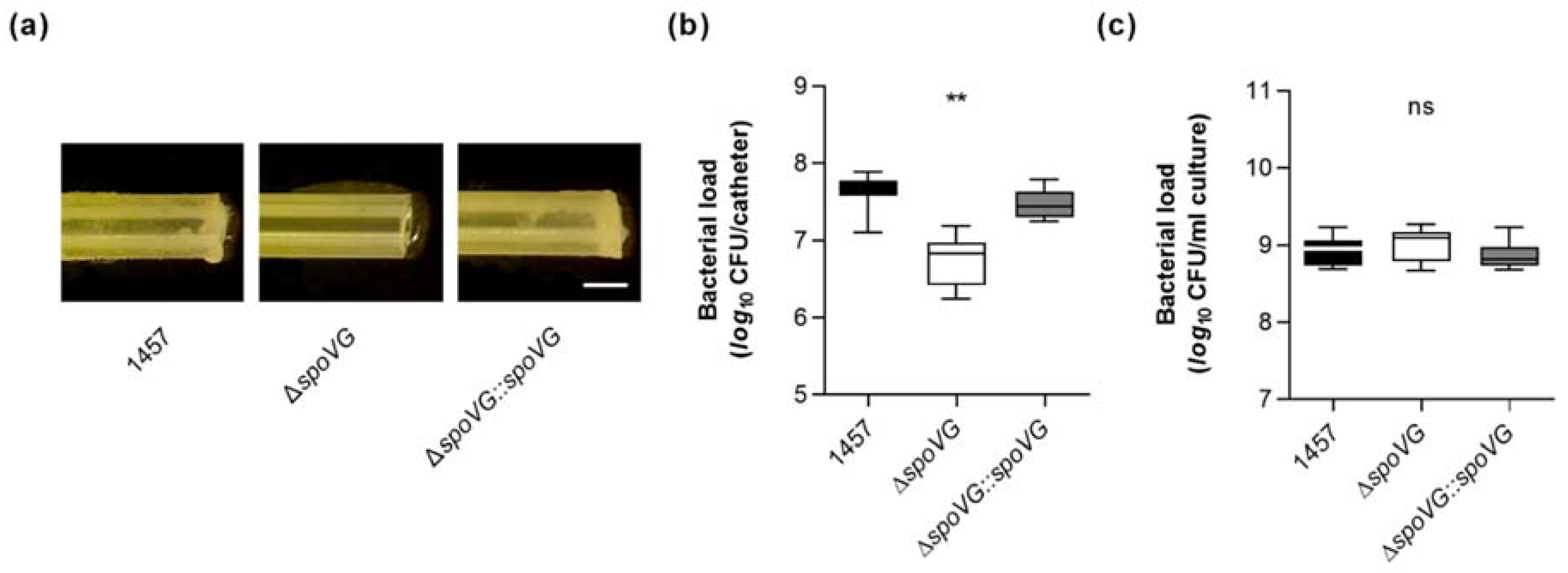
Quantification of the biomasses that attached to the glass walls by safranin staining revealed, in line with the macroscopic observations, about 10-times higher absorption rates at 530 nm (A530) for the biofilms formed by strains 1457 and 1457 ΔspoVG::spoVG, if compared to A530 values obtained with the isogenic spoVG deletion mutant in this assay (Figure 4d). To approach more closely the situation encountered by S. epidermidis in the host setting, the biofilm formation capacities of the strain triplet were next tested on peripheral venous catheter (PVC) fragments under dynamic conditions [41]. After five days of cultivation under non-limited nutrient conditions, cultures of strains 1457 and 1457 ΔspoVG::spoVG produced macroscopically visible biofilms on the catheter surfaces, while no such biofilm formation was observed with the 1457 ΔspoVG cultures (Figure 5a). Quantification of the bacterial cell numbers that attached to the catheter surfaces revealed, in line with the macroscopic observations, about 10-times higher colony forming unit (CFU) rates for the wild-type when compared to the 1457 ΔspoVG mutant, and this phenotype was again fully complemented by introducing a functional spoVG into the spoVG deletion mutant (Figure 5b). Notably, albeit of the fact that no clear biofilm was visible on the outer surfaces of the catheter tubing co-cultured with the 1457 ΔspoVG mutant, about 5 × 106 CFU/catheter were detached from the catheter surfaces, indicating that the spoVG deletion mutant did not fully lose its ability to attach to the PVC surface.
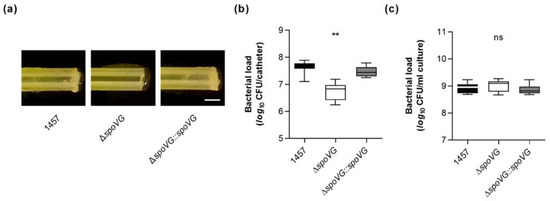
Figure 5.
Effect of the spoVG deletion on in vitro biofilm formation of S. epidermidis strain 1457 on catheter tubing. (a) Representative images of S. epidermidis-loaded catheter fragments at day 5 post inoculation (12.5-fold magnification). The results are representative of three independent experiments. Scale bar, 1 mm. (b,c) CFU rates of detached biofilms (b) and cultures (c) were determined by plate counting. The data are presented as box and whisker plot showing the interquartile range (25–75%, box), the median (horizontal line), and the standard deviation (bars) of nine independent experiments. ** p < 0.01; ns, not significant (Kruskal-Wallis test and Dunn’s multiple comparisons test, only differences between 1457 and mutants are shown).
However, we cannot exclude that at least part of the CFUs obtained from the 1457 ΔspoVG co-incubated catheter tubing originated from the bacterial cell population, which grew planktonically in the culture medium (Figure 5c), although catheter fragments were carefully washed prior to the detachment of adherent bacterial cells from the catheter surfaces by sonication. It should be also noted here that CFU determinations of detached biofilms might underestimate the real number of viable cells in these biofilms, as sonication of detached biofilms does not necessarily dissolve all bacterial cells from the ECM and might leave some smaller or even larger cell aggregates [42], which would also yield, similar to individual cells, in single colonies (if cultured on agar plates).
2.4. Impact of the spoVG Deletion on ica Transcription of S. epidermidis 1457
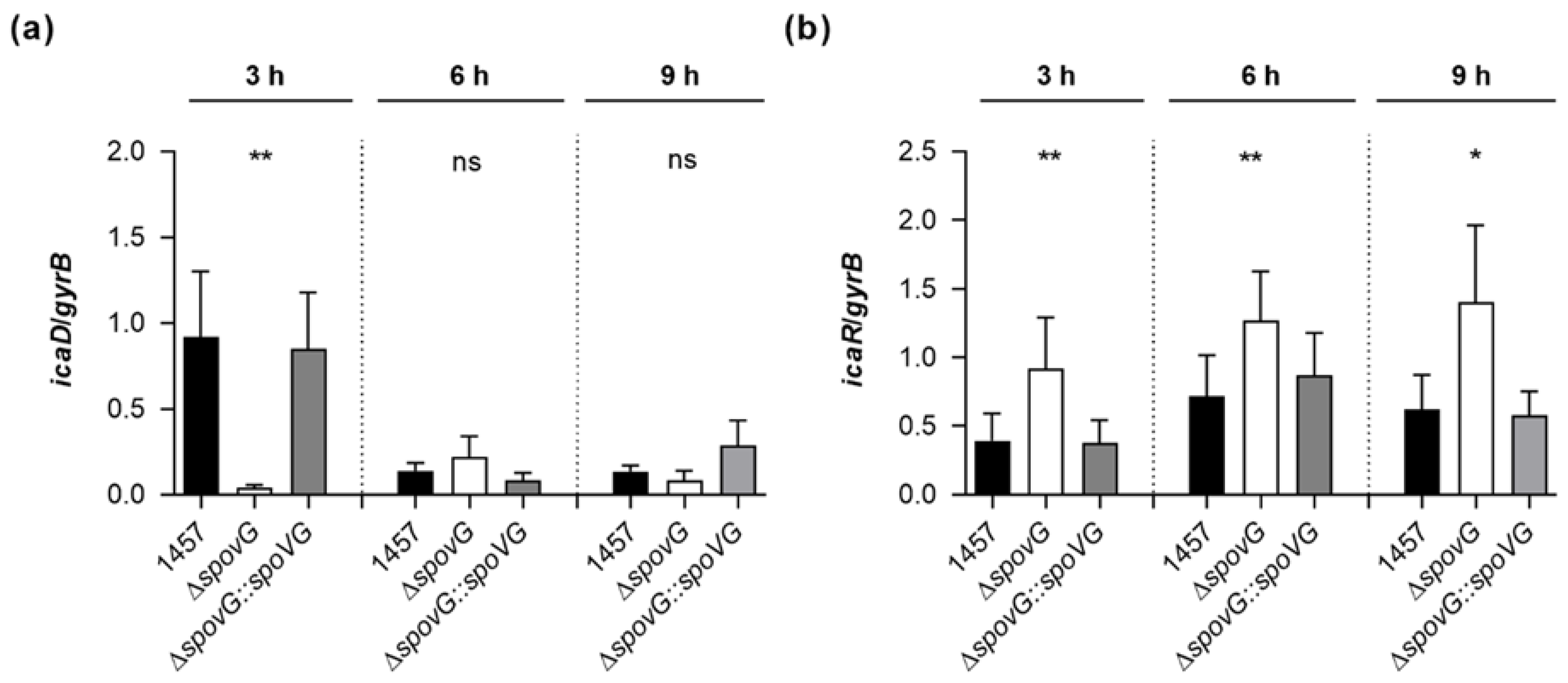
S. epidermidis strain 1457 is known to form a PIA-dependent biofilm under in vitro conditions [37]. Given the clear impact of the spoVG deletion on the in vitro biofilm formation capacities of strain 1457, we assumed that SpoVG might influence PIA production via the transcriptional control of the ica locus. To test this hypothesis, we quantified the transcription rates of icaD, which is part of the icaADBC polycistronic transcript encoding the protein machinery required to synthesize PIA, and of icaR, encoding a repressor of icaADBC [43], in TSB cultured cells by qRT-PCR (Figure 6). In line with our earlier observations [22], we detected the highest icaD transcript rates during the early growth phase (i.e., 3 h of growth) in TSB cultured 1457 cells, underlining the growth phase-dependent transcription of the icaADBC polycistronic transcript in glucose-containing medium [44]. Deletion of spoVG in strain 1457 strongly reduced the icaD transcript rates of the mutant at this early growth stage by a factor of ~20-fold, while cis-complementation of the 1457 ΔspoVG mutant with a functional spoVG created a derivative that produced icaD transcript levels comparable to the ones seen with the wild-type (Figure 6a). At the later growth stages monitored (i.e., after 6 and 9 h of growth), all three strains produced comparable icaD transcript rates, suggesting that SpoVG affects ica expression predominantly during the early growth stages. Notably, deletion of spoVG in strain 1457 also rendered the transcription of icaR (Figure 6b).
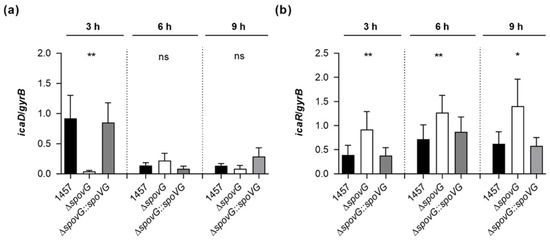
Figure 6.
Effect of the spoVG deletion on ica transcription of S. epidermidis strain 1457. Bacterial cells were cultured aerobically in TSB, as outlined in Materials and Methods. Cells were sampled at the time points indicated, total RNAs isolated, and qRT-PCRs performed for icaD (a) and icaR (b). Transcripts were quantified in reference to the transcription of gyrase B (gyrB). Data are presented as mean + SD of eight biological replicates. * p < 0.05; ** p < 0.01; ns, not significant (Kruskal-Wallis test and Dunn’s multiple comparisons test, only differences between 1457 and mutants are shown).
While the wild-type and 1457 ΔspoVG::spoVG cell populations produced icaR transcript rates that were comparable throughout growth, significantly higher icaR transcript rates were observed with the 1457 ΔspoVG cell populations at all three time points analyzed, if compared to the transcript levels seen with the wild-type cultures. These findings suggest that SpoVG might augment icaADBC transcription via the modulation of icaR transcription.
2.5. Interaction of SpoVG with the icaA-icaR Intergenic Region in S. epidermidis 1457
Animated by our icaD/icaR transcriptional findings, we conducted a series of electrophoresis mobility shift assays (EMSAs) to test whether SpoVG might directly interact with the icaA-icaR intergenic region (Figure 7).
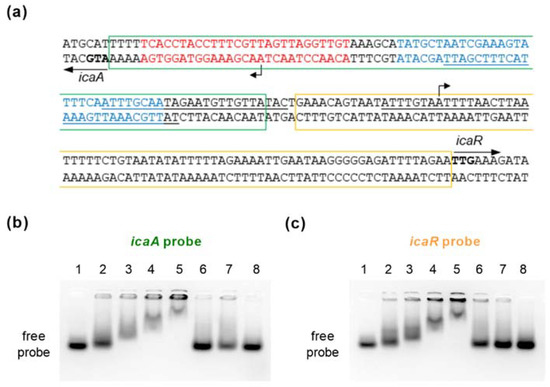
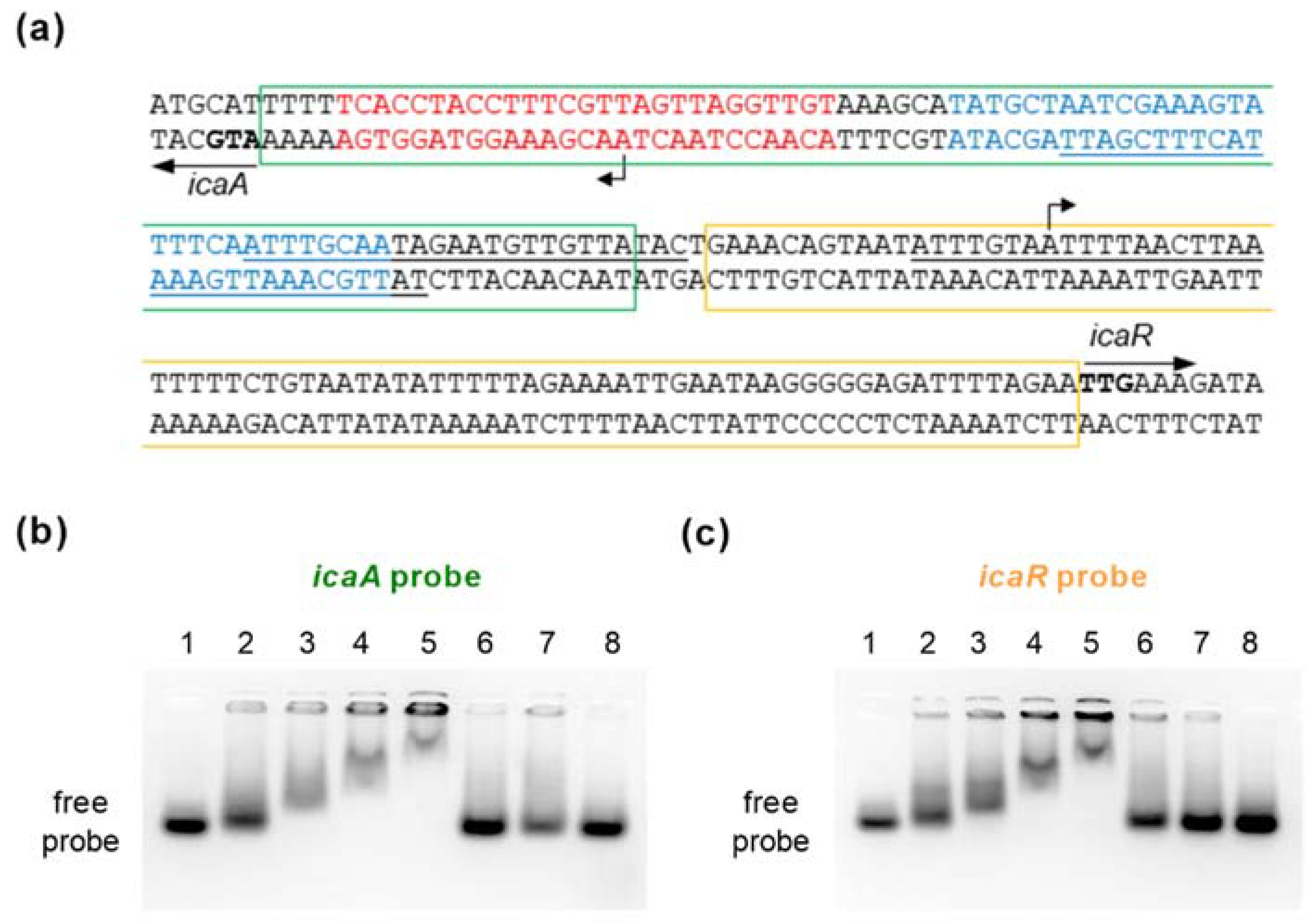
Figure 7.
EMSA using purified SpoVG and double-stranded DNA probes generated from the icaA-icaR intergenic region. (a) Nucleotide sequences of the icaA-icaR intergenic region of S. epidermidis 1457 (NCBI Reference Sequence NZ_CP020463.1). Start codons of the icaA and icaR ORFs are highlighted in bold. The highest affinity TcaR binding site proposed by [24] is indicated by blue letters. Putative SarA binding sites proposed by [19] are underlined. The IcaR binding site in front of icaA is depicted in red letters [45]. Regions covered by the icaA and icaR probes used for EMSAs are framed by green and orange rectangles, respectively. TSS of icaA [46] and icaR [47] are indicated by bent arrows (b,c) EMSAs were performed using purified recombinant N-terminally HAT-HA-tagged SpoVG and a 80-mer labeled dsDNA fragments covering the icaA (b) and icaR (c) TSS, respectively. Controls with no HAT-HA-SpoVG protein added are shown in the first lane, or with 2, 4, 8, and 12 µM of purified HAT-HA-SpoVG, respectively, in lanes 2 to 5. Labeled probes were also tested in presence of an excess of 20× or 10× unlabeled icaA (b) and icaR (c) probes (lanes 6 and 7; order switched between (b,c). A non-related 80-mer labeled dsDNA fragment in combination with 12 µM of HAT-HA-SpoVG served as specificity control (lane 8). Data are representative of at least two independent experiments.
This 165-bp spanning region harbors the transcriptional start sites (TSS) of icaADBC and icaR, respectively [46,47], and is a known binding site for regulators such as SarA [19], SarX [23], TcaR [24,25,48], and IcaR itself [45]. Two 80-nt spanning double-stranded DNA probes were generated that covered either the icaA or icaR TSS (termed icaA and icaR probe, respectively; Figure 7a). Both double-stranded DNA probes shifted with SpoVG in a dose-dependent manner (Figure 7b,c), while the control dsDNA fragment was not shifted in presence of the HAT-HA-tagged DNA/RNA-binding protein. Shifting of the labeled icaA and icaR probes was furthermore competed out by adding unlabeled icaA and icaR probes in excess, respectively, indicating that SpoVG can bind directly to the icaA and icaR promoter regions in S. epidermidis.
2.6. Impact of the spoVG Deletion on Infectivity of S. epidermidis 1457 in a Murine Foreign Body Infection Model
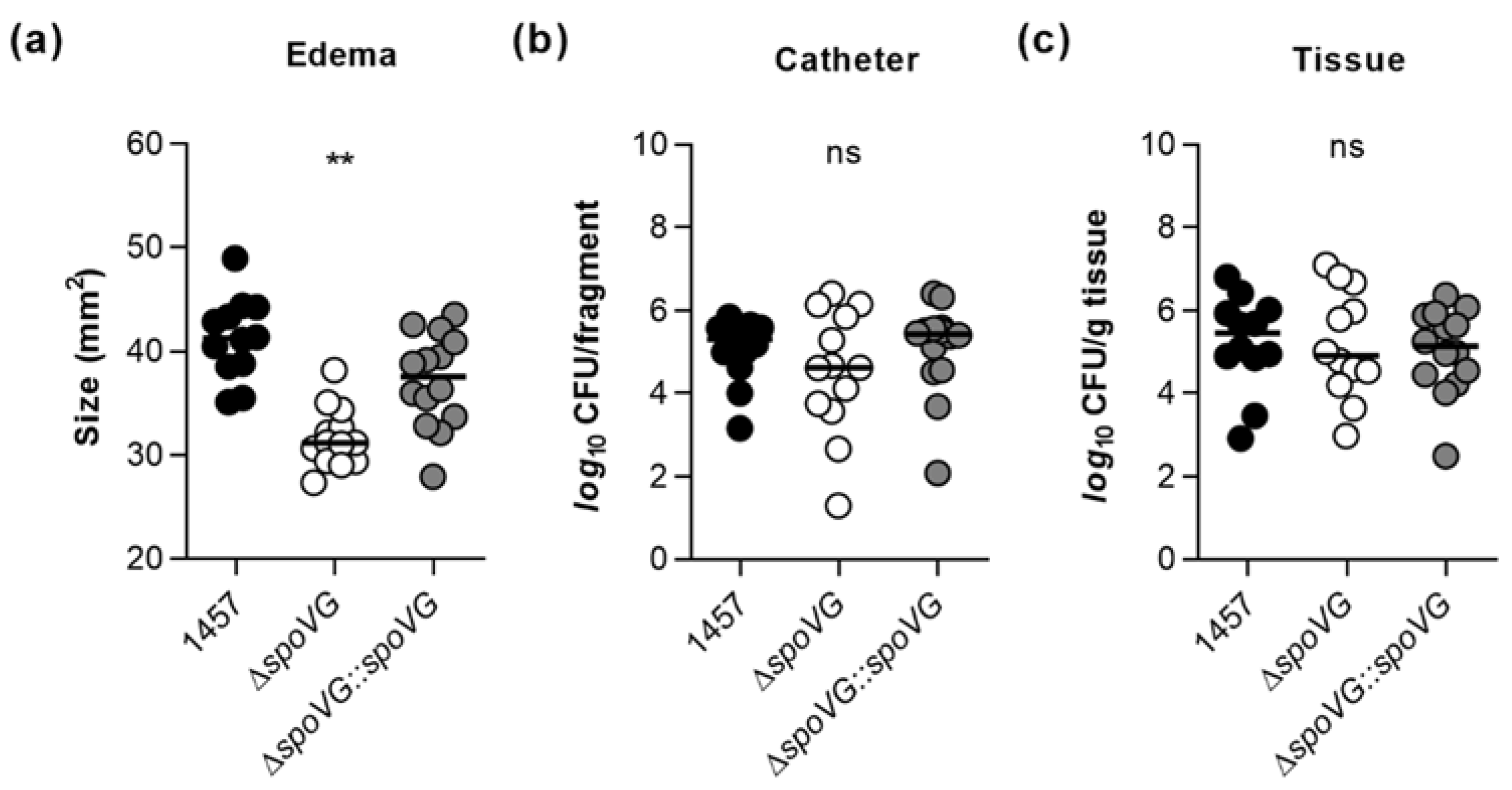
Earlier work demonstrated that inactivation of the catabolite control protein A encoding gene ccpA in S. epidermidis 1457 exerts a strong impact on the in vitro biofilm formation capacity of the mutant, but is negligible for the in vivo biofilm formation of S. epidermidis in a murine foreign body infection model [22], indicating that in vitro observed phenotypes are not necessarily predictive for the in vivo situation. Driven by the strong impact of the spoVG deletion on the biofilm formation capacity of strain 1457 seen here under in vitro conditions (Figure 4 and Figure 5), we wondered now whether and how this deletion might affect the biofilm formation capacity and infectivity of S. epidermidis under in vivo conditions. In order to address this question, we utilized a modified version of the murine foreign body infection model described by Rupp and colleagues [49], in which ~1 cm PVC fragments are inserted into the subcutis of both flanks of a mouse and the catheter lumen subsequently infected with a defined bacterial solution. Mice challenged with the ΔspoVG mutant displayed a clear reduction in edema sizes around the implanted catheter fragments, if compared to the edema sizes seen with mice infected with the wild-type (Figure 8a). However, only minor differences in bacterial loads on the implanted catheter fragments and the surrounding tissues were detected between wild-type and ΔspoVG mutant challenged mice (Figure 8b,c). Mice infected with the ΔspoVG mutant produced more heterogeneous CFU numbers on the catheter tubing than mice infected with the wild-type and the cis-complemented derivative (Figure 8b), while in infected tissues surrounding the catheter, rather comparable CFU rates were observed for all three mouse groups (Figure 8c). These findings indicate that SpoVG, albeit of being of major importance for PIA production and biofilm formation of S. epidermidis 1457 under in vitro conditions, is largely dispensable for biofilm formation on implantable devices under in vivo conditions but does affect the inflammatory response of the host at the infection site.
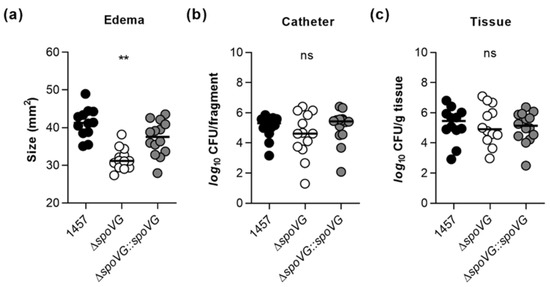
Figure 8.
Effect of the spoVG deletion on infectivity of S. epidermidis strain 1457 in a murine foreign body infection model. Catheter fragments were implanted subcutaneously into the back of normoglycemic mice and inoculated with cells of S. epidermidis strains 1457 (black symbols), 1457 ΔspoVG (white symbols) and 1457 ΔspoVG::spoVG (grey symbols), respectively. (a–c) Ten days post infection, animals were euthanized, edema sizes around the implanted catheters were measured (a), and the catheters and surrounding tissues were explanted. Bacterial loads from catheter detached biofilms (b) and in surrounding tissue homogenates (c) were determined by CFU counting (n = 12–14 per group). The data represent the values of every individual edema/catheter/tissue (symbols) and the median (horizontal line). **, p < 0.01; ns, not significant (Kruskal-Wallis test and Dunn’s multiple comparisons test, only differences between 1457 and mutants are shown).
2.7. Impact of the spoVG Deletion on Chemokine Expression in S. epidermidis 1457 Infected Tissue
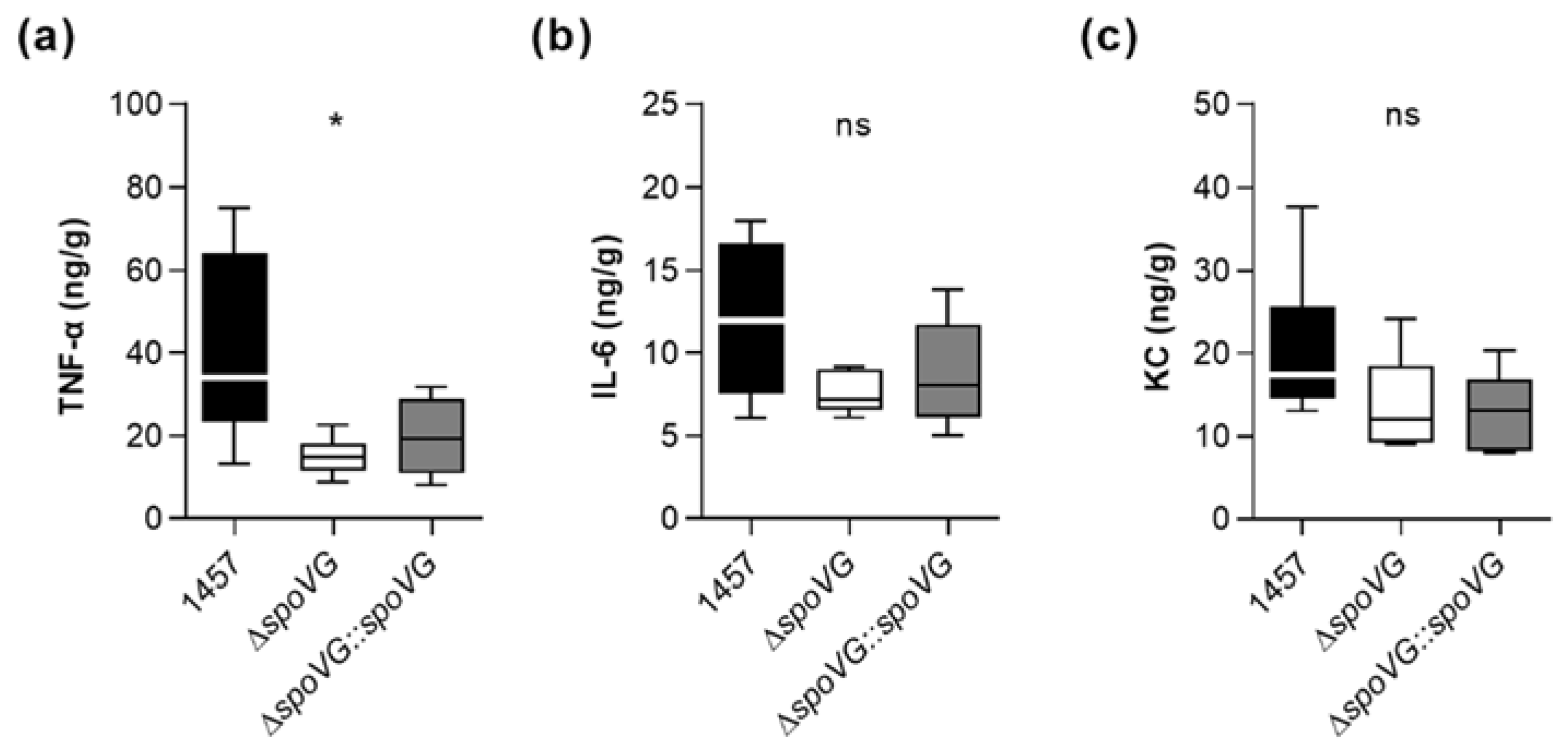
To test our hypothesis that SpoVG alters the inflammatory response of the host at the infection site, we assayed the contents of the pro-inflammatory cytokines interleukin-6 (IL-6) and tumor necrosis factor α (TNF-α), and of the neutrophil chemoattractant keratinocyte-derived chemokine (KC) in tissue homogenates obtained from excised tissues surrounding the catheter fragments of S. epidermidis-infected mice at day 10 post infection. In line with the augmented edema sizes found around the implanted catheter fragments in mice infected with the wild-type (Figure 8a), we observed significantly increased TNF-α contents in the tissue homogenates obtained from wild-type-infected mice.
Notably, mice infected with strain 1457 contained at this late stage of infection still more than twice as much of TNF-α in the infected tissue as mice infected with the isogenic ΔspoVG mutant (Figure 9a). Mice infected with the wild-type strain also possessed higher IL-6 and KC level in the infected tissue than mice infected with the ΔspoVG mutant (Figure 9b,c), however, these differences were not statistically significant. In combination with the CFU data obtained with this animal model at the infection sites (Figure 8b,c), these cytokine expression data suggest that SpoVG expressing S. epidermidis isolates elicit a long-lasting enhanced inflammatory response at the infection site leading to an increased tissue swelling, albeit without markedly altering the bacterial burden at the infection site.
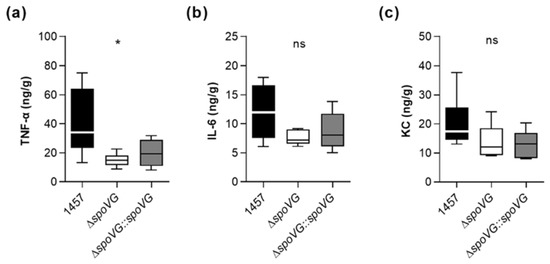
Figure 9.
Effect of the spoVG deletion on pro-inflammatory cytokine level in S. epidermidis 1457 infected tissue. Catheter fragments were implanted subcutaneously into the back of normoglycemic mice and inoculated with cells of S. epidermidis strains 1457 (black symbols), 1457 ΔspoVG (white symbols), and 1457 ΔspoVG::spoVG (grey symbols), respectively. (a–c) Concentrations of TNF-α (a), IL-6 (b), and KC (c) in tissue homogenates surrounding the infected catheter collected at day 10 post infection (n = 6–7 per group). Data are presented as box and whisker plot showing the interquartile range (25–75%, box), the median (horizontal line) and the standard deviation (bars). *, p < 0.05; ns, not significant (Kruskal-Wallis test and Dunn’s multiple comparisons test, only differences between 1457 and mutants are shown).
3. Discussion
The ability of S. epidermidis to adhere to implanted medical devices and to form biofilms on these artificial surfaces is considered as the primary virulence mechanism of this skin commensal [50]. Here, we demonstrate that the DNA/RNA-binding protein SpoVG strongly affects biofilm formation of the PIA-producing S. epidermidis strain 1457 under in vitro conditions, presumably by enhancing the transcription of icaADBC via down-regulation of IcaR expression. However, we could not identify a major impact of SpoVG on the in vivo biofilm formation capacity of strain 1457 in a murine foreign body infection model, although S. epidermidis cells harboring a functional spoVG locus elicited a stronger tissue swelling at the infection site in this model, which was accompanied by enhanced amounts of the pro-inflammatory cytokine TNF-α. These findings concur with earlier observations made with a ccpA mutant of this S. epidermidis strain, in which a strong effect of the ccpA mutation on the in vitro biofilm formation capacity was observed which was not accompanied by a clear effect in the in vivo infection model [22]. Given that inactivation of the ica locus in strain 1457 is reported to strongly decrease the biofilm formation capacity of the mutant in vitro [46,51,52], and the bacterial load at the infection site in the murine foreign body infection model [49,53,54], one can assume that the regulatory circuits leading to ica expression and PIA production between growth in vitro and in vivo are differing. While the σB-dependent transcription factor SpoVG is crucial for PIA production under in vitro conditions, it seems to be dispensable for PIA production of S. epidermidis under in vivo conditions, although we cannot exclude that the spoVG deletion mutant of strain 1457 might be able to produce a PIA-independent biofilm in this murine infection model, which yields in comparable CFU rates at the infection sites. Support for the latter hypothesis is given by recent findings demonstrating that host factors can compensate for PIA in cell-cell aggregation and biofilm formation of S. epidermidis [55].
Notably, strain 1457 was reported to elicit more severe skin and soft tissue inflammatory changes at the catheter insertion sites than the isogenic ica-negative isolate M10 [54], a phenotype that was also observed here with the spoVG deletion mutant, suggesting that 1457 ΔspoVG might produce lower levels of PIA than the wild-type in this infection model. However, the fact that 1457 cells grown in a PIA-dependent biofilm were reported to decrease the level of pro-inflammatory cytokines in a whole blood assay, if compared to M10 cells grown in a biofilm [52], suggests rather that strain 1457 does not produce high levels of PIA in the murine foreign body infection model, given the elevated TNF-α levels seen with the tissues infected with the wild-type strain.
Earlier studies highlighted the impact of the sigB operon on PIA-dependent biofilm formation in S. epidermidis [18,56,57] and suggested that the alternative σ-factor σB is likely to promote biofilm formation of strain 1457 by repressing IcaR expression [18]. However, the lack of an apparent σB consensus sequence in the ica locus suggested that σB is likely to modulate ica expression in an indirect manner [58]. Given that the promoter region of the yabJ-spoVG locus in S. epidermidis strain 1457 harbors a σB binding motif that is virtually identical to the σB binding site of the yabJ-spoVG locus of S. aureus [33], it is tempting to speculate that the impact of σB on icaR transcription in S. epidermidis is, at least in part, mediated via the σB-dependent transcriptional regulator SpoVG. Another factor mediating the σB effect on ica expression might be SarA, which positively affects icaADBC transcription [19,58], and is in part controlled by σB [18], although overexpression of the σB-dependent sarA P1 transcript in a 1457 sigB mutant was found to have, if at all, only little impact on PIA production [58]. However, regulators such as SarA, SarX, and TcaR, which directly control icaADBC transcription [19,23,24,48], might explain the discrepancy noticed in our study that deletion of spoVG significantly altered the icaR transcription at all three growth phases analyzed, while icaADBC transcription was altered significantly only in the early growth stage by the mutation. Another factor contributing to this phenotype might be Stk1, a serine/threonine protein kinase reported to enhance the DNA-binding properties of SpoVG towards its target sequences in S. aureus [36]. Experiments are ongoing in our laboratory to address this hypothesis.
4. Materials and Methods
4.1. Bacterial Strains and Plasmids
The bacterial strains and plasmids used in this study are listed in Table 2. All mutant strains generated for this study were confirmed by assessing gene transcription by quantitative real-time reverse-transcription PCR (qRT-PCR).

Table 2.
Strains and plasmids used in this study.
4.2. Bacterial Growth Conditions
E. coli strains were grown at 37 °C and 150 rpm in Luria Bertani (LB) medium (BD, Heidelberg, Germany) or on LB-agar plates supplemented with antibiotics when appropriate. Antibiotics were used at the following concentrations: erythromycin, 10 μg/mL; chloramphenicol, 10 μg/mL; ampicillin, 100 μg/mL. S. epidermidis strains were routinely grown in tryptic soy broth (TSB; BD) or on TSB plates containing 1.5% agar supplemented with antibiotics when appropriate. For growth kinetics studies, bacterial cells from overnight cultures were diluted into pre-warmed medium to an optical density at 600 nm (OD600) of 0.05. Cell suspensions were then either incubated under static conditions in 96-well microtiter plates at 37 °C and 5% CO2 in an oCelloScope (BioSense Solutions, Farum, Denmark), or aerobically in flasks at 37 °C and 225 rpm (flask-to-medium ratio 10:1). Growth kinetics of the static cultures were determined using the Background Corrected Absorption (BCA) algorithm of the UniExplorer software (BioSense Solutions, version 9.0). Growth kinetics of aerated cultures were monitored by taking samples at every hour and measuring the OD600 values using a Gene Quant 1300 spectrophotometer (Biochrom, Berlin, Germany). Cell suspensions reaching an OD600 > 0.8 were diluted 1:10 or 1:20 with phosphate buffered saline (PBS, pH 7.2), and the measured absorbance values were multiplied with the dilution factor. The generation times of S. epidermidis strains cultivated in TSB under aerobic conditions were determined as described in [41].
4.3. Mutant Construction
Construction of the 1457 spoVG deletion mutant was carried out essentially as described in [18]. DNA fragments covering the regions flanking spoVG were amplified by PCR using the Expand High Fidelity PCR system (Roche, Mannheim, Germany) with oligonucleotides shown in Table 3 and S. epidermidis 1457 DNA (NCBI Reference Sequence: NZ_CP020463.1) as template. The erythromycin resistance cassette ermB was cloned from a transposon TN917 harboring S. epidermidis strain using primer pair ermB forward/reverse. Amplified fragments were cloned into the DONR vectors of the Multisite Gateway Three-Fragment Vector Construction Kit according to manufactures instructions (Invitrogen, ThermoFisher Scientific, Schwerte, Germany).

Table 3.
Primers used in this study.
Resulting plasmids pENTRY_spoVG_up, pENTRY_spoVG_do and pENTRY_ermB were then combined with the Gateway destination vector pDEST R4-R3 (Invitrogen) in a MultiSite Gateway® LR recombination reaction to create plasmid pDEST_spoVG_ko. The spoVG up-ermB-spoVG do DNA region of the latter plasmid was next PCR amplified with primer pair spoVG_ko forward/reverse, the amplification product digested with PstI/BamHI, and ligated into the temperature-sensitive E. coli/Staphylococcus shuttle-vector pBT2 [63] digested with the same enzymes. The resulting plasmid pBT2_spoVG_KO was propagated in E. coli TOP10 (Invitrogen) and subsequently transformed into the restriction-deficient staphylococcal genetic background of S. aureus RN4220 by electroporation. pBT2_spoVG_KO isolated from this staphylococcal host was then transformed by electroporation into S. epidermis mutant M15 [56] and subsequently phage-transduced into the recipient strain, S. epidermidis 1457 with phage 71. 1457 cells positive for pBT2_spoVG_KO were grown at non-permissive temperatures, and clones were screened for double crossover with an erythromycin-resistant but chloramphenicol-sensitive phenotype. The correct chromosomal insertion of the cassette into the respective mutant was demonstrated by PCR with an ermB-specific primer paired with a primer flanking the genetically manipulated site and subsequent sequencing of the resulting fragment (data not shown).
For construction of the spoVG cis-complemented derivative of the 1457 ∆spoVG, the primer pair MBH603/MBH607 was used to amplify a ~3.0 kb fragment including the yabJ-spoVG locus from genomic DNA of the strain 1457. The amplification product was digested with SacI/BglII and ligated into SacI/BglII-digested and dephosphorylated E. coli-S. epidermidis shuttle-plasmid pBASE6 [62] to produce plasmid pBASE6_spoVG_comp. The temperature-sensitive plasmid was propagated in E. coli strain IM08B [61] and plasmids isolated from this E. coli strain were directly electroporated into 1457 ∆spoVG and cultured at 30 °C on TSA plates supplemented with chloramphenicol (10 µg/mL). Replacement of the ermB-tagged spoVG deletion by the functional yabJ-spoVG locus was carried out as previously described [64].
4.4. RNA Isolation and Purification, cDNA Synthesis and qRT-PCR
S. epidermidis strains were cultivated in TSB at 37 °C and 225 rpm. Bacterial pellets were collected after 3 h, 6 h, and 9 h of incubation by centrifugation at 5000 rpm at 4 °C for 5 min, and immediately resuspended in 100 µL ice-cold TE-buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8). Bacteria were disrupted, total RNA isolated, transcribed into cDNA, and qRT-PCRs carried out essentially as described previously [65] using the primers listed in Table 3. Transcriptional levels of target genes were normalized against the mRNA concentration of the housekeeping gene gyrB according to the 2−∆CT method [66].
4.5. Biofilm Assays
The impact of the spoVG deletion in strain 1457 on in vitro biofilm formation was assayed in different biofilm models under static and shaking conditions.
Biofilm formation under static conditions was assessed as described in [40]. Briefly, overnight cultures were diluted to an OD600 of 0.05 in fresh TSB medium, and 200 µL of the cell suspension was used per well to inoculate sterile Nunc™ MicroWell™ 96-Well, Nunclon Delta-Treated, Flat-Bottom Microplates (ThermoFisher). After incubation for 18 h at 37 °C without shaking, the plate wells were washed thrice with PBS and dried at 65 °C in an inverted position. To apply comparable shear forces to the biofilms, supernatants/wash solutions were removed with the help of a glass capillary attached to a vacuum pump system (KNF, Freiburg, Germany), and wash solutions were supplied with a multichannel pipette very slowly via the lateral planes of the plate wells. Adherent cells were safranin-stained (2 min with 0.1% safranin; Merck, Darmstadt, Germany) and the absorbance of stained biofilms was measured at 530 nm after resolving the stain with 200 µL 30% (v/v) acetic acid, using a microtiter plate reader (EnSight; Perkin Elmer, Rodgau, Germany).
The assessment of biofilm formation on peripheral venous catheter (PVC, Venflon Pro Safety 14 G; BD) fragments under dynamic conditions was carried out as describe in [49]. PVC fragments of 1 cm length were placed into reaction tubes filled with 1 mL of TSB and inoculated with 1 × 107 CFU of PBS-washed bacterial cells obtained from exponential growth phase. The PVC fragments were incubated under non-limited nutrient conditions for five days at 37 °C and 150 rpm, and the media were replaced with fresh media every 24 h. Five days post inoculation, PVC fragments were placed into fresh reaction tubes filled with 1 mL of TSB. The biofilms were detached from the catheter surface and resolved by sonication (50 watt for 5 min) followed by 1 min of vortexing. CFU rates and biomasses of resolved biofilms and culture supernatants at day five post inoculation were determined by plate counting and OD600 measurements, respectively.
Biofilm formation was additionally monitored and quantified in a glass tube assay. Briefly, 3 mL pre-warmed TSB were inoculated to an OD600 of 0.05 in glass tubes, and cultures were incubated at 37 °C and 150 rpm for 18 h. Biofilms formed at the air-liquid interface and bottom of the glass tubes were rinsed two times with PBS (pH 7.2) and air-dried for one hour. After staining with 0.1% safranin (Merck) for two minutes, glass tubes were rinsed three times with ddH2O, and stained biofilms documented photographically. Safranin bound to the biofilm was subsequently released by adding 1 mL of 30% acetic acid. 200 µL of the solution was then transferred to a 96-well plate (flat-bottom; Sarstedt, Nümbrecht, Germany) and the color intensity quantified at an absorbance of 530 nm (A530).
4.6. Cloning, Expression and Purification of Recombinant SpoVG
A synthetic DNA fragment covering the S. epidermidis 1457 spoVG (B4U56_11365) ORF with tags (ATTB1 recombination site–Shine-Dalgarno sequence–Kozac sequence– Histidine Affinity Tag [HAT]–Human influenza hemagglutinin [HA] tag–linker–spoVG- ATTB1 site) was ordered by IDT (Integrated DNA Technologies Germany GmbH, Munich, Germany). The DNA fragment was integrated in pDONR 221 to generate HAT-HA-SpoVG_pDONR 221. The sequence was recombined into the pRSF RfA Destination vector (MGC Platform; Montpellier, France), thus generating pRSF-HAT-HA-SpoVG harboring an ORF encoding the protein sequence HAT-HA-SpoVG (139 aa, 15.91 kD). E. coli BL21 Star cells (Invitrogen) transformed with pRSF-HAT-HA-SpoVG were grown in LB medium containing 1 mg/mL of glucose and 100 µg/mL of ampicillin.
For protein expression, pRSF-HAT-HA-SpoVG harboring BL21 Star cells were grown in LB at 37 °C until the culture reached an OD600 of 0.8. Cultures with this OD were supplemented with 0.5 mM isopropyl-β-D-thiogalactopyranoside and grown overnight at 20 °C. Bacterial cells were disrupted in a French pressure cell and centrifuged at 14,000 rpm for 25 min. Purifications were performed using a TALON® metal affinity resin (Takara, Saint-Germain-en-Laye, France) according to manufacturer’s instructions and eluted in 200 mM imidazole, 50 mM Tris-HCl [pH 7.4], 300 mM NaCl, and 10% [v/v] glycerol. Purity of the HAT-HA-SpoVG fractions was confirmed by Coomassie-staining of SDS-PAGEs loaded with the fractions.
4.7. Electrophoresis Mobility Shift Assay
The icaA and icaR dsDNA probes corresponding to the 80-bp regions upstream of the icaA and icaR ORFs (Figure 7a) were prepared by annealing complementary DYE682-labeled primers (Eurofins Genomics, Table 3) in 10 mM Tris–HCl (pH 8.0) and 20 mM NaCl. Mixtures were incubated at 95 °C for 5 min, and allowed to cool down to 25 °C. Indicated amounts of purified recombinant HAT-HA-SpoVG were mixed with 200 fmoles of the dsDNA probes in EMSA buffer (20 mM HEPES [pH 8.0], 10 mM KCl, 2 mM MgCl2, 0.1 mM EDTA, 0.1 mM dithiothreitol, 50 μg/mL bovine serum albumin, and 10% glycerol) in 20 μL final volume reactions. After incubation at room temperature for 30 min, the reactions were run on a 2.5% agarose gel in cold TAE (Tris-Acetate-EDTA) buffer. Gels were imaged using the 700 nm channel of the Odyssey Infrared Imaging System (LI-COR).
4.8. Murine Foreign Body Infection Model
Animal experiments were performed with approval of the local State Review Board of Saarland, Germany (project identification code 11/2019 [approved 30 April 2019]), and conducted following the national and European guidelines for the ethical and human treatment of animals. Infection of the animals was carried out as described [41], except that implanted catheter fragments were inoculated with 1 × 107 CFU of S. epidermidis strains 1457, its ∆spoVG mutant and the cis-complemented spoVG derivative, respectively. For some of the infected catheter fragments (9/48), co-infections with other bacterial species were noticed. Data obtained from these infection sites were excluded from the study.
4.9. Cytokine Determinations
Tissue homogenates obtained from excised tissues surrounding the catheter fragments of S. epidermidis-infected mice at day 10 post infection were centrifuged at 4 °C and 2500× g for 10 min. Concentrations of cytokines were measured in supernatants using ELISAs for IL-6, KC, and TNF-α (R&D Systems, Minneapolis, MN, USA) as described by the manufacturer.
4.10. Statistical Analyses
The statistical significance of changes between groups was determined using the Graph-Pad software package Prism 9.3.1. Identified p-values < 0.05 were considered statistically significant. Comparisons between groups were analyzed by the Kruskal-Wallis test and Dunn’s multiple comparisons test.
Author Contributions
Conceptualization, M.B. and J.K.K.; methodology, L.P., V.M., C.B., J.K.K. and M.B.; investigation, H.B., L.P., B.F., M.I.E., S.H.-B., M.B. and V.M.; writing—original draft preparation, M.B.; resources, S.L.B., C.B., V.M. and G.K.-C.; writing—review and editing, M.B., V.M., H.B., L.P. and J.K.K.; visualization, M.B.; supervision, M.B. and J.K.K.; project administration, M.B. and J.K.K.; funding acquisition, M.I.E. and M.B. All authors have read and agreed to the published version of the manuscript.
Funding
The APC was funded by DFG and Saarland University within the funding program Open Access Publishing. M.I.E. acknowledges support by the German Egyptian Research Long-Term Scholarship Program (GERLS).
Institutional Review Board Statement
The animal study protocol was approved by the local State Review Board of Saarland, Germany (project identification code 11/2019 [approved 30 April 2019]).
Informed Consent Statement
Not applicable.
Data Availability Statement
The datasets generated and analyzed during the current study are available from the corresponding author on reasonable request.
Acknowledgments
The authors are grateful to Karin Hilgert and Anja Honecker for excellent technical assistance.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Vanepps, J.S.; Younger, J.G. Implantable device-related infection. Shock 2016, 46, 597–608. [Google Scholar] [CrossRef]
- de Oliveira, W.F.; Silva, P.M.S.; Silva, R.C.S.; Silva, G.M.M.; Machado, G.; Coelho, L.; Correia, M.T.S. Staphylococcus aureus and Staphylococcus epidermidis infections on implants. J. Hosp. Infect. 2018, 98, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Brown, M.M.; Horswill, A.R. Staphylococcus epidermidis—Skin friend or foe? PLoS Pathog. 2020, 16, e1009026. [Google Scholar] [CrossRef] [PubMed]
- Kleinschmidt, S.; Huygens, F.; Faoagali, J.; Rathnayake, I.U.; Hafner, L.M. Staphylococcus epidermidis as a cause of bacteremia. Futur. Microbiol. 2015, 10, 1859–1879. [Google Scholar] [CrossRef] [PubMed]
- Joubert, I.A.; Otto, M.; Strunk, T.; Currie, A.J. Look who’s talking: Host and pathogen drivers of Staphylococcus epidermidis virulence in neonatal sepsis. Int. J. Mol. Sci. 2022, 23, 860. [Google Scholar] [CrossRef] [PubMed]
- Matarrese, A.N.; Ivulich, D.I.; Cesar, G.; Alaniz, F.; Ruiz, J.J.; Osatnik, J. Epidemiological analysis of catheter-related bloodstream infections in medical-surgical intensive care units. Medicina (B Aires) 2021, 81, 159–165. [Google Scholar]
- German Reference Center for Surveillance of Nosocomial Infections. STATIONS-KISS Module Reference Data 2017–2019. 2020. Available online: https://www.nrz-hygiene.de/fileadmin/nrz/module/station/infektionen/201701_201912_deviceref.pdf (accessed on 10 March 2022).
- Pinto, M.; Borges, V.; Nascimento, M.; Martins, F.; Pessanha, M.A.; Faria, I.; Rodrigues, J.; Matias, R.; Gomes, J.P.; Jordao, L. Insights on catheter-related bloodstream infections: A prospective observational study on the catheter colonization and multi-drug resistance. J. Hosp. Infect. 2022, 123, 43–51. [Google Scholar] [CrossRef]
- Strasheim, W.; Kock, M.; Ueckermann, V.; Hoosien, E.; Dreyer, A.W.; Ehlers, M.M. Surveillance of catheter-related infections: The supplementary role of the microbiology laboratory. BMC Infect. Dis. 2015, 15, 5. [Google Scholar] [CrossRef]
- Buetti, N.; Priore, E.L.; Atkinson, A.; Widmer, A.F.; Kronenberg, A.; Marschall, J. Catheter-related infections: Does the spectrum of microbial causes change over time? A nationwide surveillance study. BMJ Open 2018, 8, e023824. [Google Scholar] [CrossRef]
- Weiner-Lastinger, L.M.; Abner, S.; Edwards, J.R.; Kallen, A.J.; Karlsson, M.; Magill, S.S.; Pollock, D.; See, I.; Soe, M.M.; Walters, M.S.; et al. Antimicrobial-resistant pathogens associated with adult healthcare-associated infections: Summary of data reported to the National Healthcare Safety Network, 2015–2017. Infect. Control Hosp. Epidemiol. 2020, 41, 1–18. [Google Scholar] [CrossRef]
- Kranjec, C.; Angeles, D.M.; Mårli, M.T.; Fernández, L.; García, P.; Kjos, M.; Diep, D. Staphylococcal biofilms: Challenges and novel therapeutic perspectives. Antibiotics 2021, 10, 131. [Google Scholar] [CrossRef] [PubMed]
- Schilcher, K.; Horswill, A.R. Staphylococcal biofilm development: Structure, regulation, and treatment strategies. Microbiol. Mol. Biol. Rev. 2020, 84, e00026-19. [Google Scholar] [CrossRef] [PubMed]
- França, A.; Pérez-Cabezas, B.; Correia, A.; Pier, G.B.; Cerca, N.; Vilanova, M. Staphylococcus epidermidis biofilm-released cells induce a prompt and more marked in vivo inflammatory-type response than planktonic or biofilm cells. Front. Microbiol. 2016, 7, 1530. [Google Scholar] [CrossRef] [PubMed]
- Arciola, C.R.; Campoccia, D.; Ravaioli, S.; Montanaro, L. Polysaccharide intercellular adhesin in biofilm: Structural and regulatory aspects. Front. Cell. Infect. Microbiol. 2015, 5, 7. [Google Scholar] [CrossRef]
- Deighton, M.; Borland, R. Regulation of slime production in Staphylococcus epidermidis by iron limitation. Infect. Immun. 1993, 61, 4473–4479. [Google Scholar] [CrossRef]
- Rachid, S.; Cho, S.; Ohlsen, K.; Hacker, J.; Ziebuhr, W. Induction of Staphylococcus epidermidis biofilm formation by environmental factors: The possible involvement of the alternative transcription factor SigB. Adv. Exp. Med. Biol. 2000, 485, 159–166. [Google Scholar] [CrossRef]
- Knobloch, J.K.-M.; Jäger, S.; Horstkotte, M.A.; Rohde, H.; Mack, D. RsbU-Dependent regulation of Staphylococcus epidermidis biofilm formation is mediated via the alternative sigma factor SigB by repression of the negative regulator gene icaR. Infect. Immun. 2004, 72, 3838–3848. [Google Scholar] [CrossRef]
- Tormo, M.A.; Martí, M.; Valle, J.; Manna, A.C.; Cheung, A.L.; Lasa, I.; Penadés, J.R.; Sar, A. Is an essential positive regulator of Staphylococcus epidermidis biofilm development. J. Bacteriol. 2005, 187, 2348–2356. [Google Scholar] [CrossRef]
- Conlon, K.M.; Humphreys, H.; O’Gara, J.P. icaR encodes a transcriptional repressor involved in environmental regulation of ica operon expression and biofilm formation in Staphylococcus epidermidis. J. Bacteriol. 2002, 184, 4400–4408. [Google Scholar] [CrossRef]
- Liu, Q.; Fan, J.; Niu, C.; Wang, D.; Wang, J.; Wang, X.; Villaruz, A.E.; Li, M.; Otto, M.; Gao, Q. The Eukaryotic-type serine/threonine protein kinase Stk is required for biofilm formation and virulence in Staphylococcus epidermidis. PLoS ONE 2011, 6, e25380. [Google Scholar] [CrossRef]
- Sadykov, M.R.; Hartmann, T.; Mattes, T.A.; Hiatt, M.; Jann, N.J.; Zhu, Y.; Ledala, N.; Landmann, R.; Herrmann, M.; Rohde, H.; et al. CcpA coordinates central metabolism and biofilm formation in Staphylococcus epidermidis. Microbiology 2011, 157, 3458–3468. [Google Scholar] [CrossRef] [PubMed]
- Rowe, S.E.; Mahon, V.; Smith, S.G.; O’Gara, J.P. A novel role for SarX in Staphylococcus epidermidis biofilm regulation. Microbiology 2011, 157, 1042–1049. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.-M.; Jeng, W.-Y.; Ko, T.-P.; Yeh, Y.-J.; Chen, C.K.-M.; Wang, A.H.-J. Structural study of TcaR and its complexes with multiple antibiotics from Staphylococcus epidermidis. Proc. Natl. Acad. Sci. USA 2010, 107, 8617–8622. [Google Scholar] [CrossRef] [PubMed]
- Hoang, T.-M.; Zhou, C.; Lindgren, J.K.; Galac, M.R.; Corey, B.; Endres, J.E.; Olson, M.E.; Fey, P.D. Transcriptional regulation of icaADBC by both IcaR and TcaR in Staphylococcus epidermidis. J. Bacteriol. 2019, 201, 201. [Google Scholar] [CrossRef]
- Wu, Y.; Meng, Y.; Qian, L.; Ding, B.; Han, H.; Chen, H.; Bai, L.; Qu, D.; Wu, Y. The vancomycin resistance-associated regulatory system VraSR modulates biofilm formation of Staphylococcus epidermidis in an ica-dependent manner. mSphere 2021, 6, e0064121. [Google Scholar] [CrossRef]
- Lerch, M.F.; Schoenfelder, S.M.; Marincola, G.; Wencker, F.; Eckart, M.; Förstner, K.U.; Sharma, C.M.; Thormann, K.M.; Kucklick, M.; Engelmann, S.; et al. A non-coding RNA from the intercellular adhesion (ica) locus ofStaphylococcus epidermidiscontrols polysaccharide intercellular adhesion (PIA)-mediated biofilm formation. Mol. Microbiol. 2019, 111, 1571–1591. [Google Scholar] [CrossRef]
- Schoenfelder, S.M.K.; Lange, C.; Prakash, S.A.; Marincola, G.; Lerch, M.F.; Wencker, F.D.R.; Förstner, K.U.; Sharma, C.M.; Ziebuhr, W. The small non-coding RNA RsaE influences extracellular matrix composition in Staphylococcus epidermidis biofilm communities. PLoS Pathog. 2019, 15, e1007618. [Google Scholar] [CrossRef]
- Ziebuhr, W.; Krimmer, V.; Rachid, S.; Lossner, I.; Gotz, F.; Hacker, J. A novel mechanism of phase variation of virulence in Staphylococcus epidermidis: Evidence for control of the polysaccharide intercellular adhesin synthesis by alternating insertion and excision of the insertion sequence element IS256. Mol. Microbiol. 1999, 32, 345–356. [Google Scholar] [CrossRef]
- Jutras, B.L.; Chenail, A.M.; Rowland, C.L.; Carroll, D.; Miller, M.C.; Bykowski, T.; Stevenson, B. Eubacterial SpoVG homologs constitute a new family of site-specific DNA-binding proteins. PLoS ONE 2013, 8, e66683. [Google Scholar] [CrossRef]
- Huang, Q.; Zhang, Z.; Liu, Q.; Liu, F.; Liu, Y.; Zhang, J.; Wang, G. SpoVG is an important regulator of sporulation and affects biofilm formation by regulating Spo0A transcription in Bacillus cereus 0–9. BMC Microbiol. 2021, 21, 172. [Google Scholar] [CrossRef]
- Burke, T.P.; Portnoy, D.A. SpoVG is a conserved RNA-binding protein that regulates Listeria monocytogenes lysozyme resistance, virulence, and swarming motility. MBio 2016, 7, e00240-16. [Google Scholar] [CrossRef]
- Meier, S.; Goerke, C.; Wolz, C.; Seidl, K.; Homerova, D.; Schulthess, B.; Kormanec, J.; Berger-Bächi, B.; Bischoff, M. SigmaB and the SigmaB-dependent arlRS and yabJ-spoVG loci affect capsule formation in Staphylococcus aureus. Infect. Immun. 2007, 75, 4562–4571. [Google Scholar] [CrossRef] [PubMed]
- Schulthess, B.; Bloes, D.A.; Francois, P.; Girard, M.; Schrenzel, J.; Bischoff, M.; Berger-Bächi, B. The σB-dependent yabJ-spoVG operon is involved in the regulation of extracellular nuclease, lipase, and protease expression in Staphylococcus aureus. J. Bacteriol. 2011, 193, 4954–4962. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Liu, B.; Sun, B. SpoVG modulates cell aggregation in Staphylococcus aureus by regulating sasC expression and extracellular DNA release. Appl. Environ. Microbiol. 2020, 86, 86. [Google Scholar] [CrossRef] [PubMed]
- Bischoff, M.; Brelle, S.; Minatelli, S.; Molle, V. Stk1-mediated phosphorylation stimulates the DNA-binding properties of the Staphylococcus aureus SpoVG transcriptional factor. Biochem. Biophys. Res. Commun. 2016, 473, 1223–1228. [Google Scholar] [CrossRef] [PubMed]
- Mack, D.; Siemssen, N.; Laufs, R. Parallel induction by glucose of adherence and a polysaccharide antigen specific for plastic-adherent Staphylococcus epidermidis: Evidence for functional relation to intercellular adhesion. Infect. Immun. 1992, 60, 2048–2057. [Google Scholar] [CrossRef]
- Mäder, U.; Nicolas, P.; Depke, M.; Pané-Farré, J.; Debarbouille, M.; Van Der Kooi-Pol, M.M.; Guérin, C.; Dérozier, S.; Hiron, A.; Jarmer, H.; et al. Staphylococcus aureus transcriptome architecture: From laboratory to infection-mimicking conditions. PLoS Genet. 2016, 12, e1005962. [Google Scholar] [CrossRef]
- Ungphakorn, W.; Malmberg, C.; Lagerbäck, P.; Cars, O.; Nielsen, E.I.; Tängdén, T. Evaluation of automated time-lapse microscopy for assessment of in vitro activity of antibiotics. J. Microbiol. Methods 2017, 132, 69–75. [Google Scholar] [CrossRef]
- Seidl, K.; Goerke, C.; Wolz, C.; Mack, D.; Berger-Bächi, B.; Bischoff, M. Staphylococcus aureus CcpA affects biofilm formation. Infect. Immun. 2008, 76, 2044–2050. [Google Scholar] [CrossRef]
- Pätzold, L.; Brausch, A.-C.; Bielefeld, E.-L.; Zimmer, L.; Somerville, G.A.; Bischoff, M.; Gaupp, R. Impact of the histidine-containing phosphocarrier protein HPr on carbon metabolism and virulence in Staphylococcus aureus. Microorganisms 2021, 9, 466. [Google Scholar] [CrossRef]
- Mandakhalikar, K.D.; Rahmat, J.N.; Chiong, E.; Neoh, K.G.; Shen, L.; Tambyah, P.A. Extraction and quantification of biofilm bacteria: Method optimized for urinary catheters. Sci. Rep. 2018, 8, 8069. [Google Scholar] [CrossRef]
- O’Gara, J.P. icaand beyond: Biofilm mechanisms and regulation in Staphylococcus epidermidis and Staphylococcus aureus. FEMS Microbiol. Lett. 2007, 270, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Dobinsky, S.; Kiel, K.; Rohde, H.; Bartscht, K.; Knobloch, J.K.-M.; Horstkotte, M.A.; Mack, D. Glucose-related dissociation between icaADBC transcription and biofilm expression by Staphylococcus epidermidis: Evidence for an additional factor required for polysaccharide intercellular adhesin synthesis. J. Bacteriol. 2003, 185, 2879–2886. [Google Scholar] [CrossRef] [PubMed]
- Jeng, W.-Y.; Ko, T.-P.; Liu, C.-I.; Guo, R.-T.; Liu, C.-L.; Shr, H.-L.; Wang, A.H.-J. Crystal structure of IcaR, a repressor of the TetR family implicated in biofilm formation in Staphylococcus epidermidis. Nucleic Acids Res. 2008, 36, 1567–1577. [Google Scholar] [CrossRef] [PubMed]
- Heilmann, C.; Schweitzer, O.; Gerke, C.; Vanittanakom, N.; Mack, D.; Götz, F. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol. Microbiol. 1996, 20, 1083–1091. [Google Scholar] [CrossRef] [PubMed]
- Mozos, I.R.D.L.; Vergara-Irigaray, M.; Segura, V.; Villanueva, M.; Bitarte, N.; Saramago, M.; Domingues, S.; Arraiano, C.; Fechter, P.; Romby, P.; et al. Base Pairing interaction between 5′- and 3′-UTRs Controls icaR mRNA translation in Staphylococcus aureus. PLoS Genet. 2013, 9, e1004001. [Google Scholar] [CrossRef]
- Jefferson, K.K.; Pier, D.B.; Goldmann, D.A.; Pier, G.B. The Teicoplanin-associated locus regulator (TcaR) and the Intercellular adhesin locus regulator (IcaR) are transcriptional inhibitors of the ica locus in Staphylococcus aureus. J. Bacteriol. 2004, 186, 2449–2456. [Google Scholar] [CrossRef]
- Rupp, M.E.; Ulphani, J.S.; Fey, P.D.; Bartscht, K.; Mack, D. Characterization of the importance of polysaccharide intercellular adhesin/hemagglutinin of Staphylococcus epidermidis in the pathogenesis of biomaterial-based infection in a mouse foreign body infection model. Infect. Immun. 1999, 67, 2627–2632. [Google Scholar] [CrossRef]
- Le, K.Y.; Park, M.D.-Y.; Otto, M. Immune evasion mechanisms of Staphylococcus epidermidis biofilm infection. Front. Microbiol. 2018, 9, 359. [Google Scholar] [CrossRef]
- Mack, D.; Riedewald, J.; Rohde, H.; Magnus, T.; Feucht, H.H.; Elsner, H.-A.; Laufs, R.; Rupp, M.E. Essential functional role of the polysaccharide intercellular adhesin of Staphylococcus epidermidis in hemagglutination. Infect. Immun. 1999, 67, 1004–1008. [Google Scholar] [CrossRef]
- Fredheim, E.G.A.; Granslo, H.N.; Flaegstad, T.; Figenschau, Y.; Rohde, H.; Sadovskaya, I.; Mollnes, T.E.; Klingenberg, C. Staphylococcus epidermidis polysaccharide intercellular adhesin activates complement. FEMS Immunol. Med. Microbiol. 2011, 63, 269–280. [Google Scholar] [CrossRef] [PubMed]
- Vuong, C.; Kocianova, S.; Voyich, J.M.; Yao, Y.; Fischer, E.R.; DeLeo, F.; Otto, M. A Crucial role for exopolysaccharide modification in bacterial biofilm formation, immune evasion, and virulence. J. Biol. Chem. 2004, 279, 54881–54886. [Google Scholar] [CrossRef] [PubMed]
- Kristian, S.A.; Birkenstock, T.A.; Sauder, U.; Mack, D.; Götz, F.; Landmann, R. Biofilm Formation Induces C3a Release and protects Staphylococcus epidermidis from IgG and complement deposition and from neutrophil-dependent killing. J. Infect. Dis. 2008, 197, 1028–1035. [Google Scholar] [CrossRef] [PubMed]
- Skovdal, S.M.; Hansen, L.K.; Ivarsen, D.M.; Zeng, G.; Büttner, H.; Rohde, H.; Jørgensen, N.P.; Meyer, R.L. Host factors abolish the need for polysaccharides and extracellular matrix-binding protein in Staphylococcus epidermidis biofilm formation. J. Med. Microbiol. 2021, 70, 001287. [Google Scholar] [CrossRef]
- Mack, D.; Rohde, H.; Dobinsky, S.; Riedewald, J.; Nedelmann, M.; Knobloch, J.K.-M.; Elsner, H.-A.; Feucht, H.H. Identification of three essential regulatory gene loci governing expression of Staphylococcus epidermidis polysaccharide intercellular adhesin and biofilm formation. Infect. Immun. 2000, 68, 3799–3807. [Google Scholar] [CrossRef]
- Knobloch, J.K.-M.; Bartscht, K.; Sabottke, A.; Rohde, H.; Feucht, H.-H.; Mack, D. Biofilm formation by Staphylococcus epidermidis depends on functional RsbU, an Activator of the sigB operon: Differential activation mechanisms due to ethanol and salt stress. J. Bacteriol. 2001, 183, 2624–2633. [Google Scholar] [CrossRef]
- Handke, L.D.; Slater, S.R.; Conlon, K.M.; O’Donnell, S.T.; Olson, M.E.; Bryant, K.A.; Rupp, M.E.; O’Gara, J.; Fey, P.D. SigmaB and SarA independently regulate polysaccharide intercellular adhesin production in Staphylococcus epidermidis. Can. J. Microbiol. 2007, 53, 82–91. [Google Scholar] [CrossRef]
- Kreiswirth, B.N.; Löfdahl, S.; Betley, M.J.; O’Reilly, M.; Schlievert, P.; Bergdoll, M.S.; Novick, R. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 1983, 305, 709–712. [Google Scholar] [CrossRef]
- Nedelmann, M.; Sabottke, A.; Laufs, R.; Mack, D. Generalized transduction for genetic linkage analysis and transfer of transposon insertions in different Staphylococcus epidermidis strains. Zent. Bakteriol. 1998, 287, 85–92. [Google Scholar] [CrossRef]
- Monk, I.R.; Tree, J.; Howden, B.; Stinear, T.P.; Foster, T.J. Complete bypass of restriction systems for major Staphylococcus aureus lineages. MBio 2015, 6, e00308-15. [Google Scholar] [CrossRef]
- Geiger, T.; Francois, P.; Liebeke, M.; Fraunholz, M.; Goerke, C.; Krismer, B.; Schrenzel, J.; Lalk, M.; Wolz, C. The stringent response of Staphylococcus aureus and its impact on survival after phagocytosis through the induction of intracellular PSMs expression. PLoS Pathog. 2012, 8, e1003016. [Google Scholar] [CrossRef] [PubMed]
- Brückner, R. Gene replacement in Staphylococcus carnosus and Staphylococcus xylosus. FEMS Microbiol. Lett. 1997, 151, 1–8. [Google Scholar] [CrossRef]
- Bae, T.; Schneewind, O. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 2006, 55, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Gaupp, R.; Wirf, J.; Wonnenberg, B.; Biegel, T.; Eisenbeis, J.; Graham, J.; Herrmann, M.; Lee, C.Y.; Beisswenger, C.; Wolz, C.; et al. RpiRc is a pleiotropic effector of virulence determinant synthesis and attenuates pathogenicity in Staphylococcus aureus. Infect. Immun. 2016, 84, 2031–2041. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. Relative quantification. In Real-Time PCR; Tevfik Dorak, M., Ed.; Taylor & Francis: Abingdon, UK, 2006; pp. 63–82. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).