Mechanisms of Action of Non-Canonical ECF Sigma Factors
Abstract
1. Introduction
2. ECF Sigma Factors with a Soluble Anti-Sigma Factor
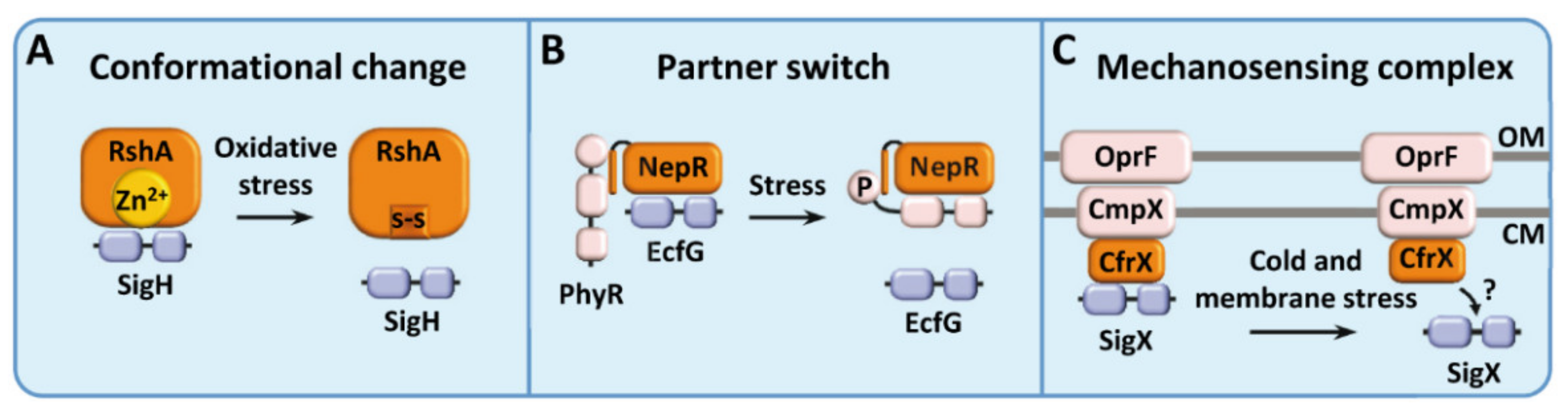
2.1. ECF Sigma Factors Regulated by Conformational Change
2.2. ECF Sigma Factors Regulated by Partner Switch
2.3. ECF Sigma Factors Regulated by a Mechanosensing Complex
3. ECF Sigma Factors Not Associated with an Anti-Sigma Factor
3.1. ECF Sigma Factors Transcriptionally Regulated
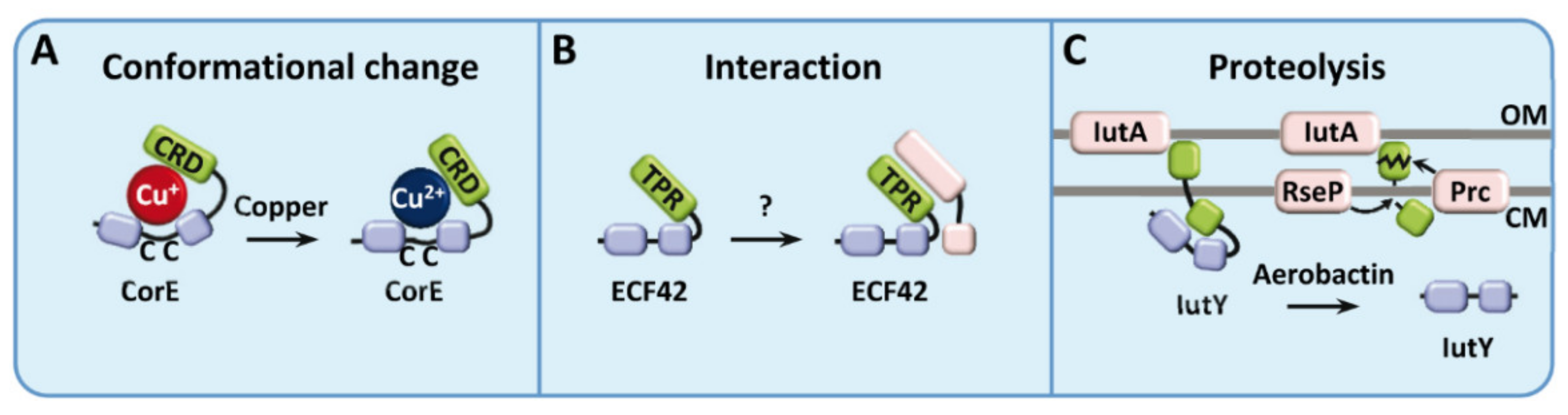
3.2. ECF Sigma Factors Regulated by Conformational Changes
3.3. ECF Sigma Factors Regulated by Proteolysis
3.4. ECF Sigma Factors Regulated by Phosphorylation
3.5. ECF Sigma Factors with Regulatory Extensions
3.5.1. Activated by Conformational Change
3.5.2. Activated by Protein Interaction
3.5.3. Activated by Proteolysis
4. Outlook and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Staroń, A.; Sofia, H.J.; Dietrich, S.; Ulrich, L.E.; Liesegang, H.; Mascher, T. The third pillar of bacterial signal transduction: Classification of the extracytoplasmic function (ECF) σ factor protein family. Mol. Microbiol. 2009, 74, 557–581. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Dorado, J.; Gómez-Santos, N.; Pérez, J. A novel mechanism of bacterial adaptation mediated by copper-dependent RNA polymerase σ factors. Transcription 2012, 3, 63–67. [Google Scholar] [CrossRef] [PubMed]
- Helmann, J.D. The extracytoplasmic function (ECF) sigma factors. Adv. Microb. Physiol. 2002, 46, 47–110. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Ammerman, N.C.; Nolan, S.; Geiman, D.E.; Lun, S.; Guo, H.; Bishai, W.R. Isoniazid resistance without a loss of fitness in Mycobacterium tuberculosis. Nat. Commun. 2012, 3, 753. [Google Scholar] [CrossRef][Green Version]
- Mascher, T. Signaling diversity and evolution of extracytoplasmic function (ECF) σ factors. Curr. Opin. Microbiol. 2013, 16, 148–155. [Google Scholar] [CrossRef]
- Pinto, D.; Mascher, T. (Actino)Bacterial “intelligence”: Using comparative genomics to unravel the information processing capacities of microbes. Curr. Genet. 2016, 62, 487–498. [Google Scholar] [CrossRef]
- Casas-Pastor, D.; Müller, R.R.; Jaenicke, S.; Brinkrolf, K.; Becker, A.; Buttner, M.J.; Gross, C.A.; Mascher, T.; Goesmann, A.; Fritz, G. Expansion and re-classification of the extracytoplasmic function (ECF) σ factor family. Nucleic Acids Res. 2021, 49, 986–1005. [Google Scholar] [CrossRef]
- Campbell, E.A.; Greenwell, R.; Anthony, J.R.; Wang, S.; Lim, L.; Das, K.; Sofia, H.J.; Donohue, T.J.; Darst, S.A. A conserved structural module regulates transcriptional responses to diverse stress signals in bacteria. Mol. Cell 2007, 27, 793–805. [Google Scholar] [CrossRef]
- Donohue, T.J. Shedding light on a Group IV (ECF11) alternative σ factor. Mol. Microbiol. 2019, 112, 374–384. [Google Scholar] [CrossRef]
- Song, T.; Dove, S.L.; Lee, K.H.; Husson, R.N. RshA an anti-sigma factor that regulates the activity of the mycobacterial stress response sigma factor SigH. Mol. Microbiol. 2003, 50, 949–959. [Google Scholar] [CrossRef]
- Boldrin, F.; Cioetto Mazzabò, L.; Anoosheh, S.; Palù, G.; Gaudreau, L.; Manganelli, R.; Provvedi, R. Assessing the role of Rv1222 (RseA) as an anti-sigma factor of the Mycobacterium tuberculosis extracytoplasmic sigma factor SigE. Sci. Rep. 2019, 9, 4513. [Google Scholar] [CrossRef]
- Huis In’t Veld, R.A.G.; Willemsen, A.M.; van Kampen, A.H.C.; Bradley, E.J.; Baas, F.; Pannekoek, Y.; van der Ende, A. Deep sequencing whole transcriptome exploration of the σE regulon in Neisseria meningitidis. PLoS ONE 2011, 6, e29002. [Google Scholar] [CrossRef]
- Francez-Charlot, A.; Kaczmarczyk, A.; Fischer, H.M.; Vorholt, J.A. The general stress response in Alphaproteobacteria. Trends Microbiol. 2015, 23, 164–171. [Google Scholar] [CrossRef]
- Chevalier, S.; Bouffartigues, E.; Bazire, A.; Tahrioui, A.; Duchesne, R.; Tortuel, D.; Maillot, O.; Clamens, T.; Orange, N.; Feuilloley, M.G.; et al. Extracytoplasmic function sigma factors in Pseudomonas aeruginosa. Biochim. Biophys. Acta-Gene Regul. Mech. 2019, 1862, 706–721. [Google Scholar] [CrossRef]
- Rajasekar, K.V.; Zdanowski, K.; Yan, J.; Hopper, J.T.S.; Francis, M.L.R.; Seepersad, C.; Sharp, C.; Pecqueur, L.; Werner, J.M.; Robinson, C.V.; et al. The anti-sigma factor RsrA responds to oxidative stress by reburying its hydrophobic core. Nat. Commun. 2016, 7, 12194. [Google Scholar] [CrossRef]
- Nam, T.W.; Ziegelhoffer, E.C.; Lemke, R.A.S.; Donohue, T.J. Proteins needed to activate a transcriptional response to the reactive oxygen species singlet oxygen. mBio 2013, 4, e00541-12. [Google Scholar] [CrossRef]
- Chabert, V.; Lebrun, V.; Lebrun, C.; Latour, J.M.; Sénèque, O. Model peptide for anti-sigma factor domain HHCC zinc fingers: High reactivity toward 1O2 leads to domain unfolding. Chem. Sci. 2019, 10, 3608–3615. [Google Scholar] [CrossRef]
- Bhat, S.A.; Singh, N.; Trivedi, A.; Kansal, P.; Gupta, P.; Kumar, A. The mechanism of redox sensing in Mycobacterium tuberculosis. Free Radic. Biol. Med. 2012, 53, 1625–1641. [Google Scholar] [CrossRef]
- Park, S.D.; Youn, J.W.; Kim, Y.J.; Lee, S.M.; Kim, Y.; Lee, H.S. Corynebacterium glutamicum σ E is involved in responses to cell surface stresses and its activity is controlled by the anti-σ factor CseE. Microbiology 2008, 154, 915–923. [Google Scholar] [CrossRef]
- Barik, S.; Sureka, K.; Mukherjee, P.; Basu, J.; Kundu, M. RseA, the SigE specific anti-sigma factor of Mycobacterium tuberculosis, is inactivated by phosphorylation-dependent ClpC1P2 proteolysis. Mol. Microbiol. 2010, 75, 592–606. [Google Scholar] [CrossRef]
- Luebke, J.L.; Eaton, D.S.; Sachleben, J.R.; Crosson, S. Allosteric control of a bacterial stress response system by an anti-σ factor. Mol. Microbiol. 2018, 107, 164–179. [Google Scholar] [CrossRef]
- Gicquel, G.; Bouffartigues, E.; Bains, M.; Oxaran, V.; Rosay, T.; Lesouhaitier, O.; Connil, N.; Bazire, A.; Maillot, O.; Bénard, M.; et al. The extra-cytoplasmic function sigma factor SigX modulates biofilm and virulence-related properties in Pseudomonas aeruginosa. PLoS ONE 2013, 8, e80407. [Google Scholar] [CrossRef]
- Otero-Asman, J.R.; Wettstadt, S.; Bernal, P.; Llamas, M.A. Diversity of extracytoplasmic function sigma (σECF) factor-dependent signaling in Pseudomonas. Mol. Microbiol. 2019, 112, 356–373. [Google Scholar] [CrossRef]
- Bouffartigues, E.; Si Hadj Mohand, I.; Maillot, O.; Tortuel, D.; Omnes, J.; David, A.; Tahrioui, A.; Duchesne, R.; Azuama, C.O.; Nusser, M.; et al. The temperature-regulation of Pseudomonas aeruginosa cmaX-cfrX-cmpX operon reveals an intriguing molecular network involving the sigma factors AlgU and SigX. Front. Microbiol. 2020, 11, 579495. [Google Scholar] [CrossRef]
- Pinto, D.; Liu, Q.; Mascher, T. ECF σ factors with regulatory extensions: The one-component systems of the σ universe. Mol. Microbiol. 2019, 112, 399–409. [Google Scholar] [CrossRef]
- Xie, Y.; Liu, W.; Shao, X.; Zhang, W.; Deng, X. Signal transduction schemes in Pseudomonas syringae. Comput. Struct. Biotechnol. J. 2020, 18, 3415–3424. [Google Scholar] [CrossRef]
- Hong, H.J. Construction of a bioassay system to identify extracellular agents targeting bacterial cell envelope. Methods Mol. Biol. 2016, 1440, 125–137. [Google Scholar] [CrossRef]
- Yanamandra, S.; Sarrafee, S.; Anaya-Bergman, C.; Jones, K.; Lewis, J. Role of the Porphyromonas gingivalis extracytoplasmic function sigma factor, SigH. Mol. Oral Microbiol. 2012, 27, 202–219. [Google Scholar] [CrossRef]
- Bilyk, B.; Kim, S.; Fazal, A.; Baker, T.A.; Seipke, R.F. Regulation of antimycin biosynthesis is controlled by the ClpXP protease. mSphere 2020, 5, e00144-20. [Google Scholar] [CrossRef]
- Iyer, S.C.; Casas-Pastor, D.; Kraus, D.; Mann, P.; Schirner, K.; Glatter, T.; Fritz, G.; Ringgaard, S. Transcriptional regulation by σ factor phosphorylation in bacteria. Nat. Microbiol. 2020, 5, 395–406. [Google Scholar] [CrossRef]
- Wu, H.; Liu, Q.; Casas-Pastor, D.; Dürr, F.; Mascher, T.; Fritz, G. The role of C-terminal extensions in controlling ECF σ factor activity in the widely conserved groups ECF41 and ECF42. Mol. Microbiol. 2019, 112, 498–514. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Santos, N.; Pérez, J.; Sánchez-Sutil, M.C.; Moraleda-Muñoz, A.; Muñoz-Dorado, J. CorE from Myxococcus xanthus is a copper-dependent RNA polymerase sigma factor. PLoS Genet. 2011, 7, e1002106. [Google Scholar] [CrossRef] [PubMed]
- Marcos-Torres, F.J.; Pérez, J.; Gómez-Santos, N.; Moraleda-Muñoz, A.; Muñoz-Dorado, J. In depth analysis of the mechanism of action of metal-dependent sigma factors: Characterization of CorE2 from Myxococcus xanthus. Nucleic Acids Res. 2016, 44, 5571–5584. [Google Scholar] [CrossRef] [PubMed]
- López-García, M.T.; Yagüe, P.; González-Quiñónez, N.; Rioseras, B.; Manteca, A. The SCO4117 ECF Sigma factor pleiotropically controls secondary metabolism and morphogenesis in Streptomyces coelicolor. Front. Microbiol. 2018, 9, 312. [Google Scholar] [CrossRef] [PubMed]
- Bastiaansen, K.C.; Civantos, C.; Bitter, W.; Llamas, M.A. New Insights into the regulation of cell-surface signaling activity acquired from a mutagenesis screen of the Pseudomonas putida IutY sigma/anti-sigma factor. Front. Microbiol. 2017, 8, 747. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Shao, X.; Deng, X. Regulation of type III secretion system in Pseudomonas syringae. Environ. Microbiol. 2019, 21, 4465–4477. [Google Scholar] [CrossRef]
- Waite, C.; Schumacher, J.; Jovanovic, M.; Bennett, M.; Buck, M. Negative autogenous control of the master type III secretion system regulator HrpL in Pseudomonas syringae. mBio 2017, 8, e02273-16. [Google Scholar] [CrossRef]
- Hutchings, M.I.; Hong, H.J.; Leibovitz, E.; Sutcliffe, I.C.; Buttner, M.J. The σ E cell envelope stress response of Streptomyces coelicolor Is influenced by a novel lipoprotein, CseA. J. Bacteriol. 2006, 188, 7222–7229. [Google Scholar] [CrossRef]
- Shu, D.; Chen, L.; Wang, W.; Yu, Z.; Ren, C.; Zhang, W.; Yang, S.; Lu, Y.; Jiang, W. afsQ1-Q2-sigQ is a pleiotropic but conditionally required signal transduction system for both secondary metabolism and morphological development in Streptomyces coelicolor. Appl. Microbiol. Biotechnol. 2009, 81, 1149–1160. [Google Scholar] [CrossRef]
- Wang, C.; Chen, W.; Xia, A.; Zhang, R.; Huang, Y.; Yang, S.; Ni, L.; Jin, F. carbon starvation induces the expression of PprB-Regulated genes in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2019, 85, e01705-19. [Google Scholar] [CrossRef]
- Moraleda-Muñoz, A.; Marcos-Torres, F.J.; Pérez, J.; Muñoz-Dorado, J. Metal-responsive RNA polymerase extracytoplasmic function (ECF) sigma factors. Mol. Microbiol. 2019, 112, 385–398. [Google Scholar] [CrossRef]
- Sachdeva, P.; Misra, R.; Tyagi, A.K.; Singh, Y. The sigma factors of Mycobacterium tuberculosis: Regulation of the regulators. FEBS J. 2009, 277, 605–626. [Google Scholar] [CrossRef]
- Grosse-Siestrup, B.T.; Gupta, T.; Helms, S.; Tucker, S.L.; Voskuil, M.I.; Quinn, F.D.; Karls, R.K. A Role for Mycobacterium tuberculosis sigma factor C in copper nutritional immunity. Int. J. Mol. Sci. 2021, 22, 2118. [Google Scholar] [CrossRef]
- Manganelli, R.; Voskuil, M.I.; Schoolnik, G.K.; Dubnau, E.; Gomez, M.; Smith, I. Role of the extracytoplasmic-function sigma factor sigmaH in Mycobacterium tuberculosis global gene expression. Mol. Microbiol. 2002, 45, 365–374. [Google Scholar] [CrossRef]
- Thakur, K.G.; Joshi, A.M.; Gopal, B. Structural and biophysical studies on two promoter recognition domains of the extra-cytoplasmic function σ factor σC from Mycobacterium tuberculosis. J. Biol. Chem. 2007, 282, 4711–4718. [Google Scholar] [CrossRef]
- Seipke, R.F.; Patrick, E.; Hutchings, M.I. Regulation of antimycin biosynthesis by the orphan ECF RNA polymerase sigma factorσAntA. PeerJ 2014, 2, e253. [Google Scholar] [CrossRef]
- Huang, X.; Pinto, D.; Fritz, G.; Mascher, T. Environmental sensing in actinobacteria: A comprehensive survey on the signaling capacity of this phylum. J. Bacteriol. 2015, 197, 2517–2535. [Google Scholar] [CrossRef]
- Bayer-Santos, E.; Lima, L.D.P.; Ceseti, L.D.M.; Ratagami, C.Y.; de Santana, E.S.; da Silva, A.M.; Farah, C.S.; Alvarez-Martinez, C.E. Xanthomonas citri T6SS mediates resistance to Dictyostelium predation and is regulated by an ECF σ factor and cognate Ser/Thr kinase. Environ. Microbiol. 2018, 20, 1562–1575. [Google Scholar] [CrossRef]
- Joo, D.M.; Ng, N.; Calendar, R. A 32 mutant with a single amino acid change in the highly conserved region 2.2 exhibits reduced core RNA polymerase affinity. Proc. Natl. Acad. Sci. USA 1997, 94, 4907–4912. [Google Scholar] [CrossRef]
- Sharp, M.M.; Chan, C.L.; Lu, C.Z.; Marr, M.T.; Nechaev, S.; Merritt, E.W.; Severinov, K.; Roberts, J.W.; Gross, C.A. The interface of sigma with core RNA polymerase is extensive, conserved, and functionally specialized. Genes Dev. 1999, 13, 3015–3026. [Google Scholar] [CrossRef]
- Wilson, M.J.; Lamont, I.L. Mutational analysis of an extracytoplasmic-function sigma factor to investigate its interactions with RNA polymerase and DNA. J. Bacteriol. 2006, 188, 1935–1942. [Google Scholar] [CrossRef]
- Lane, W.J.; Darst, S.A. Molecular evolution of multisubunit RNA polymerases: Structural analysis. J. Mol. Biol. 2010, 395, 686–704. [Google Scholar] [CrossRef]
- Wecke, T.; Halang, P.; Staroń, A.; Dufour, Y.S.; Donohue, T.J.; Mascher, T. Extracytoplasmic function σ factors of the widely distributed group ECF41 contain a fused regulatory domain. Microbiologyopen 2012, 1, 194–213. [Google Scholar] [CrossRef]
- Goutam, K.; Gupta, A.K.; Gopal, B. The fused SnoaL_2 domain in the Mycobacterium tuberculosis sigma factor σJ modulates promoter recognition. Nucleic Acids Res. 2017, 45, 9760–9772. [Google Scholar] [CrossRef][Green Version]
- Pérez, J.; Muñoz-Dorado, J.; Moraleda-Muñoz, A. The complex global response to copper in the multicellular bacterium Myxococcus xanthus. Metallomics 2018, 10, 876–886. [Google Scholar] [CrossRef]
- Mallick Gupta, A.; Mandal, S. The C-terminal domain of M. tuberculosis ECF sigma factor I (SigI) interferes in SigI-RNAP interaction. J. Mol. Model. 2020, 26, 77. [Google Scholar] [CrossRef]
- Dubey, A.P.; Pandey, P.; Singh, V.S.; Mishra, M.N.; Singh, S.; Mishra, R.; Tripathi, A.K. An ECF41 family σ factor controls motility and biogenesis of lateral flagella in Azospirillum brasilense sp245. J. Bacteriol. 2020, 202, e00231-20. [Google Scholar] [CrossRef]
- Liu, Q.; Pinto, D.; Mascher, T. Characterization of the widely distributed novel ECF42 group of extracytoplasmic function σ factors in Streptomyces venezuelae. J. Bacteriol. 2018, 200, e00437-18. [Google Scholar] [CrossRef]
- Tettmann, B.; Dötsch, A.; Armant, O.; Fjell, C.D.; Overhage, J. Knockout of extracytoplasmic function sigma factor ecf-10 affects stress resistance and biofilm formation in Pseudomonas putida KT2440. Appl. Environ. Microbiol. 2014, 80, 4911–4919. [Google Scholar] [CrossRef]
- Manteca, A.; Ye, J.; Sánchez, J.; Jensen, O.N. Phosphoproteome analysis of Streptomyces development reveals extensive protein phosphorylation accompanying bacterial differentiation. J. Proteome Res. 2011, 10, 5481–5492. [Google Scholar] [CrossRef]

| Proposed Mechanism | ECF Group | Model Sigma Factor | Reference |
|---|---|---|---|
| Conformational change | ECF11 | RpoE from Rhodobacter sphaeroides | [9] |
| ECF12 | SigH from Mycobacterium tuberculosis | [10] | |
| ECF14 | SigE from Mycobacterium tuberculosis | [11] | |
| ECF19 * | WP_016472479.1 from Streptomyces albus | [7] | |
| ECF293 * | RpoE from Neisseria meningitidis | [12] | |
| Partner switch | ECF15 | EcfG from Methylobacterium extorquens | [13] |
| Mechanosensing | ECF102 | SigX from Pseudomonas aeruginosa | [14] |
| Unknown mechanism | ECF125 | WP_044516075.1 from Mycolicibacterium septicum | [7] |
| ECF127 | EJO88542.1 from Mycobacterium colombiense | [7] | |
| ECF270 | ODS58609.1 from Acidobacteria bacterium SCN 69–37 | [7] | |
| ECF271 | OGO36537.1 from Chloroflexi bacterium RBG_16_56_8 | [7] | |
| ECF286 | WP_003983642.1 from Streptomyces rimosus | [7] | |
| ECF292 | WP_036395736.1 from Mycolicibacterium cosmeticum | [7] |
| Proposed Mechanism | ECF Group | Model Sigma Factor | References | |
|---|---|---|---|---|
| Transcriptional regulation | ECF12 * | ECF12s9 and ECF12s2 from Mycobacterium sp. | [7] | |
| ECF32 | HrpL from Pseudomonas syringae | [26] | ||
| ECF39 * | SigE from Streptomyces coelicolor | [27] | ||
| ECF114 | SigH from Porphyromonas gingivalis | [28] | ||
| ECF203 | SCD72908.1 from Streptomyces sp. DvalAA-19 | [7] | ||
| ECF234 | APQ59451.1 from Paenibacillus polymyxa | [7] | ||
| ECF293 * | PA3285 from Pseudomonas aeruginosa | [14] | ||
| Conformational changes | ECF36 * | SigC from Mycobacterium tuberculosis | [7] | |
| Proteolysis | ECF54 | SFT86700.1 from Geodermatophilus amargosae | [7] | |
| ECF282 | AntA from Streptomyces albus | [29] | ||
| Phosphorylation | ECF43 | EcfP from Vibrio parahaemolyticus | [30] | |
| ECF59 | SFI47409.1 from Planctomicrobium piriforme | [7] | ||
| ECF61 | OJW24604.1 from Planctomycetales bacterium 71–10 | [7] | ||
| ECF62 | WP_008685225.1 from Rhodopirellula sallentina | [7] | ||
| ECF217 | ELP31162.1 from Rhodopirellula baltica | [7] | ||
| ECF283 | WP_056749340.1 from Nocardioides sp. Root190 | [7] | ||
| Withregulatory extensions | Conformational change | ECF41 | SigJ from Mycobacterium tuberculosis | [31] |
| ECF238 | CorE from Myxococcus xanthus | [32,33] | ||
| Protein interaction | ECF42 | Sven_0747 from Streptomyces venezuelae | [31] | |
| ECF57 * | WP_015250107.1 from Singulisphaera acidiphila | [6] | ||
| Proteolysis | ECF36 * | KLO31890.1 from Mycolicibacter heraklionensis | [7] | |
| ECF48 | WP_048473130.1 from Mycolicibacterium chlorophenolicum | [7] | ||
| ECF52 | SCO4117 from Streptomyces coelicolor | [34] | ||
| ECF53 | WP_030276194.1 from Streptomyces purpeochromogenes | [7] | ||
| ECF115 | KOP67510.1 from Bacillus sp. FJAT-18019 | [7] | ||
| ECF243 * | IutY from Pseudomonas putida | [35] | ||
| ECF270 * | WP_011419852.1 from Anaeromyxobacter dehalogenans | [7] | ||
| Others | ECF29 | SED43577.1 from Bradyrhizobium lablabi | [7] | |
| ECF56 | WP_042440600.1 from Streptacidiphilus albus | [7] | ||
| ECF123 * | WP_028426757.1 from Streptomyces sp. TAA040 | [7] | ||
| ECF205 | WP_019068201.1 from Streptomyces hokutonensis | [7] | ||
| ECF216 * | QDE78790.1 from Myxococcus xanthus | [7] | ||
| ECF220 | WP_061622786.1 from Sorangium cellulosum | [7] | ||
| ECF237 | OLT65459.1 from Moorea producens | [7] | ||
| ECF240 * | SIO28919.1 from Chryseobacterium scophthalmum | [7] | ||
| ECF262 | SFB89493.1 from Ruminococcus albus | [7] | ||
| ECF264 | WP_037286607.1 from Saccharibacillus sacchari | [7] | ||
| ECF276 * | WP_063815919.1 from Sorangium cellulosum | [7] | ||
| ECF287 | WP_033089221.1 from Nocardia seriolae | [7] | ||
| ECF288 | WP_018594055.1 from Blautia producta | [7] | ||
| ECF294 | AKZ62584.1 from Herbaspirillum hiltneri | [7] | ||
| ECF295 | WP_063065904.1 Nocardia violaceofusca | [7] | ||
| Unknown mechanism | ECF58 | APZ92118.1 from Fuerstia marisgermanicae | [7] | |
| ECF122 | WP_057211282.1 from Cellulomonas sp. Root930 | [7] | ||
| ECF201 | CDO03659.1 from Oceanobacillus picturae | [7] | ||
| ECF248 | EOZ99538.1 from Indibacter alkaliphilus | [7] | ||
| ECF257 | WP_010287217.1 from Kurthia massiliensis | [7] | ||
| ECF265 * | AKO94994.1 from Bacillus endophyticus | [7] | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marcos-Torres, F.J.; Moraleda-Muñoz, A.; Contreras-Moreno, F.J.; Muñoz-Dorado, J.; Pérez, J. Mechanisms of Action of Non-Canonical ECF Sigma Factors. Int. J. Mol. Sci. 2022, 23, 3601. https://doi.org/10.3390/ijms23073601
Marcos-Torres FJ, Moraleda-Muñoz A, Contreras-Moreno FJ, Muñoz-Dorado J, Pérez J. Mechanisms of Action of Non-Canonical ECF Sigma Factors. International Journal of Molecular Sciences. 2022; 23(7):3601. https://doi.org/10.3390/ijms23073601
Chicago/Turabian StyleMarcos-Torres, Francisco Javier, Aurelio Moraleda-Muñoz, Francisco Javier Contreras-Moreno, José Muñoz-Dorado, and Juana Pérez. 2022. "Mechanisms of Action of Non-Canonical ECF Sigma Factors" International Journal of Molecular Sciences 23, no. 7: 3601. https://doi.org/10.3390/ijms23073601
APA StyleMarcos-Torres, F. J., Moraleda-Muñoz, A., Contreras-Moreno, F. J., Muñoz-Dorado, J., & Pérez, J. (2022). Mechanisms of Action of Non-Canonical ECF Sigma Factors. International Journal of Molecular Sciences, 23(7), 3601. https://doi.org/10.3390/ijms23073601






