A Novel Family of Winged-Helix Single-Stranded DNA-Binding Proteins from Archaea
Abstract
1. Introduction
2. Results
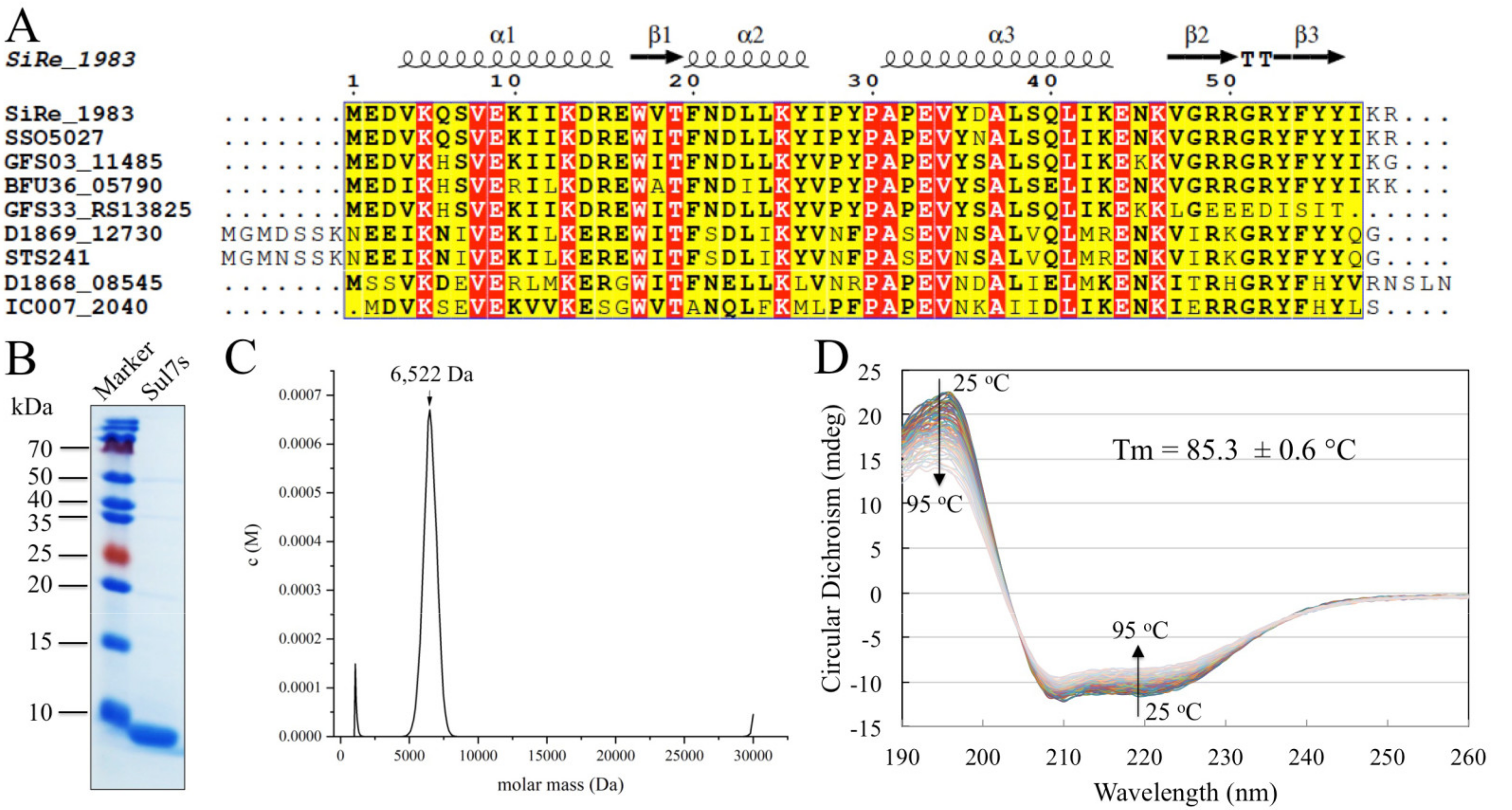
2.1. Sul7s Exists as a Thermostable Monomer in Solution
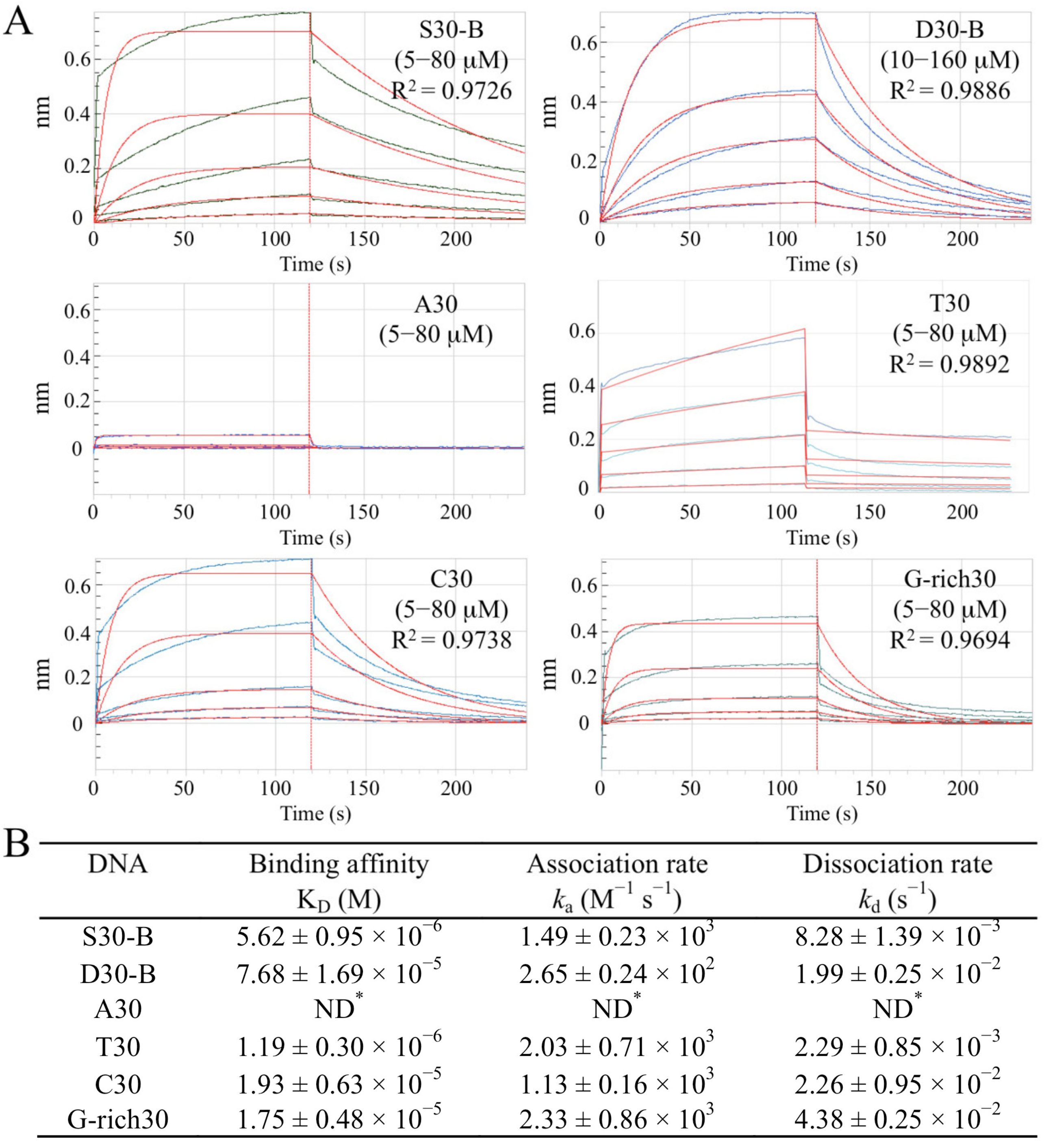
2.2. Sul7s Is an ssDNA-Binding Protein
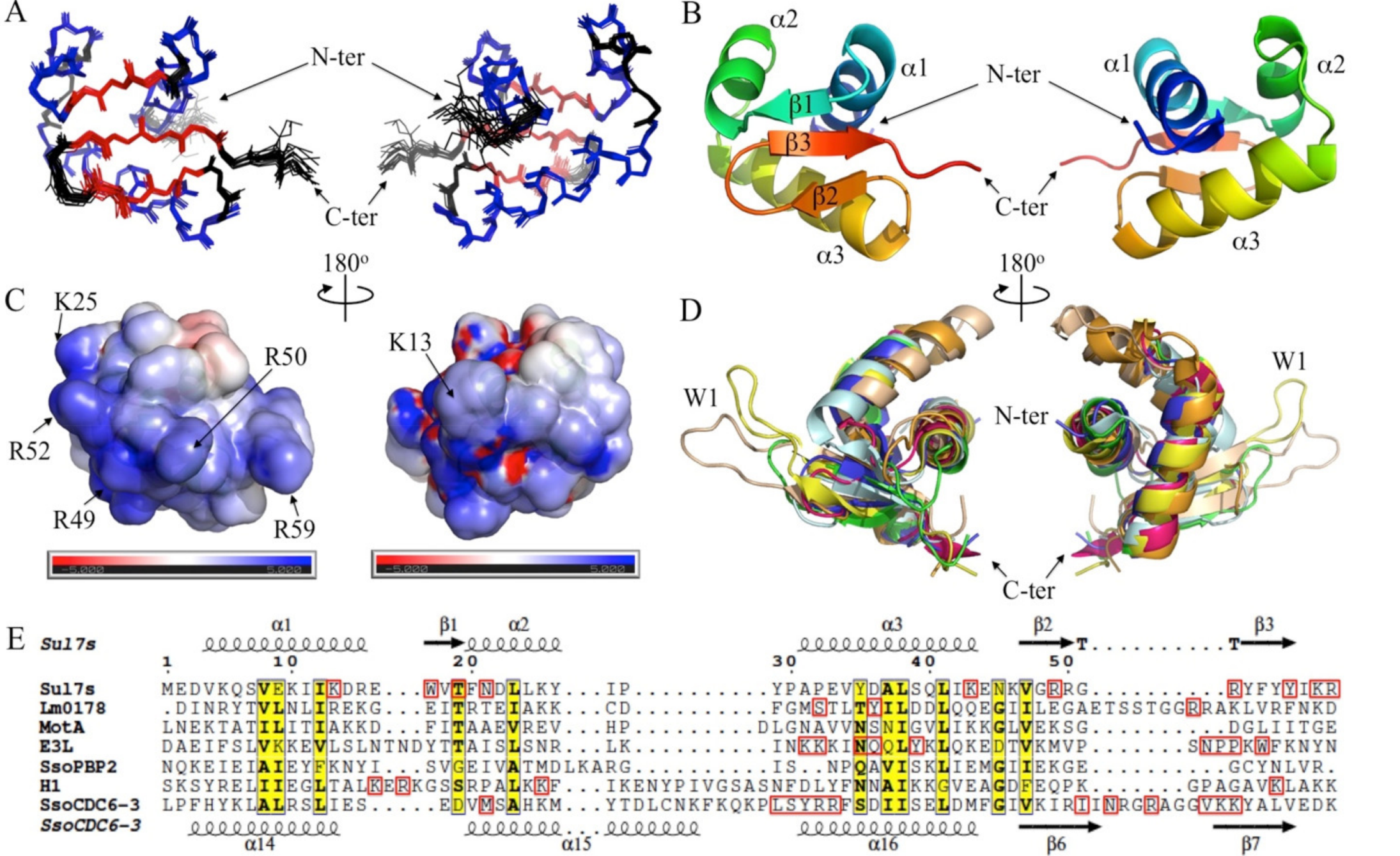
2.3. Solution Structure of Sul7s Reveals a Winged-Helix Fold
2.4. Sul7s Possesses a Unique ssDNA-Binding Surface
3. Discussion
4. Materials and Methods
4.1. Protein Overproduction and Purification
4.2. Preparation of Mutant Proteins
4.3. Oligonucleotides
- S30-F: 5′-biotin-TTTCTACCCTTTGGTGCTAATGCCCATACT
- S30-R: 5′-AGTATGGGCATTAGCACCAAAGGGTAGAAA
- A30: 5′-biotin-AAAAAAAAAAAAAAAAAAAAAAAAAAAAAA
- T30: 5′-biotin-TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTT
- C30: 5′-biotin-AAAAAAAAAAAAAAAAAAAAAAAAAAAAAA
- G-rich30: 5′-biotin-CTGGGGGCTGGGGGCTGGGGGCTGGGGGCT
- AT26: 5′-ATATATATATATATATATATATATAT
- S32-F: 5′-6-FAM-AGGGTTCTTTGTGGCGGCGTCATCTGTGCTTC
- S32-R: 5′-GAAGCACAGATGACGCCGCCACAAAGAACCCT-BHQ1
- S20-F: 5′-GTAGTCAGACACAGTAGTTC
4.4. Analytical Ultracentrifugation
4.5. Circular Dichroism (CD)
4.6. Biolayer Interferometry (BLI) Assays
4.7. Strand-Annealing Assay
4.8. Thermal Denaturation of dsDNA
4.9. Nick Closure Assays
4.10. NMR Spectroscopy
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mushegian, A.; Koonin, E.V. A minimal gene set for cellular life derived by comparison of complete bacterial genomes. Proc. Natl. Acad. Sci. USA 1996, 93, 10268–10273. [Google Scholar] [CrossRef] [PubMed]
- Antony, E.; Lohman, T.M. Dynamics of E. coli single stranded DNA binding (SSB) protein-DNA complexes. Semin. Cell Dev. Biol. 2019, 86, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Byrne, B.M.; Oakley, G.G. Replication protein A, the laxative that keeps DNA regular: The importance of RPA phosphorylation in maintaining genome stability. Semin. Cell Dev. Biol. 2019, 86, 112–120. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Shamoo, Y. Biochemical Characterization of Interactions between DNA Polymerase and Single-stranded DNA-binding Protein in Bacteriophage RB69. J. Biol. Chem. 2003, 278, 3876–3881. [Google Scholar] [CrossRef] [PubMed]
- Shamoo, Y.; Friedman, A.M.; Parsons, M.R.; Konigsberg, W.H.; Steitz, T.A. Crystal structure of a replication fork single-stranded DNA binding protein (T4 gp32) complexed to DNA. Nature 1995, 376, 362–366. [Google Scholar] [CrossRef]
- Hollis, T.; Stattel, J.M.; Walther, D.S.; Richardson, C.C.; Ellenberger, T. Structure of the gene 2.5 protein, a single-stranded DNA binding protein encoded by bacteriophage T7. Proc. Natl. Acad. Sci. USA 2001, 98, 9557–9562. [Google Scholar] [CrossRef]
- Meyer, R.R.; Laine, P.S. The single-stranded DNA-binding protein of Escherichia coli. Microbiol. Rev. 1990, 54, 342–380. [Google Scholar] [CrossRef]
- Wold, M.S. Replication Protein A: A Heterotrimeric, Single-Stranded DNA-Binding Protein Required for Eukaryotic DNA Metabolism. Annu. Rev. Biochem. 1997, 66, 61–92. [Google Scholar] [CrossRef]
- Shereda, R.D.; Kozlov, A.G.; Lohman, T.M.; Cox, M.M.; Keck, J.L. SSB as an Organizer/Mobilizer of Genome Maintenance Complexes. Crit. Rev. Biochem. Mol. 2008, 43, 289–318. [Google Scholar] [CrossRef]
- Zou, Y.; Liu, Y.; Wu, X.; Shell, S.M. Functions of human replication protein A (RPA): From DNA replication to DNA damage and stress responses. J. Cell. Physiol. 2006, 208, 267–273. [Google Scholar] [CrossRef]
- Fanning, E.; Klimovich, V.; Nager, A.R. A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 2006, 34, 4126–4137. [Google Scholar] [CrossRef]
- Suck, D. Common fold, common function, common origin? Nat. Struct. Biol. 1997, 4, 161–165. [Google Scholar] [CrossRef] [PubMed]
- Theobald, D.L.; Mitton-Fry, R.M.; Wuttke, D.S. Nucleic Acid Recognition by OB-Fold Proteins. Annu. Rev. Biophys. Biomol. Struct. 2003, 32, 115–133. [Google Scholar] [CrossRef]
- Raghunathan, S.; Kozlov, A.G.; Lohman, T.M.; Waksman, G. Structure of the DNA binding domain of E. coli SSB bound to ssDNA. Nat. Struct. Biol. 2000, 7, 648–652. [Google Scholar] [CrossRef] [PubMed]
- Bochkarev, A.; Bochkareva, E.; Frappier, L.; Edwards, A.M. The crystal structure of the complex of replication protein A subunits RPA32 and RPA14 reveals a mechanism for single-stranded DNA binding. EMBO J. 1999, 18, 4498–4504. [Google Scholar] [CrossRef] [PubMed]
- Bochkarev, A.; Pfuetzner, R.A.; Edwards, A.M.; Frappier, L. Structure of the single-stranded-DNA-binding domain of replication protein A bound to DNA. Nature 1997, 385, 176–181. [Google Scholar] [CrossRef]
- Komori, K.; Ishino, Y. Replication Protein A in Pyrococcus furiosus Is Involved in Homologous DNA Recombination. J. Biol. Chem. 2001, 276, 25654–25660. [Google Scholar] [CrossRef]
- White, M. Archaeal DNA repair: Paradigms and puzzles. Biochem. Soc. Trans. 2003, 31, 690–693. [Google Scholar] [CrossRef]
- Wadsworth, R.I.; White, M.F. Identification and properties of the crenarchaeal single-stranded DNA binding protein from Sulfolobus solfataricus. Nucleic Acids Res. 2001, 29, 914–920. [Google Scholar] [CrossRef][Green Version]
- Touma, C.; Kariawasam, R.; Gimenez, A.X.; Bernardo, R.E.; Ashton, N.W.; Adams, M.N.; Paquet, N.; Croll, T.I.; O’Byrne, K.J.; Richard, D.J.; et al. A structural analysis of DNA binding by hSSB1 (NABP2/OBFC2B) in solution. Nucleic Acids Res. 2016, 44, 7963–7973. [Google Scholar] [CrossRef]
- Kerr, I.D.; Wadsworth, R.I.M.; Cubeddu, L.; Blankenfeldt, W.; Naismith, J.H.; White, M.F. Insights into ssDNA recognition by the OB fold from a structural and thermodynamic study of Sulfolobus SSB protein. EMBO J. 2003, 22, 2561–2570. [Google Scholar] [CrossRef] [PubMed]
- Paytubi, S.; McMahon, S.A.; Graham, S.; Liu, H.; Botting, C.H.; Makarova, K.S.; Koonin, E.V.; Naismith, J.H.; White, M.F. Displacement of the canonical single-stranded DNA-binding protein in the Thermoproteales. Proc. Natl. Acad. Sci. USA 2012, 109, E398–E405. [Google Scholar] [CrossRef] [PubMed]
- Ghalei, H.; Von Moeller, H.; Eppers, D.; Sohmen, D.; Wilson, D.; Loll, B.; Wahl, M.C. Entrapment of DNA in an intersubunit tunnel system of a single-stranded DNA-binding protein. Nucleic Acids Res. 2014, 42, 6698–6708. [Google Scholar] [CrossRef]
- Zhang, C.; Phillips, A.P.R.; Wipfler, R.L.; Olsen, G.J.; Whitaker, R.J. The essential genome of the crenarchaeal model Sulfolobus islandicus. Nat. Commun. 2018, 9, 4908. [Google Scholar] [CrossRef]
- Suzuki, S.; Kurosawa, N. Robust growth of archaeal cells lacking a canonical single-stranded DNA-binding protein. FEMS Microbiol. Lett. 2019, 366, 366. [Google Scholar] [CrossRef] [PubMed]
- Van Dijk, M.; Mulders, J.; Poutsma, A.; Könst, A.A.M.; Lachmeijer, A.A.M.; Dekker, A.G.; Blankenstein, M.; Oudejans, C.B.M. Maternal segregation of the Dutch preeclampsia locus at 10q22 with a new member of the winged helix gene family. Nat. Genet. 2005, 37, 514–519. [Google Scholar] [CrossRef] [PubMed]
- Rieping, W.; Habeck, M.; Bardiaux, B.; Bernard, A.; Malliavin, T.E.; Nilges, M. ARIA2: Automated NOE assignment and data integration in NMR structure calculation. Bioinformatics 2007, 23, 381–382. [Google Scholar] [CrossRef]
- Vranken, W.; Boucher, W.; Stevens, T.J.; Fogh, R.H.; Pajon, A.; Llinas, M.; Ulrich, E.L.; Markley, J.L.; Ionides, J.; Laue, E.D. The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins 2005, 59, 687–696. [Google Scholar] [CrossRef]
- Chen, V.B.; Arendall, W.B., III; Headd, J.J.; Keedy, D.A.; Immormino, R.M.; Kapral, G.J.; Murray, L.W.; Richardson, J.S.; Richardson, D.C. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 2010, 66, 12–21. [Google Scholar] [CrossRef]
- Gajiwala, K.S.; Burley, S.K. Winged helix proteins. Curr. Opin. Struct. Biol. 2000, 10, 110–116. [Google Scholar] [CrossRef]
- Clark, K.L.; Halay, E.D.; Lai, E.; Burley, S.K. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature 1993, 364, 412–420. [Google Scholar] [CrossRef]
- Hasegawa, H.; Holm, L. Advances and pitfalls of protein structural alignment. Curr. Opin. Struct. Biol. 2009, 19, 341–348. [Google Scholar] [CrossRef] [PubMed]
- Light, S.H.; Cahoon, L.A.; Halavaty, A.S.; Freitag, N.E.; Anderson, W.F. Structure to function of an alpha-glucan metabolic pathway that promotes Listeria monocytogenes pathogenesis. Nat. Microbiol. 2016, 2, 16202. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Wen, A.; Zhao, M.; You, L.; Zhang, Y.; Feng, Y. Structural basis of sigma appropriation. Nucleic Acids Res. 2019, 47, 9423–9432. [Google Scholar] [CrossRef] [PubMed]
- Ha, S.C.; Lokanath, N.K.; Van Quyen, D.; Wu, C.A.; Lowenhaupt, K.; Rich, A.; Kim, Y.-G.; Kim, K.K. A poxvirus protein forms a complex with left-handed Z-DNA: Crystal structure of a Yatapoxvirus Zalpha bound to DNA. Proc. Natl. Acad. Sci. USA 2004, 101, 14367–14372. [Google Scholar] [CrossRef]
- Yan, J.; Beattie, T.R.; Rojas, A.L.; Schermerhorn, K.; Gristwood, T.; Trinidad, J.C.; Albers, S.V.; Roversi, P.; Gardner, A.F.; Abrescia, N.G.A.; et al. Identification and characterization of a heterotrimeric archaeal DNA polymerase holoenzyme. Nat. Commun. 2017, 8, 15075. [Google Scholar] [CrossRef]
- Ali, T.; Coles, P.; Stevens, T.J.; Stott, K.; Thomas, J.O. Two Homologous Domains of Similar Structure but Different Stability in the Yeast Linker Histone, Hho1p. J. Mol. Biol. 2004, 338, 139–148. [Google Scholar] [CrossRef]
- Dueber, E.L.C.; Corn, J.E.; Bell, S.D.; Berger, J.M. Replication Origin Recognition and Deformation by a Heterodimeric Archaeal Orc1 Complex. Science 2007, 317, 1210–1213. [Google Scholar] [CrossRef]
- Gajiwala, K.S.; Chen, H.; Cornille, F.; Roques, B.P.; Reith, W.; Mach, B.; Burley, S. Structure of the winged-helix protein hRFX1 reveals a new mode of DNA binding. Nature 2000, 403, 916–921. [Google Scholar] [CrossRef]
- Harami, G.M.; Gyimesi, M.; Kovács, M. From keys to bulldozers: Expanding roles for winged helix domains in nucleic-acid-binding proteins. Trends Biochem. Sci. 2013, 38, 364–371. [Google Scholar] [CrossRef]
- Sanchis, I.M.; Pigli, Y.Z.; Rice, P.A. Crystal Structure of an Unusual Single-Stranded DNA-Binding Protein Encoded by Staphylococcal Cassette Chromosome Elements. Structure 2018, 26, 1144–1150.e3. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, S.; Kurosawa, N. Development of the Multiple Gene Knockout System with One-Step PCR in Thermoacidophilic Crenarchaeon Sulfolobus acidocaldarius. Archaea 2017, 2017, 7459310. [Google Scholar] [CrossRef] [PubMed]
- Gellert, M.; Lipsett, M.N.; Davies, D.R. Helix formation by guanylic acid. Proc. Natl. Acad. Sci. USA 1962, 48, 2013–2018. [Google Scholar] [CrossRef] [PubMed]
- Saenger, W.; Riecke, J.; Suck, D. A structural model for the polyadenylic acid single helix. J. Mol. Biol. 1975, 93, 529–534. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhao, M.; Wang, L.; Chen, Y.; Dong, Y.; Gong, Y.; Huang, L. Roles of Leu28 side chain intercalation in the interaction between Cren7 and DNA. Biochem. J. 2017, 474, 1727–1739. [Google Scholar] [CrossRef] [PubMed]
- Ikura, M.; Kay, L.E.; Bax, A. A Novel Approach for Sequential Assignment of 1H, 13C, and 15N Spectra of Proteins: Heteronuclear Triple-Resonance Three-Dimensional NMR Spectroscopy. Application to Calmodulin. Biochemistry 1990, 29, 4659–4667. [Google Scholar] [CrossRef]
- Bax, A.; Ikura, M.; Kay, L.E.; Barbato, G.; Spera, S. Multidimensional Triple Resonance NMR Spectroscopy of Isotopically Uniformly Enriched Proteins: A Powerful New Strategy for Structure Determination. Ciba Found Symp. 1991, 161, 108–119. [Google Scholar] [CrossRef]
- Bax, A.; Ikura, M. An efficient 3D NMR technique for correlating the proton and15N backbone amide resonances with the α-carbon of the preceding residue in uniformly15N/13C enriched proteins. J. Biomol. NMR 1991, 1, 99–104. [Google Scholar] [CrossRef]
- Delaglio, F.; Grzesiek, S.; Vuister, G.W.; Zhu, G.; Pfeifer, J.; Bax, A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 1995, 6, 277–293. [Google Scholar] [CrossRef]
- Brunger, A.T.; Adams, P.; Clore, G.M.; Delano, W.L.; Gros, P.; Grosse-Kunstleve, R.W.; Jiang, J.S.; Kuszewski, J.; Nilges, M.; Pannu, N.S.; et al. Crystallography & NMR System: A New Software Suite for Macromolecular Structure Determination. Acta Crystallogr. Sect. D Biol. Crystallogr. 1998, 54, 905–921. [Google Scholar] [CrossRef]
- Brunger, A.T. Version 1.2 of the Crystallography and NMR system. Nat. Protoc. 2007, 2, 2728–2733. [Google Scholar] [CrossRef] [PubMed]
- Cheung, M.-S.; Maguire, M.L.; Stevens, T.J.; Broadhurst, R.W. DANGLE: A Bayesian inferential method for predicting protein backbone dihedral angles and secondary structure. J. Magn. Reson. 2010, 202, 223–233. [Google Scholar] [CrossRef] [PubMed]



| Total number of distance restraints 1 | 1388 |
| intra residual (|i-j| = 0) | 565 |
| sequential (|i-j| = 1) | 246 |
| medium range (1<|i-j|<5) | 134 |
| long range (|i-j|> = 5) | 264 |
| ambiguous restraints | 179 |
| Dihedral angle restraints 2 | |
| Φ | 52 |
| Ψ | 51 |
| Average number of restraints per residue | 25.3 |
| Mean rmsd from idealized covalent geometry 1 | |
| bond (Å) | 3.252 × 10−3 ± 8.766 × 10−5 |
| angle (o) | 4.781 × 10−1 ± 8.692 × 10−3 |
| improper (o) | 1.042 ± 6.632 × 10−2 |
| Mean rmsd from the experimental restraints 1 | |
| dihedral angle (o) | 3.400 × 10−1 ± 8.356 × 10−2 |
| distance (Å) | 2.078 × 10−2 ± 1.678 × 10−3 |
| Ramachandran plot 3 | |
| % residues in the most favorable regions | 93% |
| allowed regions | 7% |
| disallowed regions | 0% |
| Atomic rmsd 1 | |
| backbone | 0.426 ± 0.118 |
| heavy atoms | 0.967 ± 0.142 |
| Mutants | Binding Affinity KD (M) | Association Rate ka (M−1 s−1) | Dissociation Rate kd (s−1) |
|---|---|---|---|
| D3A | 6.01 ± 0.80 × 10−6 | 6.91 ± 0.65 × 102 | 4.09 ± 0.16 × 10−3 |
| K13A | 3.04 ± 0.54 × 10−5 | 4.28 ± 0.27 × 102 | 1.32 ± 0.31 × 10−2 |
| R15A | 4.67 ± 0.53 × 10−6 | 1.24 ± 0.40 × 102 | 5.80 ± 0.85 × 10−3 |
| W17A | 1.07 ± 0.35 × 10−5 | 5.84 ± 0.74 × 102 | 6.29 ± 1.00 × 10−3 |
| T19G | 1.41 ± 0.40 × 10−5 | 4.83 ± 0.20 × 102 | 6.74 ± 1.62 × 10−3 |
| N21A | 8.92 ± 0.29 × 10−6 | 8.39 ± 0.37 × 102 | 7.50 ± 0.57 × 10−3 |
| E33A | 1.21 ± 0.09 × 10−6 | 4.57 ± 0.59 × 103 | 5.56 ± 1.11 × 10−3 |
| D36A | 8.83 ± 1.15 × 10−5 | 3.74 ± 0.12 × 102 | 3.29 ± 0.33 × 10−2 |
| K43A | 2.79 ± 0.50 × 10−5 | 5.01 ± 0.22 × 102 | 1.41 ± 0.31 × 10−2 |
| R49A | ND 2 | ND 2 | ND 2 |
| R52A | ND 2 | ND 2 | ND 2 |
| Y56A | 1.56 ± 0.67 × 10−5 | 4.94 ± 0.78 × 102 | 7.17 ± 2.10 × 10−3 |
| K58A | 2.69 ± 0.42 × 10−5 | 3.54 ± 0.04 × 102 | 9.52 ± 1.61 × 10−3 |
| R59A | ND 2 | ND 2 | ND 2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, C.; Liu, X.; Chen, Y.; Zhou, J.; Li, W.; Ding, N.; Huang, L.; Chen, J.; Zhang, Z. A Novel Family of Winged-Helix Single-Stranded DNA-Binding Proteins from Archaea. Int. J. Mol. Sci. 2022, 23, 3455. https://doi.org/10.3390/ijms23073455
Huang C, Liu X, Chen Y, Zhou J, Li W, Ding N, Huang L, Chen J, Zhang Z. A Novel Family of Winged-Helix Single-Stranded DNA-Binding Proteins from Archaea. International Journal of Molecular Sciences. 2022; 23(7):3455. https://doi.org/10.3390/ijms23073455
Chicago/Turabian StyleHuang, Can, Xuehui Liu, Yuanyuan Chen, Junshi Zhou, Wenqian Li, Niannian Ding, Li Huang, Jingyu Chen, and Zhenfeng Zhang. 2022. "A Novel Family of Winged-Helix Single-Stranded DNA-Binding Proteins from Archaea" International Journal of Molecular Sciences 23, no. 7: 3455. https://doi.org/10.3390/ijms23073455
APA StyleHuang, C., Liu, X., Chen, Y., Zhou, J., Li, W., Ding, N., Huang, L., Chen, J., & Zhang, Z. (2022). A Novel Family of Winged-Helix Single-Stranded DNA-Binding Proteins from Archaea. International Journal of Molecular Sciences, 23(7), 3455. https://doi.org/10.3390/ijms23073455








