Structural and Biophysical Insights into SPINK1 Bound to Human Cationic Trypsin
Abstract
1. Introduction
2. Results
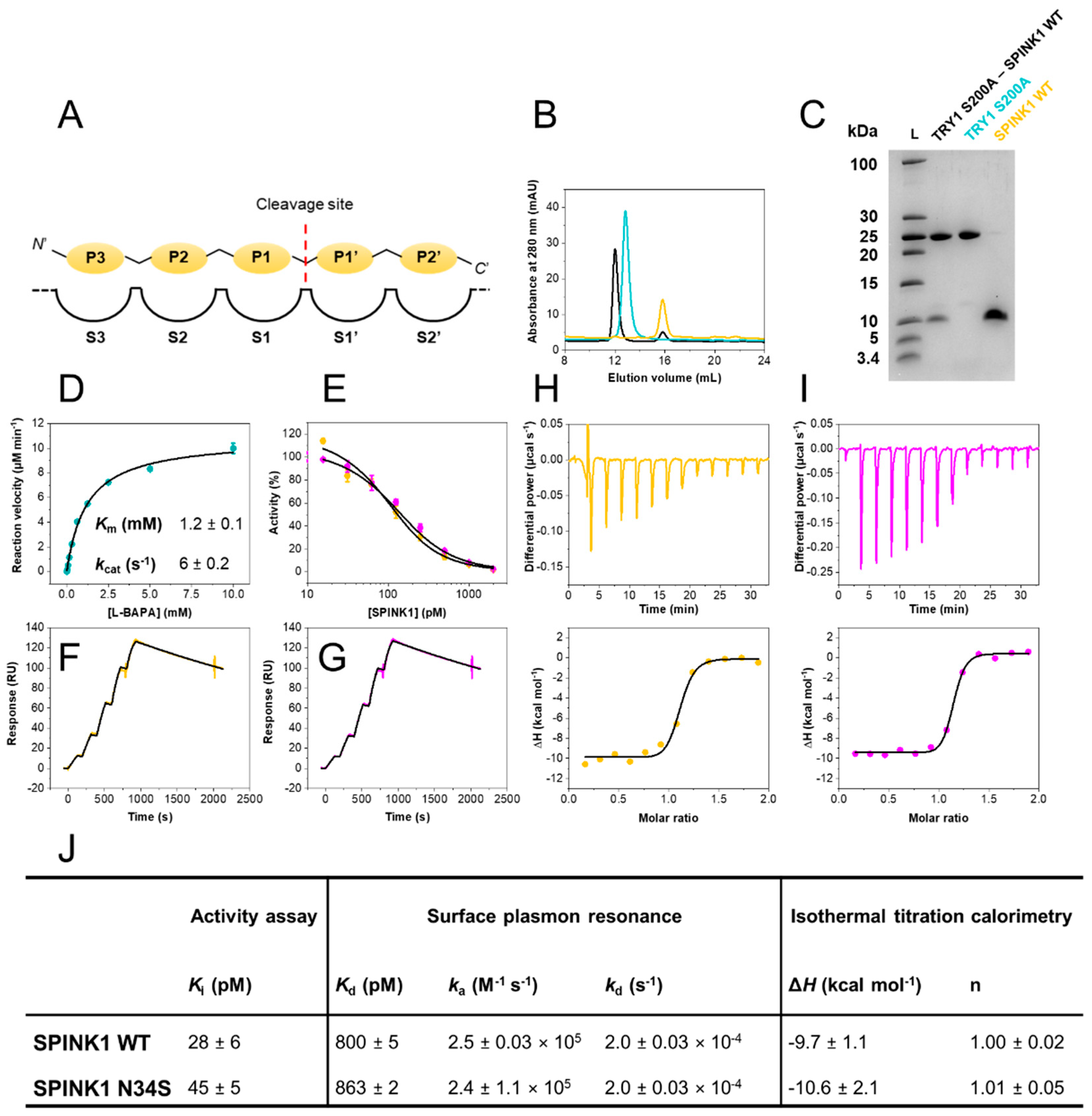
2.1. Inhibitory Activities of SPINK1 WT and p.N34S Are Similar
2.2. SPINK1 WT and p.N34S Show Similar Kinetics for Their Interaction with TRY1
2.3. SPINK1 WT and p.N34S Display Similar Thermodynamic Profiles for Their Interaction with TRY1
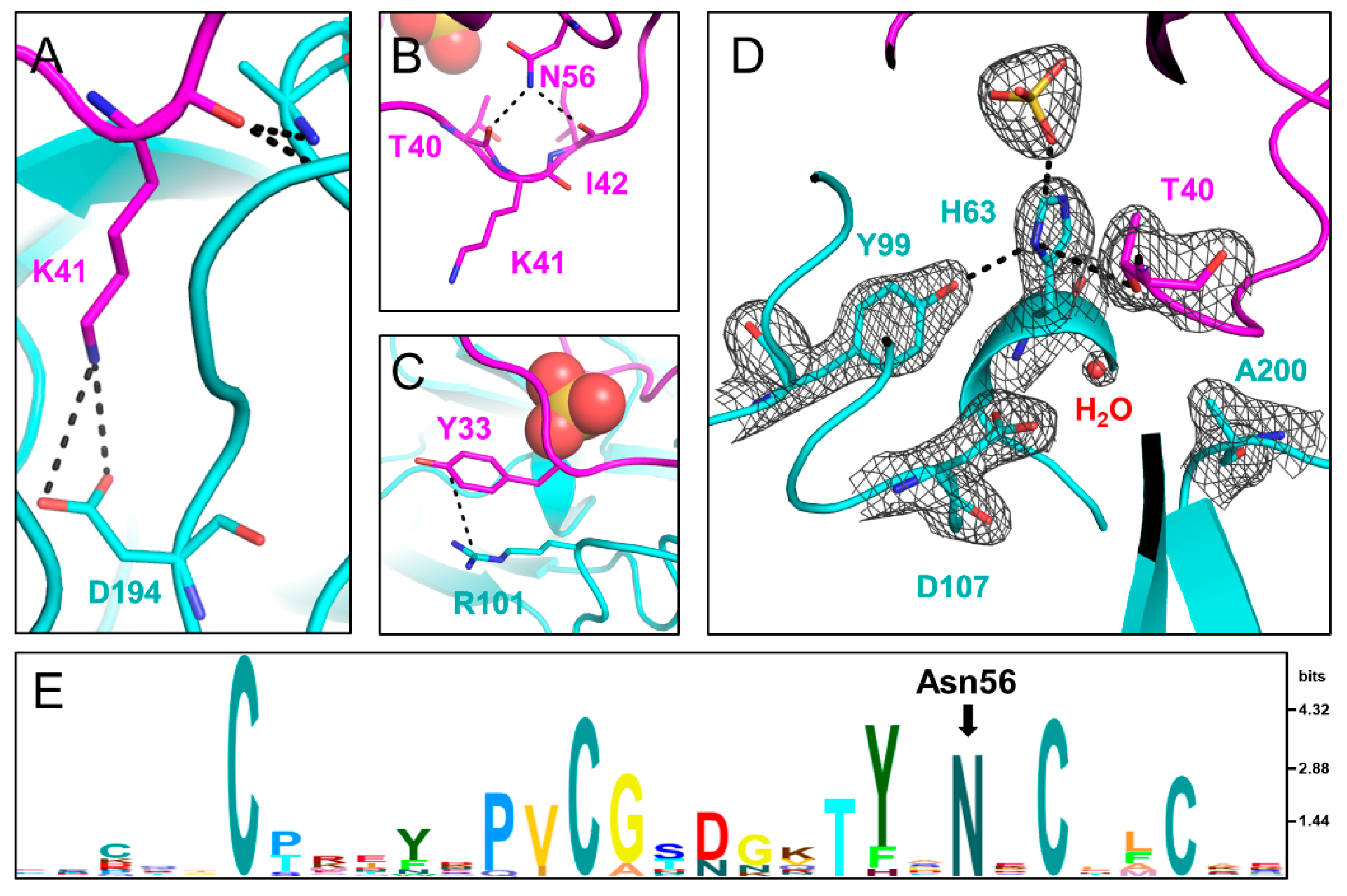
2.4. Structure of SPINK1 Variants Bound to TRY1
2.5. Interactions of SPINK1 Variants with TRY1 p.S200A
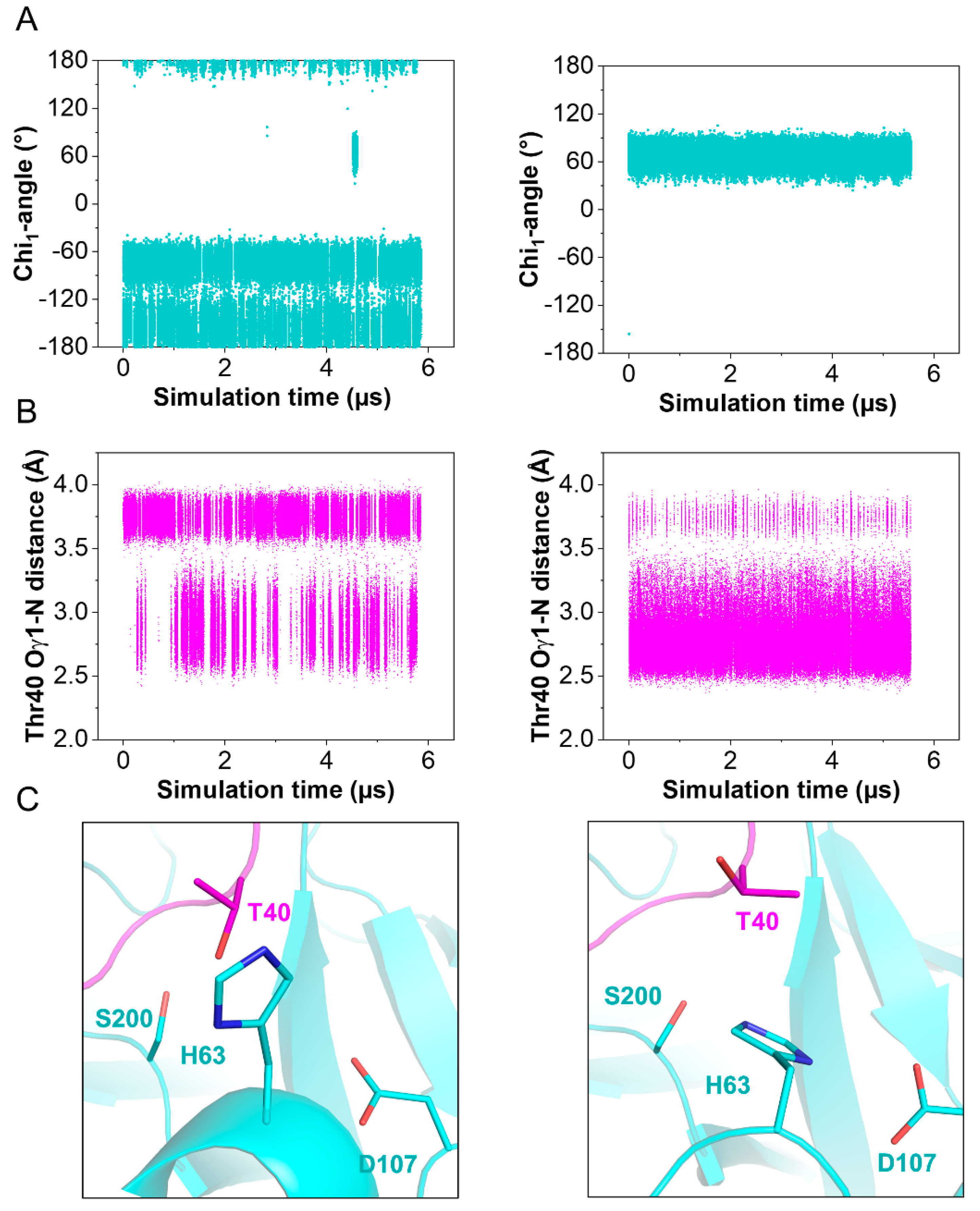
2.6. Conformational Change in the TRY1 Catalytic Triad
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Protein Expression and Purification
5.2. Enzyme Inhibition Studies
5.3. Surface Plasmon Resonance
5.4. Isothermal Titration Calorimetry
5.5. Crystallization and Data Collection
5.6. Structure Determination
5.7. Molecular Dynamics Simulations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef] [PubMed]
- Rawlings, N.D.; Barrett, A.J.; Bateman, A. MEROPS: The peptidase database. Nucleic Acids Res. 2010, 38, D227–D233. [Google Scholar] [CrossRef] [PubMed]
- Papamokos, E.; Weber, E.; Bode, W.; Huber, R.; Empie, M.W.; Kato, I.; Laskowski, M. Crystallographic refinement of Japanese quail ovomucoid, a Kazal-type inhibitor, and model building studies of complexes with serine proteases. J. Mol. Biol. 1982, 158, 515–537. [Google Scholar] [CrossRef]
- Laskowski, J.M.; Kato, I. Protein inhibitors of proteinases. Annu. Rev. Biochem. 1980, 49, 593–626. [Google Scholar] [CrossRef]
- Schechter, I.; Berger, A. On the size of the active site in proteases. I. Papain. Biochem. Biophys. Res. Commun. 1967, 27, 157–162. [Google Scholar] [CrossRef]
- Bode, W.; Huber, R. Natural protein proteinase inhibitors and their interaction with proteinases. Eur. J. Biochem. 1992, 204, 433–451. [Google Scholar] [CrossRef]
- Otlewski, J.; Jelen, F.; Zakrzewska, M.; Oleksy, A. The many faces of protease-protein inhibitor interaction. EMBO J. 2005, 24, 1303–1310. [Google Scholar] [CrossRef]
- Rawlings, N.D.; Tolle, D.P.; Barrett, A.J. Evolutionary families of peptidase inhibitors. Biochem. J. 2004, 378, 705–716. [Google Scholar] [CrossRef]
- Zakharova, E.; Horvath, M.P.; Goldenberg, D.P. Structure of a serine protease poised to resynthesize a peptide bond. Proc. Natl. Acad. Sci. USA 2009, 106, 11034–11039. [Google Scholar] [CrossRef]
- Longstaff, C.; Campbell, A.F.; Fersht, A.R. Recombinant chymotrypsin inhibitor 2: Expression, kinetic analysis of inhibition with alpha-chymotrypsin and wild-type and mutant subtilisin BPN’, and protein engineering to investigate inhibitory specificity and mechanism. Biochemistry 1990, 29, 7339–7347. [Google Scholar] [CrossRef]
- Lu, W.Y.; Starovasnik, M.A.; Dwyer, J.J.; Kossiakoff, A.A.; Kent, S.B.; Lu, W. Deciphering the role of the electrostatic interactions involving Gly70 in eglin C by total chemical protein synthesis. Biochemistry 2000, 39, 3575–3584. [Google Scholar] [CrossRef] [PubMed]
- Peräkylä, M.; Kollman, P.A. Why Does Trypsin Cleave BPTI so Slowly? J. Am. Chem. Soc. 2000, 122, 3436–3444. [Google Scholar] [CrossRef]
- Radisky, E.S.; Koshland, D.E.J. A clogged gutter mechanism for protease inhibitors. Proc. Natl. Acad. Sci. USA 2002, 99, 10316–10321. [Google Scholar] [CrossRef] [PubMed]
- Witt, H.; Luck, W.; Hennies, H.C.; Classen, M.; Kage, A.; Lass, U.; Landt, O.; Becker, M. Mutations in the gene encoding the serine protease inhibitor, Kazal type 1 are associated with chronic pancreatitis. Nat. Genet. 2000, 25, 213–216. [Google Scholar] [CrossRef]
- Pfützer, R.H.; Barmada, M.M.; Brunskill, A.P.J.; Finch, R.; Hart, P.S.; Neoptolemos, J.; Furey, W.F.; Whitcomb, D.C. SPINK1/PSTI polymorphisms act as disease modifiers in familial and idiopathic chronic pancreatitis. Gastroenterology 2000, 119, 615–623. [Google Scholar] [CrossRef] [PubMed]
- Aoun, E.; Chang, C.C.H.; Greer, J.B.; Papachristou, G.I.; Barmada, M.M.; Whitcomb, D.C. Pathways to injury in chronic pancreatitis: Decoding the role of the high-risk SPINK1 N34S haplotype using meta-analysis. PLoS ONE 2008, 3, e2003. [Google Scholar] [CrossRef] [PubMed]
- Chandak, G.R.; Idris, M.M.; Reddy, D.N.; Mani, K.R.; Bhaskar, S.; Rao, G.V.; Singh, L. Absence of PRSS1 mutations and association of SPINK1 trypsin inhibitor mutations in hereditary and non-hereditary chronic pancreatitis. Gut 2004, 53, 723–728. [Google Scholar] [CrossRef]
- Kereszturi, E.; Király, O.; Sahin-Tóth, M. Minigene analysis of intronic variants in common SPINK1 haplotypes associated with chronic pancreatitis. Gut 2009, 58, 545–549. [Google Scholar] [CrossRef]
- Lukas, J.; Pospech, J.; Oppermann, C.; Hund, C.; Iwanov, K.; Pantoom, S.; Petters, J.; Frech, M.; Seemann, S.; Thiel, F.-G.; et al. Role of endoplasmic reticulum stress and protein misfolding in disorders of the liver and pancreas. Adv. Med. Sci. 2019, 64, 315–323. [Google Scholar] [CrossRef]
- Threadgold, J.; Greenhalf, W.; Ellis, I.; Howes, N.; Lerch, M.M.; Simon, P.; Jansen, J.; Charnley, R.; Laugier, R.; Frulloni, L.; et al. The N34S mutation of SPINK1 (PSTI) is associated with a familial pattern of idiopathic chronic pancreatitis but does not cause the disease. Gut 2002, 50, 675–681. [Google Scholar] [CrossRef]
- Chen, J.-M.; Férec, C. The true culprit within the SPINK1 p.N34S-containing haplotype is still at large. Gut 2009, 58, 478–480. [Google Scholar] [CrossRef] [PubMed]
- Pu, N.; Masson, E.; Cooper, D.N.; Génin, E.; Férec, C.; Chen, J.-M. Chronic Pancreatitis: The True Pathogenic Culprit within the SPINK1 N34S-Containing Haplotype Is No Longer at Large. Genes 2021, 12, 1683. [Google Scholar] [CrossRef]
- Mehner, C.; Radisky, E.S. Bad Tumors Made Worse: SPINK1. Front. Cell Dev. Biol. 2019, 7, 10. [Google Scholar] [CrossRef] [PubMed]
- Räsänen, K.; Itkonen, O.; Koistinen, H.; Stenman, U.-H. Emerging Roles of SPINK1 in Cancer. Clin. Chem. 2016, 62, 449–457. [Google Scholar] [CrossRef]
- Lin, T.-C. Functional Roles of SPINK1 in Cancers. Int. J. Mol. Sci. 2021, 22, 3814. [Google Scholar] [CrossRef] [PubMed]
- Ozaki, N.; Ohmuraya, M.; Hirota, M.; Ida, S.; Wang, J.; Takamori, H.; Higashiyama, S.; Baba, H.; Yamamura, K. Serine Protease Inhibitor Kazal Type 1 Promotes Proliferation of Pancreatic Cancer Cells through the Epidermal Growth Factor Receptor. Mol. Cancer Res. 2009, 7, 1572–1581. [Google Scholar] [CrossRef]
- Lu, F.; Lamontagne, J.; Sun, A.; Pinkerton, M.; Block, T.; Lu, X. Role of the inflammatory protein serine protease inhibitor Kazal in preventing cytolytic granule granzyme A-mediated apoptosis. Immunology 2011, 134, 398–408. [Google Scholar] [CrossRef]
- Kulke, M.; Nagel, F.; Schulig, L.; Geist, N.; Gabor, M.; Mayerle, J.; Lerch, M.M.; Link, A.; Delcea, M. A Hypothesized Mechanism for Chronic Pancreatitis Caused by the N34S Mutation of Serine Protease Inhibitor Kazal-Type 1 Based on Conformational Studies. J. Inflamm. Res. 2021, 14, 2111. [Google Scholar] [CrossRef]
- Sun, Z.; Kolossváry, I.; Kozakov, D.; Sahin-Tóth, M.; Vajda, S. The N34S mutation of SPINK1 may impact the kinetics of trypsinogen activation to cause early trypsin release in the pancreas. bioRxiv 2020. [Google Scholar] [CrossRef]
- Ohmuraya, M.; Sugano, A.; Hirota, M.; Takaoka, Y.; Yamamura, K.I. Role of intrapancreatic SPINK1/Spink3 expression in the development of pancreatitis. Front. Physiol. 2012, 3, 126. [Google Scholar] [CrossRef]
- Szabó, A.; Toldi, V.; Gazda, L.D.; Demcsák, A.; Tőzsér, J.; Sahin-Tóth, M. Defective binding of SPINK1 variants is an uncommon mechanism for impaired trypsin inhibition in chronic pancreatitis. J. Biol. Chem. 2021, 296, 100343. [Google Scholar] [CrossRef] [PubMed]
- Buchholz, I.; Nagel, F.; Klein, A.; Wagh, P.R.; Mahajan, U.M.; Greinacher, A.; Lerch, M.M.; Mayerle, J.; Delcea, M. The impact of physiological stress conditions on protein structure and trypsin inhibition of serine protease inhibitor Kazal type 1 (SPINK1) and its N34S variant. Biochim. Biophys. Acta Proteins Proteom. 2020, 1868, 140281. [Google Scholar] [CrossRef] [PubMed]
- Boros, E.; Sebák, F.; Héja, D.; Szakács, D.; Zboray, K.; Schlosser, G.; Micsonai, A.; Kardos, J.; Bodor, A.; Pál, G. Directed Evolution of Canonical Loops and Their Swapping between Unrelated Serine Proteinase Inhibitors Disprove the Interscaffolding Additivity Model. J. Mol. Biol. 2019, 431, 557–575. [Google Scholar] [CrossRef] [PubMed]
- McGrath, M.E.; Haymore, B.L.; Summers, N.L.; Craik, C.S.; Fletterick, R.J. Structure of an engineered, metal-actuated switch in trypsin. Biochemistry 1993, 32, 1914–1919. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, T.J.; Clements, J.; Finn, R.D. Skylign: A tool for creating informative, interactive logos representing sequence alignments and profile hidden Markov models. BMC Bioinform. 2014, 15, 7. [Google Scholar] [CrossRef] [PubMed]
- Salameh, M.A.; Soares, A.S.; Navaneetham, D.; Sinha, D.; Walsh, P.N.; Radisky, E.S. Determinants of Affinity and Proteolytic Stability in Interactions of Kunitz Family Protease Inhibitors with Mesotrypsin. J. Biol. Chem. 2010, 285, 36884–36896. [Google Scholar] [CrossRef]
- Salameh, M.A.; Soares, A.S.; Hockla, A.; Radisky, E.S. Structural Basis for Accelerated Cleavage of Bovine Pancreatic Trypsin Inhibitor (BPTI) by Human Mesotrypsin. J. Biol. Chem. 2008, 283, 4115–4123. [Google Scholar] [CrossRef]
- Király, O.; Wartmann, T.; Sahin-Tóth, M. Missense mutations in pancreatic secretory trypsin inhibitor (SPINK1) cause intracellular retention and degradation. Gut 2007, 56, 1433–1438. [Google Scholar] [CrossRef]
- Kuwata, K.; Hirota, M.; Shimizu, H.; Nakae, M.; Nishihara, S.; Takimoto, A.; Mitsushima, K.; Kikuchi, N.; Endo, K.; Inoue, M.; et al. Functional analysis of recombinant pancreatic secretory trypsin inhibitor protein with amino-acid substitution. J. Gastroenterol. 2002, 37, 928–934. [Google Scholar] [CrossRef]
- Yadav, D.; Lowenfels, A.B. The epidemiology of pancreatitis and pancreatic cancer. Gastroenterology 2013, 144, 1252–1261. [Google Scholar] [CrossRef]
- Lazim, R.; Suh, D.; Choi, S. Advances in molecular dynamics simulations and enhanced sampling methods for the study of protein systems. Int. J. Mol. Sci. 2020, 21, 6339. [Google Scholar] [CrossRef] [PubMed]
- Jing, H.; Babu, Y.S.; Moore, D.; Kilpatrick, J.M.; Liu, X.-Y.; Volanakis, J.E.; Narayana, S.V.L. Structures of native and complexed complement factor D: Implications of the atypical his57 conformation and self-inhibitory loop in the regulation of specific serine protease activity11. J. Mol. Biol. 1998, 282, 1061–1081. [Google Scholar] [CrossRef] [PubMed]
- Bolognesi, M.; Gatti, G.; Menegatti, E.; Guarneri, M.; Marquart, M.; Papamokos, E.; Huber, R. Three-dimensional structure of the complex between pancreatic secretory trypsin inhibitor (Kazal type) and trypsinogen at 1.8 A resolution: Structure solution, crystallographic refinement and preliminary structural interpretation. J. Mol. Biol. 1982, 162, 839–868. [Google Scholar] [CrossRef]
- Perona, J.J.; Craik, C.S. Evolutionary divergence of substrate specificity within the chymotrypsin-like serine protease fold. J. Biol. Chem. 1997, 272, 29987–29990. [Google Scholar] [CrossRef]
- Perona, J.J.; Craik, C.S. Structural basis of substrate specificity in the serine proteases. Protein Sci. 1995, 4, 337–360. [Google Scholar] [CrossRef]
- Corey, D.R.; Craik, C.S. An investigation into the minimum requirements for peptide hydrolysis by mutation of the catalytic triad of trypsin. J. Am. Chem. Soc. 1992, 114, 1784–1790. [Google Scholar] [CrossRef]
- Corey, D.R.; Willett, W.S.; Coombs, G.S.; Craik, C.S. Trypsin specificity increased through substrate-assisted catalysis. Biochemistry 1995, 34, 11521–11527. [Google Scholar] [CrossRef]
- Sahin-Tóth, M.; Szabó, A. Expression, purification and activity measurements of pancreatic proteases. Pancreapedia Exocrine Pancreas Knowl. Base 2011. [Google Scholar] [CrossRef][Green Version]
- Kabsch, W. XDS. Acta Crystallogr. Sect. D 2010, 66, 125–132. [Google Scholar] [CrossRef]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef]
- Gaboriaud, C.; Serre, L.; Guy-Crotte, O.; Forest, E.; Fontecilla-Camps, J.-C. Crystal Structure of Human Trypsin 1: Unexpected Phosphorylation of Tyr151. J. Mol. Biol. 1996, 259, 995–1010. [Google Scholar] [CrossRef] [PubMed]
- Hecht, H.J.; Szardenings, M.; Collins, J.; Schomburg, D. Three-dimensional structure of the complexes between bovine chymotrypsinogen A and two recombinant variants of human pancreatic secretory trypsin inhibitor (Kazal-type). J. Mol. Biol. 1991, 220, 711–722. [Google Scholar] [CrossRef]
- Murshudov, G.N.; Vagin, A.A.; Dodson, E.J. Refinement of Macromolecular Structures by the Maximum-Likelihood Method. Acta Crystallogr. Sect. D 1997, 53, 240–255. [Google Scholar] [CrossRef]
- Liebschner, D.; Afonine, P.V.; Baker, M.L.; Bunkoczi, G.; Chen, V.B.; Croll, T.I.; Hintze, B.; Hung, L.-W.; Jain, S.; McCoy, A.J.; et al. Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. Sect. D 2019, 75, 861–877. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kalé, L.; Schulten, K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; MacKerell, J.A.D. CHARMM36 all-atom additive protein force field: Validation based on comparison to NMR data. J. Comput. Chem. 2013, 34, 2135–2145. [Google Scholar] [CrossRef]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Hopkins, C.W.; Le Grand, S.; Walker, R.C.; Roitberg, A.E. Long-Time-Step Molecular Dynamics through Hydrogen Mass Repartitioning. J. Chem. Theory Comput. 2015, 11, 1864–1874. [Google Scholar] [CrossRef]

| SPINK1 WT–TRY1 p.S200A | SPINK1 p.N34S–TRY1 p.S200A | |
|---|---|---|
| Data collection | ||
| Beamline | 14.1 at BESSY | 14.2 at BESSY |
| Wavelength (Å) | 0.9184 | 0.9184 |
| Unit-cell parameters (Å) a, c in space group P3121 | 77.63, 187.35 | 76.55, 189.72 |
| Resolution (Å) | 50–2.90 (3.08–2.90) | 50–2.10 (2.22–2.10) |
| No. of unique reflections (Friedel pairs merged) | 15,112 (2371) | 38,593 (6081) |
| Redundancy | 19.1 (19.4) | 19.7 (19.4) |
| Completeness (%) | 99.8 (99.0) | 99.8 (98.7) |
| Rmerge | 0.166 (2.651) | 0.174 (3.172) |
| cc1/2 | 0.999 (0.550) | 0.999 (0.411) |
| <I/σ(I)> | 16.3 (1.2) | 14.8 (1.0) |
| Wilson B-factor (Å2) | 85.6 | 50.1 |
| Refinement | ||
| Resolution range (Å) | 50–2.90 (2.99–2.90) | 50–2.10 (2.15–2.10) |
| Completeness (%) | 99.7 (98.0) | 99.8 (97.1) |
| No. of reflections, working set | 13589 (1191) | 36688 (2600) |
| No. of reflections, test set | 1509 (133) | 1904 (134) |
| Final Rwork | 0.222 (0.365) | 0.197 (0.334) |
| Final Rfree | 0.243 (0.374) | 0.233 (0.421) |
| No. of non-H atoms | ||
| Protein | 4219 | 4269 |
| Solvent | 16 | 236 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.011 | 0.008 |
| Angles (°) | 1.619 | 1.505 |
| Average B factors (Å2) | ||
| Protein | 100.01 | 36.73 |
| Solvent | 138.68 | 51.09 |
| Molprobity analysis | ||
| Ramachandran most favored (%) | 96.17 | 96.75 |
| Ramachandran outliers (%) | 0.0 | 0.0 |
| Overall score | 1.90 | 1.80 |
| Clash score | 13.48 | 3.80 |
| PDB entry | 7QE8 | 7QE9 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nagel, F.; Palm, G.J.; Geist, N.; McDonnell, T.C.R.; Susemihl, A.; Girbardt, B.; Mayerle, J.; Lerch, M.M.; Lammers, M.; Delcea, M. Structural and Biophysical Insights into SPINK1 Bound to Human Cationic Trypsin. Int. J. Mol. Sci. 2022, 23, 3468. https://doi.org/10.3390/ijms23073468
Nagel F, Palm GJ, Geist N, McDonnell TCR, Susemihl A, Girbardt B, Mayerle J, Lerch MM, Lammers M, Delcea M. Structural and Biophysical Insights into SPINK1 Bound to Human Cationic Trypsin. International Journal of Molecular Sciences. 2022; 23(7):3468. https://doi.org/10.3390/ijms23073468
Chicago/Turabian StyleNagel, Felix, Gottfried J. Palm, Norman Geist, Thomas C. R. McDonnell, Anne Susemihl, Britta Girbardt, Julia Mayerle, Markus M. Lerch, Michael Lammers, and Mihaela Delcea. 2022. "Structural and Biophysical Insights into SPINK1 Bound to Human Cationic Trypsin" International Journal of Molecular Sciences 23, no. 7: 3468. https://doi.org/10.3390/ijms23073468
APA StyleNagel, F., Palm, G. J., Geist, N., McDonnell, T. C. R., Susemihl, A., Girbardt, B., Mayerle, J., Lerch, M. M., Lammers, M., & Delcea, M. (2022). Structural and Biophysical Insights into SPINK1 Bound to Human Cationic Trypsin. International Journal of Molecular Sciences, 23(7), 3468. https://doi.org/10.3390/ijms23073468







