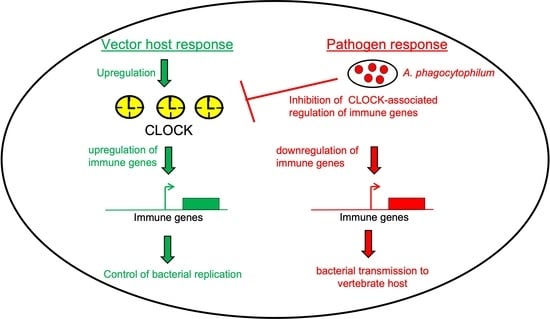
Rickettsial Pathogen Perturbs Tick Circadian Gene to Infect the Vertebrate Host
Abstract
:1. Introduction
2. Results
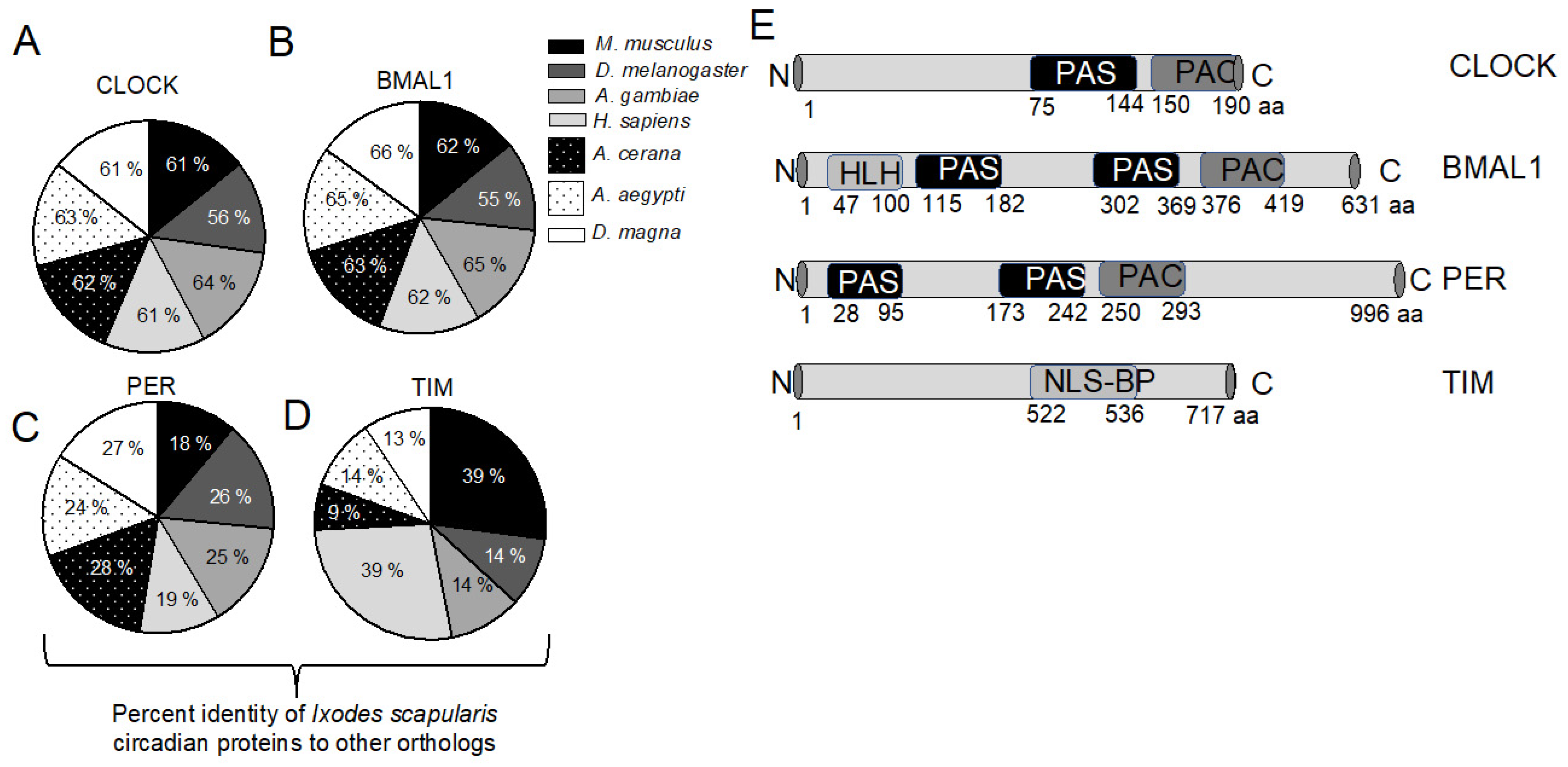
2.1. Identification and Phylogenetic Analysis of Circadian Genes in Ixodes scapularis Genome
2.2. Tick Circadian Genes Are Developmentally Regulated
2.3. Anaplasma phagocytophilum Perturb Oscillation of Circadian Genes in Ticks and Tick Cells
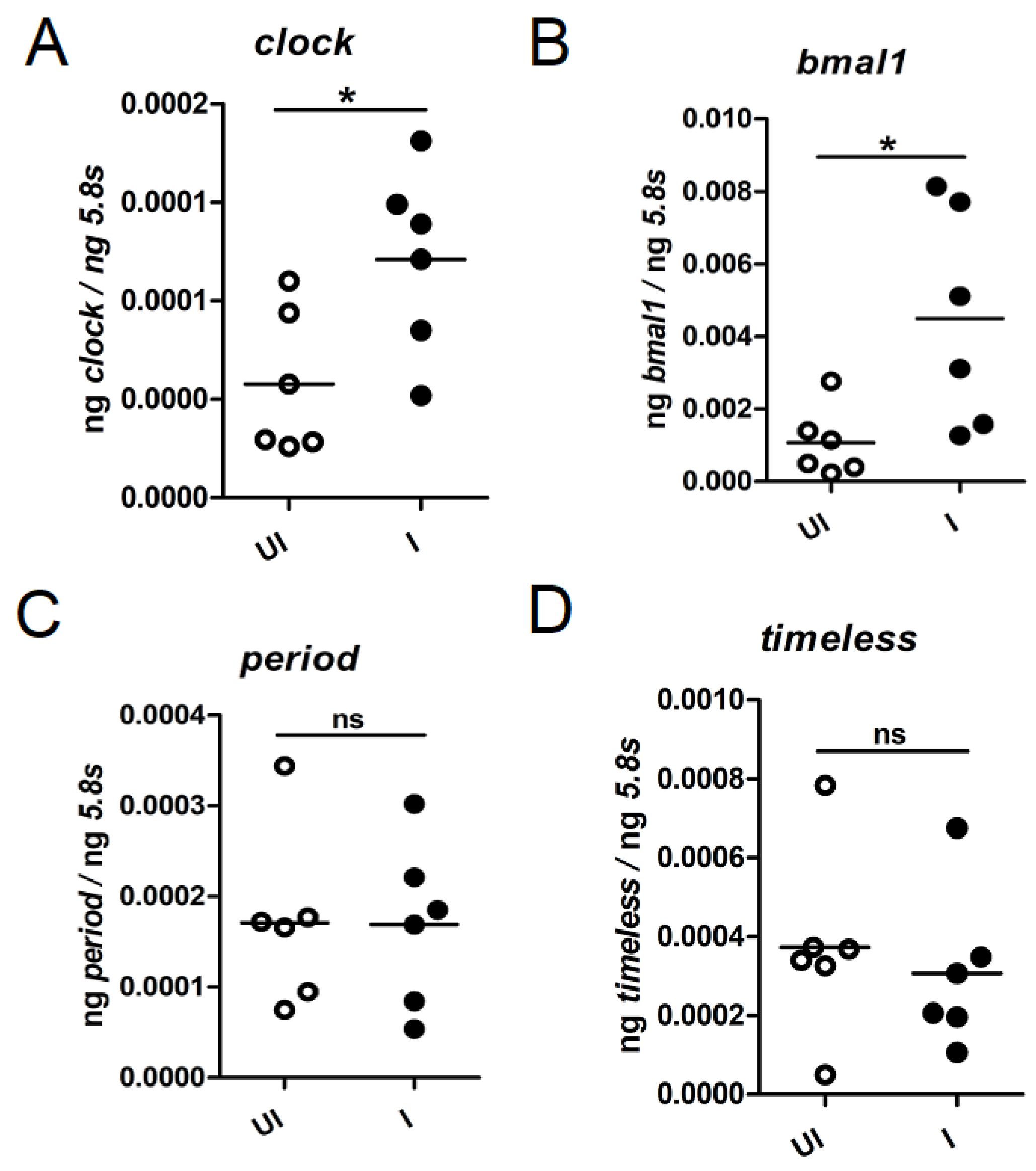
2.4. Tick clock and bmal1 Are Upregulated during Transmission of A. phagocytophilum to the Vertebrate Host
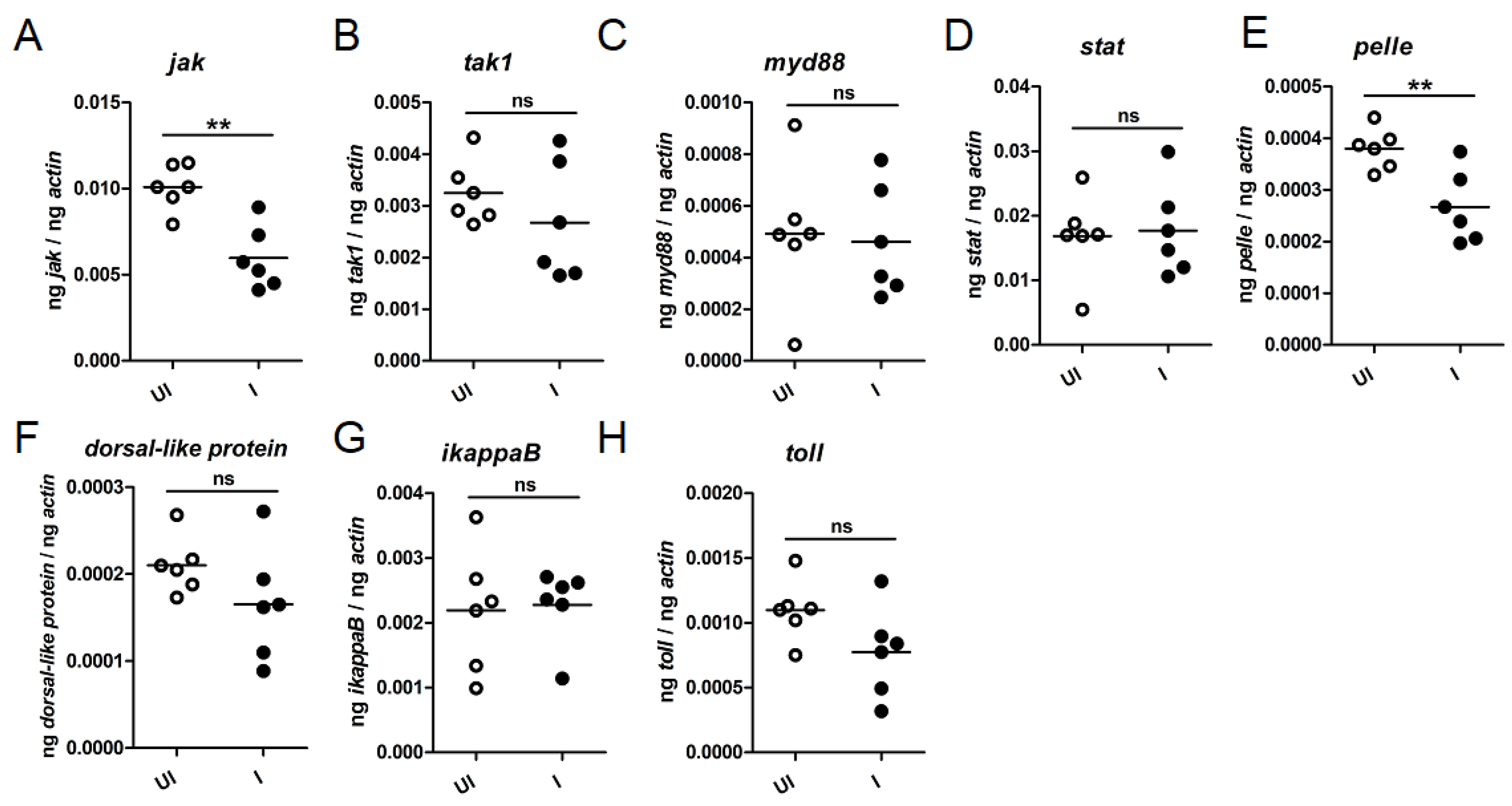
2.5. Anaplasma phagocytophilum Downregulates Tick Immune Genes during Transmission to the Vertebrate Host
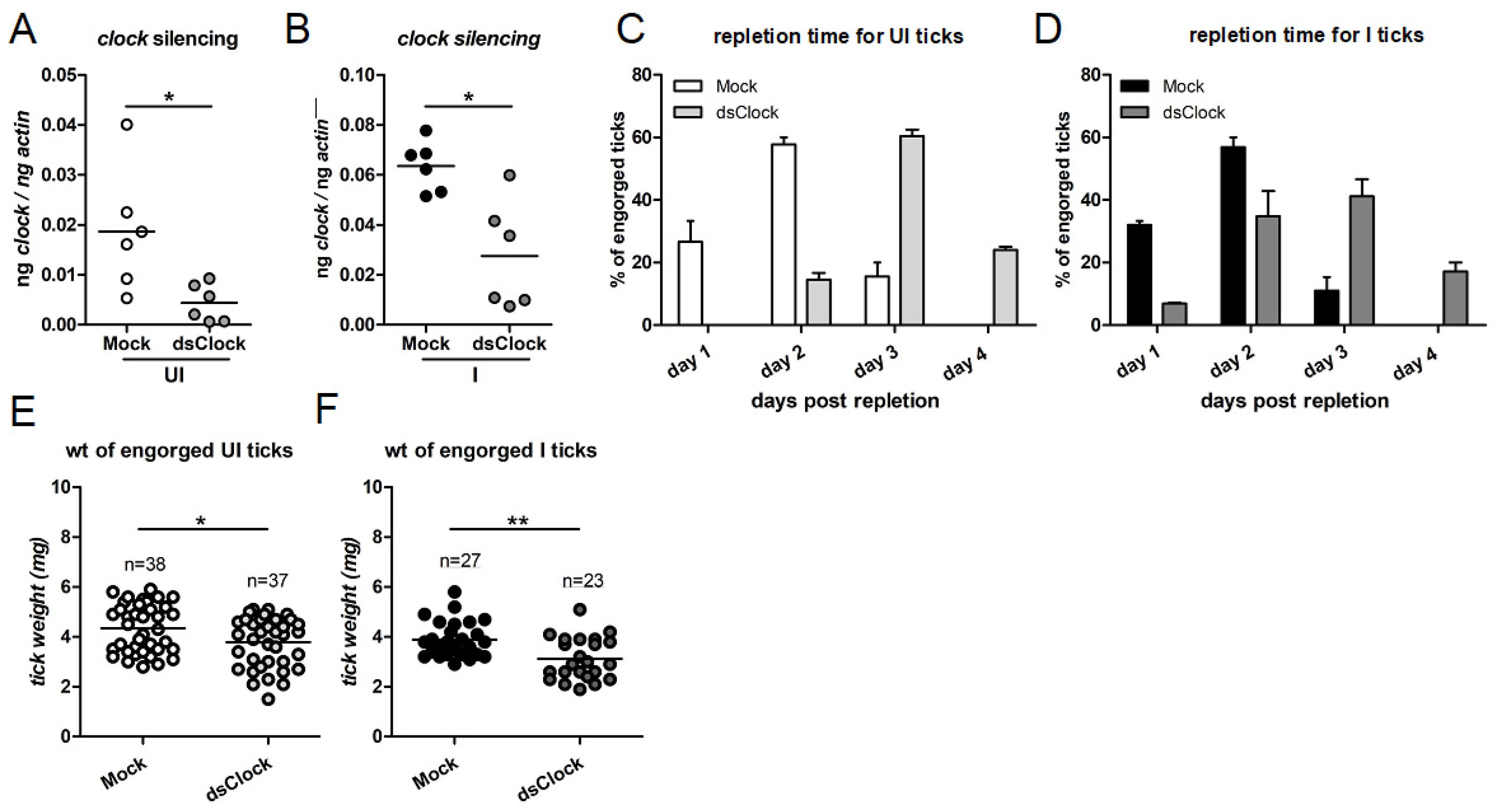
2.6. RNAi-Mediated Silencing of Clock Expression Affects Tick Blood Feeding
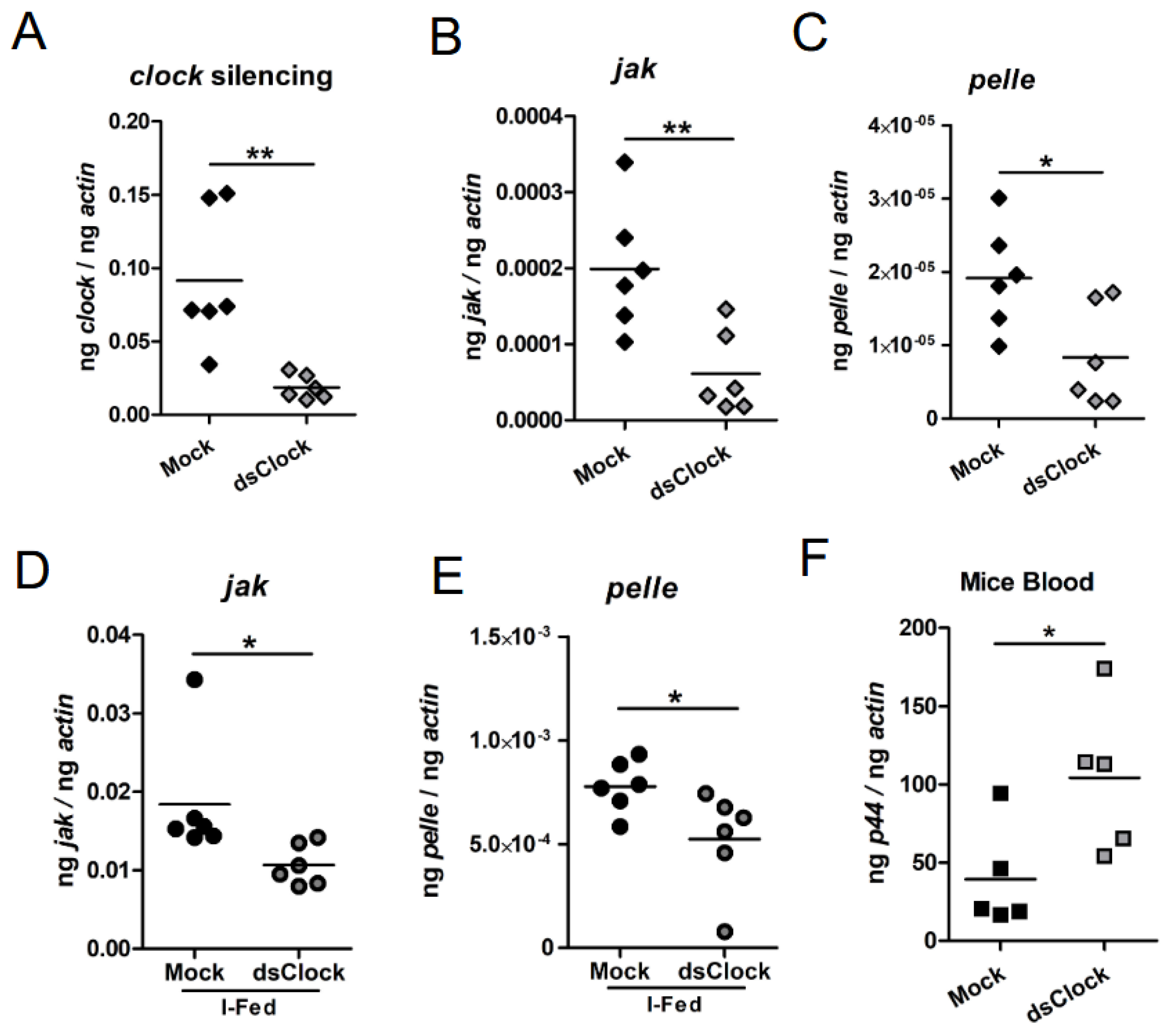
2.7. RNAi-Mediated Silencing of clock Expression Affects Immune Gene Expression in Ticks That Facilitates Increased Bacterial Transmission to the Murine Host
3. Discussion
4. Materials and Methods
4.1. Ticks and Bacterial Isolates
4.2. Mice and Tick Feeding
4.3. Ethics Statement
4.4. Identification of Circadian Genes from Tick Genome
4.5. Sequence Alignment and Phylogenetic Analysis
4.6. Circadian Oscillation Experiments
4.7. RNA/DNA Extractions and Gene Expression Analysis
4.8. dsRNA Synthesis and Silencing Experiments in Ticks and Tick Cells
4.9. Anaplasma phagocytophilum Transmission Experiments from Ticks to Mice
4.10. Statistics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wikel, S.K. Ticks and Tick-Borne Infections: Complex Ecology, Agents, and Host Interactions. Vet. Sci. 2018, 5, 60. [Google Scholar] [CrossRef] [Green Version]
- Neelakanta, G.; Sultana, H.; Fish, D.; Anderson, J.F.; Fikrig, E. Anaplasma phagocytophilum induces Ixodes scapularis ticks to express an antifreeze glycoprotein gene that enhances their survival in the cold. J. Clin. Investig. 2010, 120, 3179–3190. [Google Scholar] [CrossRef] [Green Version]
- Anderson, J.F.; Magnarelli, L.A. Biology of ticks. Infect. Dis. Clin. N. Am. 2008, 22, 195–215. [Google Scholar] [CrossRef]
- Keirans, J.E.; Hutcheson, H.J.; Durden, L.A.; Klompen, J. Ixodes (Ixodes) scapularis (Acari:Ixodidae): Redescription of all active stages, distribution, hosts, geographical variation, and medical and veterinary importance. J. Med. Entomol. 1996, 33, 297–318. [Google Scholar] [CrossRef]
- Wilson, M.L.; Spielman, A. Seasonal activity of immature Ixodes dammini (Acari: Ixodidae). J. Med. Entomol. 1985, 22, 408–414. [Google Scholar] [CrossRef]
- Rikihisa, Y. Mechanisms of obligatory intracellular infection with Anaplasma phagocytophilum. Clin. Microbiol. Rev. 2011, 24, 469–489. [Google Scholar] [CrossRef] [Green Version]
- Jaarsma, R.I.; Sprong, H.; Takumi, K.; Kazimirova, M.; Silaghi, C.; Mysterud, A.; Rudolf, I.; Beck, R.; Földvári, G.; Tomassone, L.; et al. Anaplasma phagocytophilum evolves in geographical and biotic niches of vertebrates and ticks. Parasit. Vectors 2019, 12, 328. [Google Scholar] [CrossRef]
- Khanal, S.; Taank, V.; Anderson, J.F.; Sultana, H.; Neelakanta, G. Arthropod transcriptional activator protein-1 (AP-1) aids tick-rickettsial pathogen survival in the cold. Sci. Rep. 2018, 8, 11409. [Google Scholar] [CrossRef] [Green Version]
- Villar, M.; López, V.; Ayllón, N.; Cabezas-Cruz, A.; Lopez, J.A.; Vázquez, J.; Alberdi, P.; De La Fuente, J. The intracellular bacterium Anaplasma phagocytophilum selectively manipulates the levels of vertebrate host proteins in the tick vector Ixodes scapularis. Parasit. Vectors 2016, 9, 467. [Google Scholar] [CrossRef] [Green Version]
- Dahmani, M.; Anderson, J.F.; Sultana, H.; Neelakanta, G. Rickettsial pathogen uses arthropod tryptophan pathway metabolites to evade reactive oxygen species in tick cells. Cell Microbiol. 2020, 22, e13237. [Google Scholar] [CrossRef]
- Ramasamy, E.; Taank, V.; Anderson, J.F.; Sultana, H.; Neelakanta, G. Repression of tick microRNA-133 induces organic anion transporting polypeptide expression critical for Anaplasma phagocytophilum survival in the vector and transmission to the vertebrate host. PLoS Genet. 2020, 16, e1008856. [Google Scholar] [CrossRef]
- Sultana, H.; Neelakanta, G.; Kantor, F.S.; Malawista, S.E.; Fish, D.; Montgomery, R.R.; Fikrig, E. Anaplasma phagocytophilum induces actin phosphorylation to selectively regulate gene transcription in Ixodes scapularis ticks. J. Exp. Med. 2010, 207, 1727–1743. [Google Scholar] [CrossRef] [Green Version]
- Taank, V.; Dutta, S.; Dasgupta, A.; Steeves, T.K.; Fish, D.; Anderson, J.F.; Sultana, H.; Neelakanta, G. Human rickettsial pathogen modulates arthropod organic anion transporting polypeptide and tryptophan pathway for its survival in ticks. Sci. Rep. 2017, 7, 13256. [Google Scholar] [CrossRef]
- Turck, J.W.; Taank, V.; Neelakanta, G.; Sultana, H. Ixodes scapularis Src tyrosine kinase facilitates Anaplasma phagocytophilum survival in its arthropod vector. Ticks Tick Borne Dis. 2019, 10, 838–847. [Google Scholar] [CrossRef]
- Cabezas-Cruz, A.; Espinosa, P.; Alberdi, P.; de la Fuente, J. Tick-Pathogen Interactions: The Metabolic Perspective. Trends Parasitol. 2019, 35, 316–328. [Google Scholar] [CrossRef]
- Cabezas-Cruz, A.; Estrada-Peña, A.; Rego, R.O.M.; De la Fuente, J. Tick-Pathogen Ensembles: Do Molecular Interactions Lead Ecological Innovation? Front Cell Infect Microbiol. 2017, 7, 74. [Google Scholar] [CrossRef] [Green Version]
- de la Fuente, J.; Peña, A.E.; Cabezas-Cruz, A.; Kocan, K.M. Anaplasma phagocytophilum Uses Common Strategies for Infection of Ticks and Vertebrate Hosts. Trends Microbiol. 2016, 24, 173–180. [Google Scholar] [CrossRef]
- Chávez, A.S.O.; Fairman, J.W.; Felsheim, R.F.; Nelson, C.M.; Herron, M.J.; Higgins, L.; Burkhardt, N.Y.; Oliver, J.D.; Markowski, T.W.; Kurtti, T.J.; et al. An O-Methyltransferase Is Required for Infection of Tick Cells by Anaplasma phagocytophilum. PLoS Pathog. 2015, 11, e1005248. [Google Scholar] [CrossRef] [Green Version]
- Carroll, E.E.M.; Wang, X.; Shaw, D.K.; O’Neal, A.J.; Chávez, A.S.O.; Brown, L.J.; Boradia, V.M.; Hammond, H.L.; Pedra, J.H.F. p47 licenses activation of the immune deficiency pathway in the tick Ixodes scapularis. Proc. Natl. Acad. Sci. USA 2019, 116, 205–210. [Google Scholar] [CrossRef] [Green Version]
- Burtis, J.C.; Yavitt, J.B.; Fahey, T.J.; Ostfeld, R. Ticks as Soil-Dwelling Arthropods: An Intersection between Disease and Soil Ecology. J. Med. Entomol. 2019, 56, 1555–1564. [Google Scholar] [CrossRef]
- De La Fuente, J.; Villar, M.; Cabezas-Cruz, A.; Peña, A.E.; Ayllón, N.; Alberdi, P. Tick-Host-Pathogen Interactions: Conflict and Cooperation. PLoS Pathog. 2016, 12, e1005488. [Google Scholar] [CrossRef]
- Edgar, R.S.; Green, E.W.; Zhao, Y.; van Ooijen, G.; Olmedo, M.; Qin, X.; Xu, Y.; Pan, M.; Valekunja, U.K.; Feeney, K.A.; et al. Peroxiredoxins are conserved markers of circadian rhythms. Nature 2012, 485, 459–464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Curtis, A.M.; Bellet, M.M.; Sassone-Corsi, P.; O’Neill, L.A. Circadian clock proteins and immunity. Immunity 2014, 40, 178–186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meireles-Filho, A.; Kyriacou, C.P. Circadian rhythms in insect disease vectors. Mem. Inst. Oswaldo Cruz 2013, 108 (Suppl. 1), 48–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mendoza-Viveros, L.; Bouchard-Cannon, P.; Hegazi, S.; Cheng, A.H.; Pastore, S.; Cheng, H.-Y.M. Molecular modulators of the circadian clock: Lessons from flies and mice. Cell Mol. Life Sci. 2017, 74, 1035–1059. [Google Scholar] [CrossRef] [PubMed]
- Ozkaya, O.; Rosato, E. The circadian clock of the fly: A neurogenetics journey through time. Adv. Genet. 2012, 77, 79–123. [Google Scholar] [CrossRef]
- Helfrich-Forster, C. Neurobiology of the fruit fly’s circadian clock. Genes Brain Behav. 2005, 4, 65–76. [Google Scholar] [CrossRef]
- Reppert, S.M.; Weaver, D.R. Coordination of circadian timing in mammals. Nature 2002, 418, 935–941. [Google Scholar] [CrossRef] [PubMed]
- Sehgal, A.; Price, J.L.; Man, B.; Young, M.W. Loss of circadian behavioral rhythms and per RNA oscillations in the Drosophila mutant timeless. Science 1994, 263, 1603–1606. [Google Scholar] [CrossRef]
- Hardin, P.E. Molecular genetic analysis of circadian timekeeping in Drosophila. Adv. Genet. 2011, 74, 141–173. [Google Scholar] [CrossRef] [Green Version]
- Patke, A.; Young, M.W.; Axelrod, S. Molecular mechanisms and physiological importance of circadian rhythms. Nat. Rev. Mol. Cell Biol. 2020, 21, 67–84. [Google Scholar] [CrossRef] [PubMed]
- Ceriani, M.F.; Darlington, T.K.; Staknis, D.; Más, P.; Petti, A.A.; Weitz, C.J.; Kay, S.A. Light-dependent sequestration of TIMELESS by CRYPTOCHROME. Science 1999, 285, 553–556. [Google Scholar] [CrossRef] [PubMed]
- Price, J.L.; Blau, J.; Rothenfluh, A.; Abodeely, M.; Kloss, B.; Young, M.W. double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell 1998, 94, 83–95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rund, S.S.C.; O’Donnell, A.J.; Gentile, J.E.; Reece, S.E. Daily Rhythms in Mosquitoes and Their Consequences for Malaria Transmission. Insects 2016, 7, 14. [Google Scholar] [CrossRef] [Green Version]
- Lima-Camara, T.N.; Bruno, R.V.; Luz, P.; Castro, M.G.; Lourenço-De-Oliveira, R.; Sorgine, M.H.F.; Peixoto, A.A. Dengue infection increases the locomotor activity of Aedes aegypti females. PLoS ONE 2011, 6, e17690. [Google Scholar] [CrossRef] [Green Version]
- Rund, S.S.; Gentile, J.E.; Duffield, G.E. Extensive circadian and light regulation of the transcriptome in the malaria mosquito Anopheles gambiae. BMC Genom. 2013, 14, 218. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.-Y.; Liu, Y.; Teng, H.-J.; Sauman, I.; Sehnal, F.; Lee, H.-J. Circadian control of permethrin-resistance in the mosquito Aedes aegypti. J. Insect. Physiol. 2010, 56, 1219–1223. [Google Scholar] [CrossRef]
- Nagel, D.H.; Kay, S.A. Complexity in the wiring and regulation of plant circadian networks. Curr. Biol. 2012, 22, R648–R657. [Google Scholar] [CrossRef] [Green Version]
- Tomioka, K.; Uryu, O.; Kamae, Y.; Umezaki, Y.; Yoshii, T. Peripheral circadian rhythms and their regulatory mechanism in insects and some other arthropods: A review. J. Comp. Physiol. B 2012, 182, 729–740. [Google Scholar] [CrossRef]
- Ponting, C.P.; Aravind, L. PAS: A multifunctional domain family comes to light. Curr. Biol. 1997, 7, R674–R677. [Google Scholar] [CrossRef] [Green Version]
- Mitchell, R.D.; Zhu, J.; Carr, A.L.; Dhammi, A.; Cave, G.; Sonenshine, D.E.; Roe, R.M. Infrared light detection by the haller’s organ of adult american dog ticks, Dermacentor variabilis (Ixodida: Ixodidae). Ticks Tick Borne Dis. 2017, 8, 764–771. [Google Scholar] [CrossRef] [PubMed]
- Taank, V.; Ramasamy, E.; Sultana, H.; Neelakanta, G. An efficient microinjection method to generate human anaplasmosis agent Anaplasma phagocytophilum-infected ticks. Sci. Rep. 2020, 10, 15994. [Google Scholar] [CrossRef] [PubMed]
- Hergenhan, S.; Holtkamp, S.; Scheiermann, C. Molecular Interactions between Components of the Circadian Clock and the Immune System. J. Mol. Biol. 2020, 432, 3700–3713. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.A.; Pal, U. Immunity-related genes in Ixodes scapularis--perspectives from genome information. Front Cell Infect. Microbiol. 2014, 4, 116. [Google Scholar] [CrossRef] [Green Version]
- Neelakanta, G.; Li, X.; Pal, U.; Liu, X.; Beck, D.S.; DePonte, K.; Fish, D.; Kantor, F.S.; Fikrig, E. Outer surface protein B is critical for Borrelia burgdorferi adherence and survival within Ixodes ticks. PLoS Pathogens. 2007, 3, e33. [Google Scholar] [CrossRef]
- Baker, C.L.; Loros, J.J.; Dunlap, J.C. The circadian clock of Neurospora crassa. FEMS Microbiol. Rev. 2012, 36, 95–110. [Google Scholar] [CrossRef] [Green Version]
- Dong, G.; Kim, Y.-I.; Golden, S.S. Simplicity and complexity in the cyanobacterial circadian clock mechanism. Curr. Opin. Genet. Dev. 2010, 20, 619–625. [Google Scholar] [CrossRef] [Green Version]
- Gulia-Nuss, M.; Nuss, A.; Meyer, J.M.; Sonenshine, D.E.; Roe, R.M.; Waterhouse, R.M.; Sattelle, D.B.; De La Fuente, J.; Ribeiro, J.; Megy, K.; et al. Genomic insights into the Ixodes scapularis tick vector of Lyme disease. Nat. Commun. 2016, 7, 10507. [Google Scholar] [CrossRef] [Green Version]
- Kaltenrieder, M.; Labhart, T.; Hess, E. Spectral sensitivity, absolute threshold, and visual field of two tick species, Hyalomma dromedarii and Amblyomma variegatum. J. Comp. Physiol. A 1989, 165, 155–164. [Google Scholar] [CrossRef]
- Leuterer, G.; Gothe, R. On the reaction of adult Rhipicephalus evertsi mimeticus and Hyalomma truncatum to horizontally incidenting optical radiation of various wavelengths ranges and different irradiances and to optical radiation of a sun-simulating wavelength spectrum. Parasitol. Res. 1991, 77, 353–358. [Google Scholar] [CrossRef]
- Perret, J.-L.; Guerin, P.M.; Diehl, P.A.; Vlimant, M.; Gern, L. Darkness induces mobility, and saturation deficit limits questing duration, in the tick Ixodes ricinus. J. Exp. Biol. 2003, 206, 1809–1815. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dubie, T.R.; Turner, J.; Noden, B.H. Questing Behavior and Analysis of Tick-Borne Bacteria in Ixodes scapularis (Acari: Ixodidae) in Oklahoma. J. Med. Entomol. 2018, 55, 1569–1574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Madden, S.C.; Madden, R.C. Seasonality in diurnal locomotory patterns of adult blacklegged ticks (Acari: Ixodidae). J. Med. Entomol. 2005, 42, 582–588. [Google Scholar] [CrossRef] [PubMed]
- Carroll, J.F.; Mills, G.D., Jr.; Schmidtmann, E.T. Patterns of activity in host-seeking adult Ixodes scapularis (Acari: Ixodidae) and host-produced kairomones. J. Med. Entomol. 1998, 35, 11–15. [Google Scholar] [CrossRef] [PubMed]
- Durden, L.A.; Vogel, G.N.; Oliver, J.H., Jr. Nocturnal questing by adult blacklegged ticks, Ixodes scapularis (Acari: Ixodidae). J. Parasitol. 1996, 82, 174–175. [Google Scholar] [CrossRef]
- Das, S.; Dimopoulos, G. Molecular analysis of photic inhibition of blood-feeding in Anopheles gambiae. BMC Physiol. 2008, 8, 23. [Google Scholar] [CrossRef] [Green Version]
- Baxter, R.H.; Contet, A.; Krueger, K. Arthropod Innate Immune Systems and Vector-Borne Diseases. Biochemistry 2017, 56, 907–918. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Dai, J.; Zhao, Y.O.; Narasimhan, S.; Yang, Y.; Zhang, L.; Fikrig, E. Ixodes scapularis JAK-STAT pathway regulates tick antimicrobial peptides, thereby controlling the agent of human granulocytic anaplasmosis. J. Infect. Dis. 2012, 206, 1233–1241. [Google Scholar] [CrossRef]
- Vimonish, R.; Johnson, W.C.; Mousel, M.R.; Brayton, K.A.; Scoles, G.A.; Noh, S.M.; Ueti, M.W. Quantitative analysis of Anaplasma marginale acquisition and transmission by Dermacentor andersoni fed in vitro. Sci. Rep. 2020, 10, 470. [Google Scholar] [CrossRef] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khanal, S.; Taank, V.; Anderson, J.F.; Sultana, H.; Neelakanta, G. Rickettsial Pathogen Perturbs Tick Circadian Gene to Infect the Vertebrate Host. Int. J. Mol. Sci. 2022, 23, 3545. https://doi.org/10.3390/ijms23073545
Khanal S, Taank V, Anderson JF, Sultana H, Neelakanta G. Rickettsial Pathogen Perturbs Tick Circadian Gene to Infect the Vertebrate Host. International Journal of Molecular Sciences. 2022; 23(7):3545. https://doi.org/10.3390/ijms23073545
Chicago/Turabian StyleKhanal, Supreet, Vikas Taank, John F. Anderson, Hameeda Sultana, and Girish Neelakanta. 2022. "Rickettsial Pathogen Perturbs Tick Circadian Gene to Infect the Vertebrate Host" International Journal of Molecular Sciences 23, no. 7: 3545. https://doi.org/10.3390/ijms23073545
APA StyleKhanal, S., Taank, V., Anderson, J. F., Sultana, H., & Neelakanta, G. (2022). Rickettsial Pathogen Perturbs Tick Circadian Gene to Infect the Vertebrate Host. International Journal of Molecular Sciences, 23(7), 3545. https://doi.org/10.3390/ijms23073545