TKTL1 Knockdown Impairs Hypoxia-Induced Glucose-6-phosphate Dehydrogenase and Glyceraldehyde-3-phosphate Dehydrogenase Overexpression
Abstract
:1. Introduction
2. Results
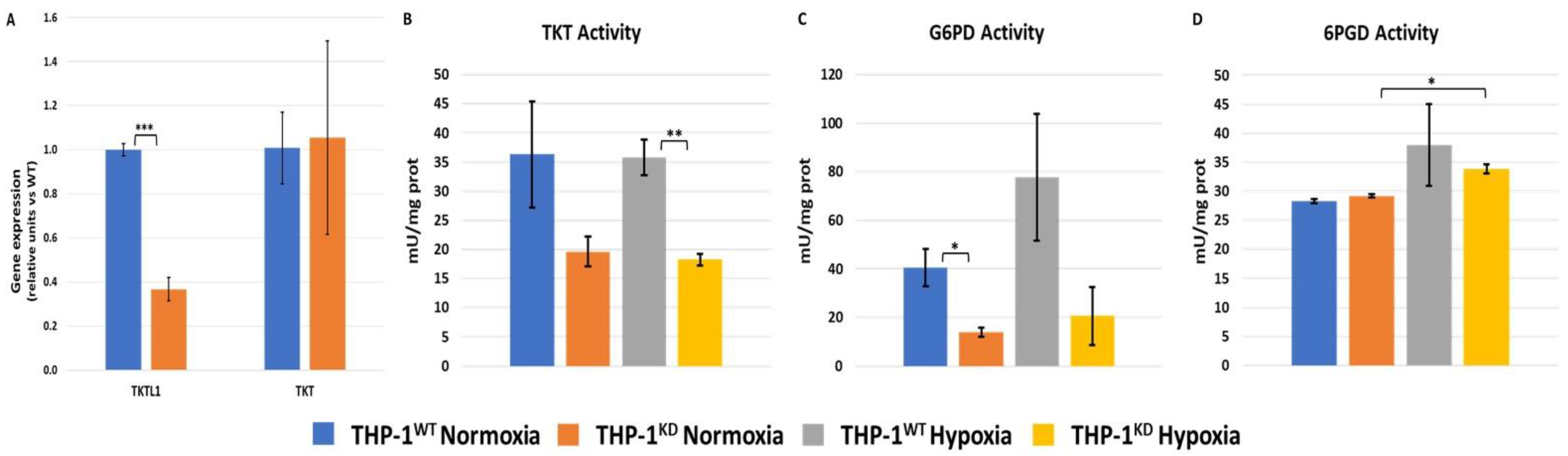
2.1. TKTL1 Contributes to Cell Growth and Pentose Phosphate Pathway in Normoxia and Hypoxia
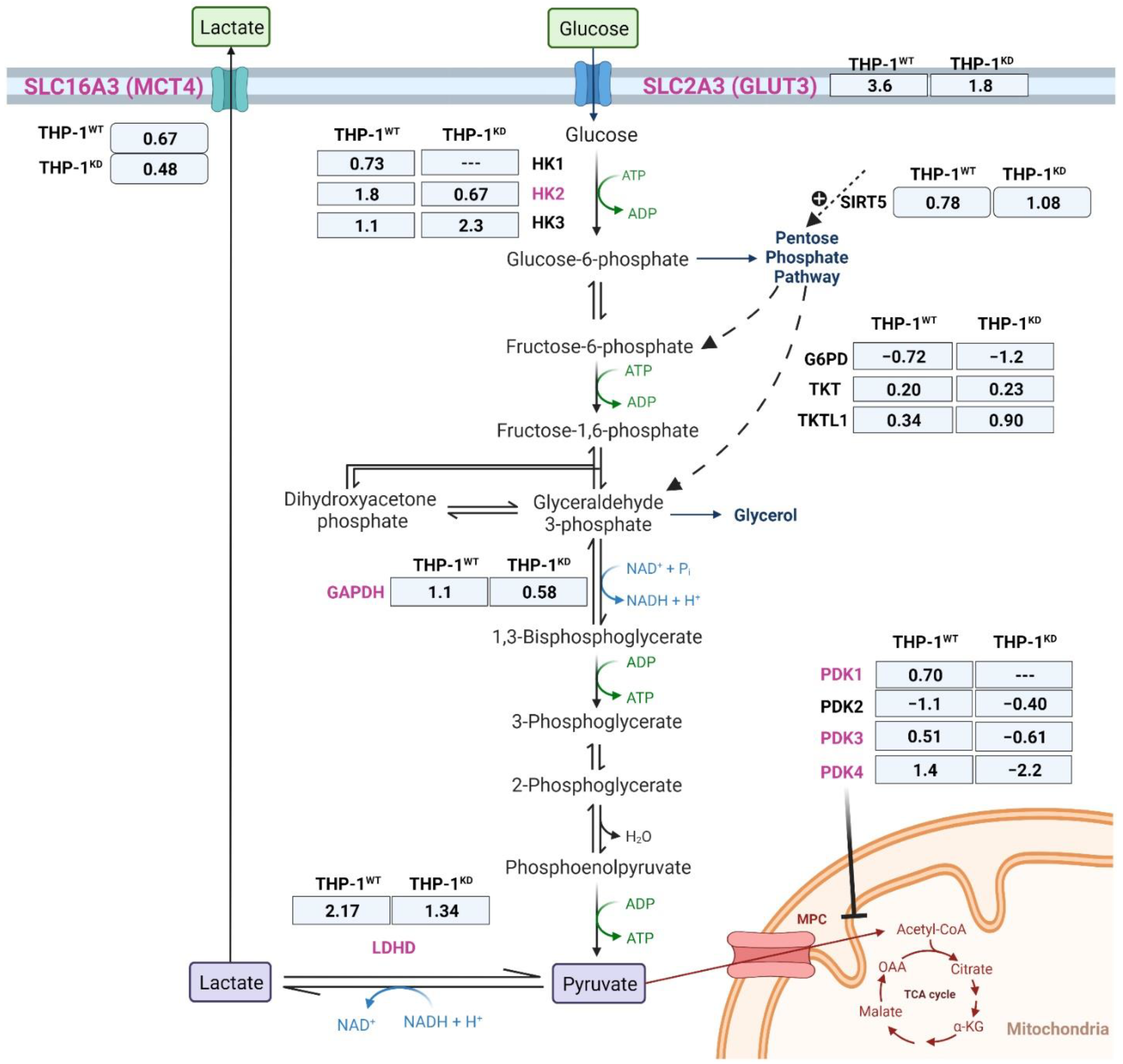
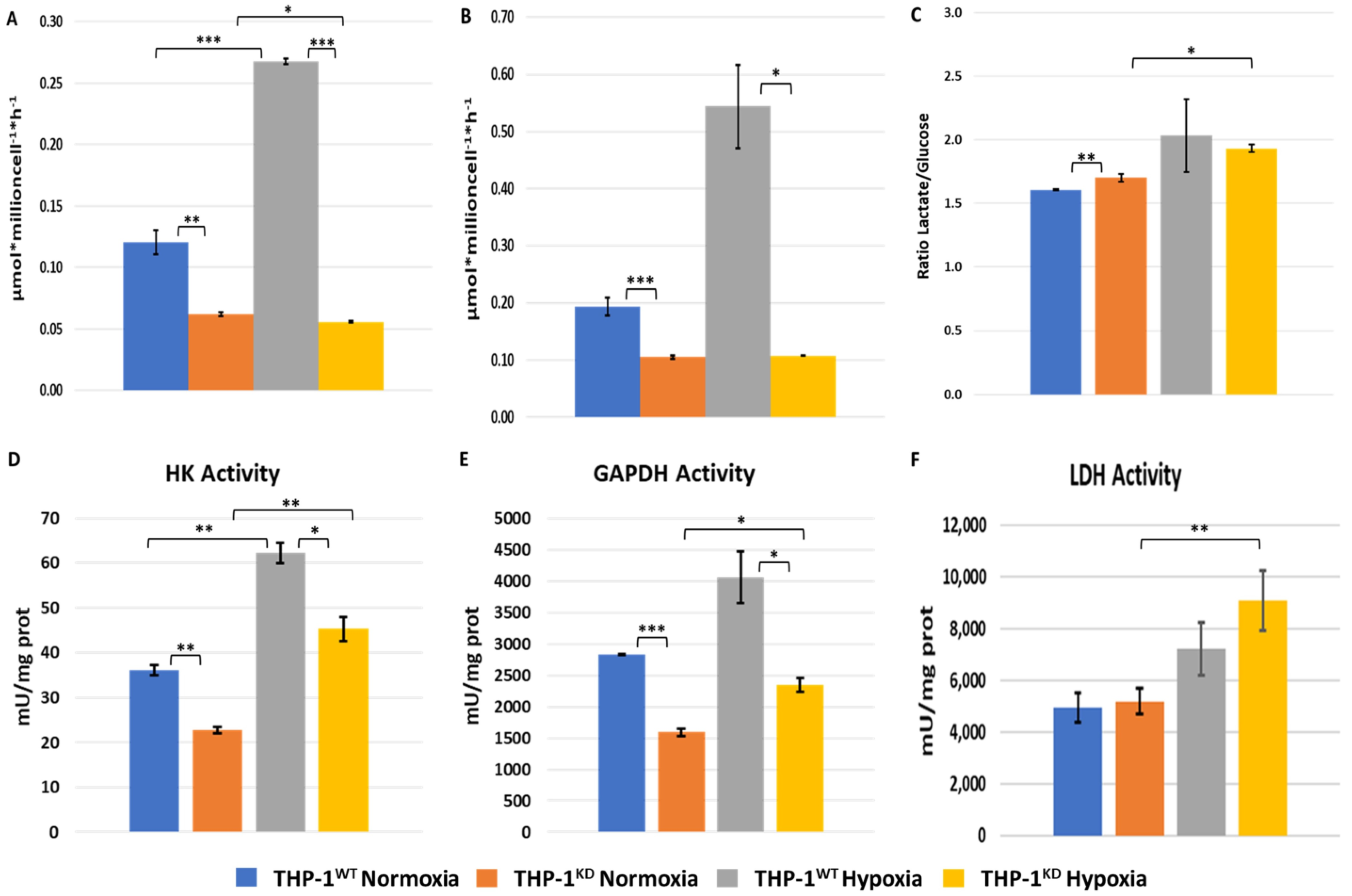
2.2. TKTL1 Is Essential to Trigger Hypoxia-Induced Changes for the Fate of Glucose in THP-1 Cells
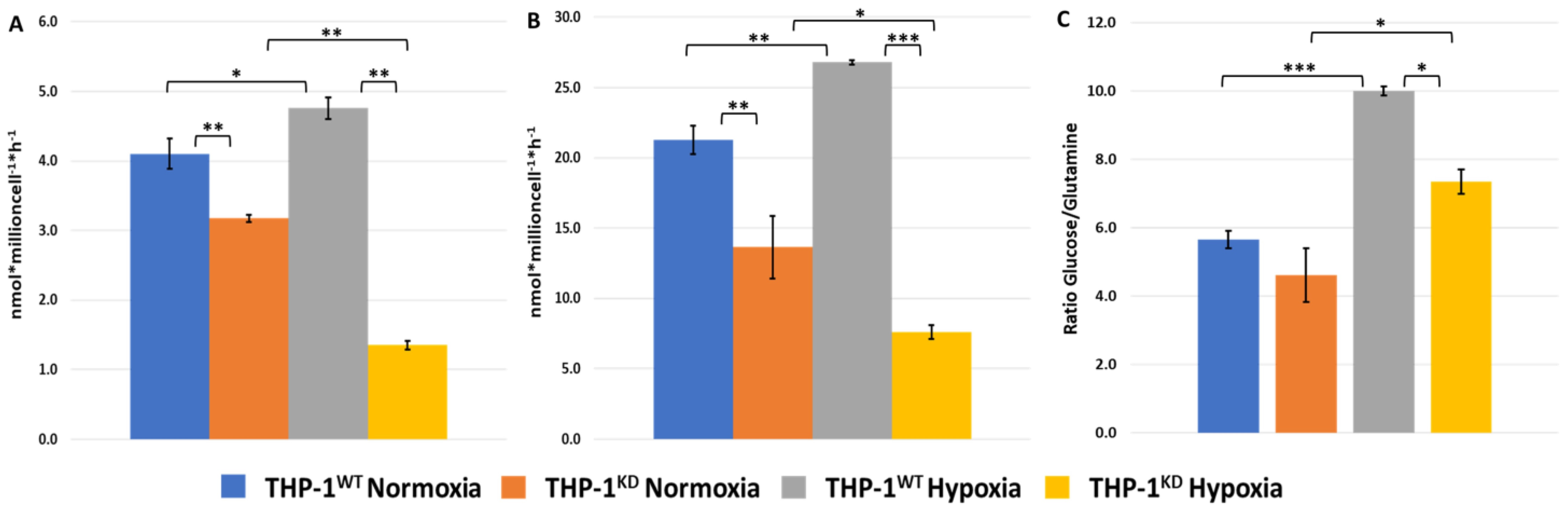
2.3. TKTL1 Is Essential to Trigger Hypoxia-Induced Changes to the Fate of Glutamine in THP-1 Cells
2.4. TKTL1 Is Essential to Trigger Hypoxia-Induced Changes in the Fate of Amino Acids in THP-1 Cells
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Cell Viability Assays
4.3. Measurement of Enzyme Activities, Concentration of Metabolites in Incubation Medium, and Intracellular Concentrations of Glucose, Lactate, Amino Acids, and Biogenic Amines
4.4. Real-Time PCR
4.5. Transcriptomics
4.5.1. RNA Extraction
4.5.2. RNA-Seq Library Preparation, Sequencing and Generation of FastQ Files
4.5.3. Analysis of the Raw RNA Sequencing Data
4.5.4. Differential Expression Analysis
4.6. Statistical Analysis
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| Gene Symbol | Description | Log2 Fold Change | lfcSE 1 | p-Value | p-adj. 2 |
|---|---|---|---|---|---|
| G6PD | glucose-6-phosphate dehydrogenase | −0.72 | 0.09 | 1.45 × 10−15 | 1.41 × 10−14 |
| SIRT5 | sirtuin 5 | 0.78 | 1.10 | 7.01 × 10−13 | 5.32 × 10−12 |
| TKTL1 | transketolase-like 1 | 0.34 | 0.11 | 2.4 × 10−3 | 5.8 × 10−3 |
| TKT | transketolase | 0.20 | 0.07 | 2.7 × 10−3 | 6.5 × 10−3 |
| HK1 | hexokinase 1 | 0.73 | 0.12 | 1.49 × 10−9 | 8.00 × 10−9 |
| HK2 | hexokinase 2 | 1.82 | 0.17 | 2.71 × 10−27 | 6.56 × 10−26 |
| HK3 | hexokinase 3 | 1.09 | 0.17 | 2.27 × 10−10 | 1.32 × 10−9 |
| PDK1 | pyruvate dehydrogenase kinase 1 | 0.70 | 0.21 | 7.6 × 10−4 | 2.0 × 10−3 |
| PDK3 | pyruvate dehydrogenase kinase 3 | 0.51 | 0.21 | 1.6 × 10−2 | 3.1 × 10−2 |
| PDK4 | pyruvate dehydrogenase kinase 4 | 1.42 | 0.49 | 4.0 × 10−3 | 9.1 × 10−3 |
| GAPDH | glyceraldehyde-3-phosphate dehydrogenase | 1.05 | 0.11 | 9.71 × 10−22 | 1.59 × 10−20 |
| LDHD | lactate dehydrogenase D | 2.18 | 0.25 | 7.55 × 10−18 | 8.89 × 10−17 |
| SLC2A3 | solute carrier family 2 member 3 | 0.67 | 0.32 | 3.11 × 10−8 | 1.44 × 10−7 |
| SLC16A3 | solute carrier family 16 member 3 | 3.60 | 0.12 | 6.36 × 10−30 | 1.81 × 10−28 |
| ESRRB | estrogen related receptor beta | 4.64 | 1.4 | 1.2 × 10−3 | 3.0 × 10−3 |
| GLUL | glutamate-ammonia ligase | 1.38 | 0.12 | 2.16 × 10−30 | 6.37 × 10−29 |
| SLC1A3 | solute carrier family 1 member 3 | 0.80 | 0.29 | 5.9 × 10−3 | 1.3 × 10−2 |
| SLC17A7 | solute carrier family 17 member 7 | 3.31 | 0.61 | 5.31 × 10−8 | 2.40 × 10−7 |
| SLC38A2 | solute carrier family 38 member 2 | 0.28 | 0.10 | 2.6 × 10−2 | 4.9 × 10−2 |
| IDO2 | indoleamine 2,3-dioxygenase 2 | 4.55 | 1.75 | 9.4 × 10−3 | 2.0 × 10−2 |
| PHGDH | phosphoglycerate dehydrogenase | 0.77 | 0.09 | 3.11 × 10−19 | 4.15 × 10−18 |
| SHMT2 | serine hydroxymethyltransferase 2 | 0.75 | 0.04 | 7.34 × 10−65 | 1.46 × 10−6 |
| PCK1 | phosphoenolpyruvate carboxykinase 1 | 5.03 | 1.51 | 8.8 × 10−4 | 2.3 × 10−3 |
| TPH2 | tryptophan hydroxylase 2 | -- | -- | -- | -- |
| BCAT1 | Branched chain amino acid transaminase 1 | −1.56 | 0.14 | 9.14 × 10−31 | 2.74 × 10−29 |
| BCKDH | Branched chain keto acid dehydrogenase E1 subunit beta | −0.38 | 0.10 | 2.3 × 10−4 | 6.7 × 10−4 |
| SLC7A7 | solute carrier family 7 member 7 | 2.28 | 0.16 | 6.47 × 10−46 | 4.86 × 10−44 |
| SLC7A8 | solute carrier family 7 member 8 | 3.21 | 0.85 | 0.00015 | 0.00044 |
| Gene Symbol | Description | Log2 Fold Change | lfcSE 1 | p-Value | p-adj. 2 |
|---|---|---|---|---|---|
| G6PD | glucose-6-phosphate dehydrogenase | −1.24 | 0.10 | 3.15 × 10−37 | 1.51 × 10−35 |
| SIRT5 | sirtuin 5 | 1.08 | 0.12 | 2.92 × 10−19 | 3.50 × 10−18 |
| TKTL1 | transketolase-like 1 | 0.90 | 0.12 | 2.14 × 10−14 | 1.66 × 10−13 |
| TKT | transketolase | 0.24 | 0.07 | 5.9 × 10−4 | 1.5 × 10−3 |
| HK2 | hexokinase 2 | 0.67 | 0.17 | 7.82 × 10−5 | 2.2 × 10−4 |
| HK3 | hexokinase 3 | 2.73 | 0.17 | 7.21 × 10−57 | 1.17 × 10−54 |
| PDK3 | pyruvate dehydrogenase kinase 3 | −0.61 | 0.21 | 3.9 × 10−3 | 8.9 × 10−3 |
| PDK4 | pyruvate dehydrogenase kinase 4 | −2.20 | 0.53 | 3.53 × 10−5 | 1.1 × 10−4 |
| GAPDH | glyceraldehyde-3-phosphate dehydrogenase | 0.57 | 0.11 | 1.86 × 10−7 | 7.30 × 10−7 |
| LDHD | lactate dehydrogenase D | 1.34 | 0.14 | 1.53 × 10−20 | 2.03 × 10−19 |
| SLC2A3 | solute carrier family 2 member 3 | 1.81 | 0.32 | 2.11 × 10−8 | 9.12 × 10−8 |
| SLC16A3 | solute carrier family 16 member 3 | 0.48 | 0.12 | 7.73 × 10−5 | 2.2 × 10−4 |
| ESRRB | estrogen related receptor beta | -- | -- | -- | -- |
| GLUL | glutamate-ammonia ligase | 0.60 | 0.12 | 2.07 × 10−7 | 8.09 × 10−7 |
| GLS | glutaminase | −0.88 | 0.13 | 1.67 × 10−11 | 9.95 × 10−11 |
| SLC1A3 | solute carrier family 1 member 3 | −0.89 | 0.32 | 5.4 × 10−3 | 1.1 × 10−2 |
| SLC38A2 | solute carrier family 38 member 2 | −0.38 | 0.10 | 2.8 × 10−4 | 7.1 × 10−4 |
| IDO2 | indoleamine 2,3-dioxygenase 2 | -- | -- | -- | -- |
| PHGDH | phosphoglycerate dehydrogenase | 0.46 | 0.09 | 1.10 × 10−7 | 4.42 × 10−7 |
| SHMT2 | serine hydroxymethyltransferase 2 | 0.37 | 0.05 | 1.81 × 10−15 | 1.53 × 10−14 |
| TPH2 | tryptophan hydroxylase 2 | 6.93 | 1.40 | 6.88 × 10−7 | 2.53 × 10−6 |
| BCAT1 | branched chain amino acid transaminase 1 | −2.41 | 0.14 | 9.87 × 10−66 | 2.61 × 10−63 |
Appendix B
| Metabolite | THP-1WT Normoxia | THP-1KD Normoxia | THP-1WT Hypoxia | THP-1KD Hypoxia | p-Value THP-1WT (Nor vs. Hyp) | p-Value THP-1KD (Nor vs. Hyp) |
|---|---|---|---|---|---|---|
| Alanine | 402.30 ± 143.45 | 473.60 ± 144.63 | 346.52 ± 5.56 | 279.01 ± 79.01 | 0.5378 | 0.1103 |
| Arginine | 273.40 ± 77.02 | 272.04 ± 150.85 | 292.70 ± 144.44 | 111.34 ± 86.22 | 0.8482 | 0.1844 |
| Asparagine | 771.52 ± 228.94 | 563.26 ± 54.48 | 474.11 ± 39.58 | 702.79 ± 395.83 | 0.0909 | 0.5779 |
| Aspartate | 118.19 ± 37.52 | 136.50 ± 46.37 | 36.98 ± 10.07 | 55.27 ± 44.06 | 0.0223 | 0.0927 |
| Citrulline | 0.24 ± 0.23 | 3.20 ± 4.66 | 2.54 ± 2.93 | 0.52 ± 0.89 | 0.2464 | 0.3824 |
| Glutamine | 1636.46 ± 205.69 | 1760.44 ± 510.50 | 1377.17 ± 295.79 | 1100.21 ± 204.71 | 0.2806 | 0.1061 |
| Glutamate | 855.50 ± 1481.77 | 1089.97 ± 943.94 | 0.00 ± 0.00 | 1609.70 ± 571.89 | 0.3739 | 0.4605 |
| Glycine | 915.60 ± 375.13 | 1075.90 ± 575.77 | 583.59 ± 285.89 | 778.96 ± 222.69 | 0.1685 | 0.4516 |
| Histidine | 72.78 ± 13.73 | 46.93 ± 17.33 | 58.96 ± 8.22 | 36.64 ± 16.40 | 0.2093 | 0.4967 |
| Isoleucine | 185.40 ± 38.58 | 200.03 ± 38.97 | 210.65 ± 76.16 | 153.94 ± 44.02 | 0.6355 | 0.2461 |
| Leucine | 236.18± 55.20 | 248.53 ± 52.65 | 249.76 ± 63.02 | 168.81 ± 66.49 | 0.7928 | 0.1789 |
| Lysine | 40.25 ± 17.59 | 53.42 ± 8.72 | 63.64 ± 23.21 | 38.99 ± 25.69 | 0.2368 | 0.4090 |
| Methionine | 71.61 ± 20.62 | 66.44 ± 25.40 | 63.10 ± 21.25 | 41.63 ± 22.51 | 0.6446 | 0.2741 |
| Ornithine | 22.30 ± 10.41 | 21.32 ± 9.00 | 35.94 ± 6.38 | 29.14 ± 16.43 | 0.1253 | 0.5096 |
| Phenylalanine | 42.47 ± 7.37 | 39.58 ± 11.15 | 59.42 ± 22.21 | 34.28 ± 9.91 | 0.2781 | 0.5709 |
| Proline | 705.55 ± 139.47 | 743.98 ± 116.06 | 249.71 ± 37.70 | 191.38 ± 38.52 | 0.0055 | 0.0014 |
| Serine | 70.15 ± 23.94 | 129.16 ± 90.81 | 353.88 ± 116.37 | 186.89 ± 34.27 | 0.0144 | 0.3612 |
| Threonine | 173.83 ± 92.88 | 142.23 ± 28.16 | 200.71 ± 89.78 | 111.91 ± 40.27 | 0.7368 | 0.3455 |
| Tryptophan | 11.02 ± 3.72 | 12.83 ± 1.75 | 16.12 ± 6.45 | 9.69 ± 1.04 | 0.3014 | 0.0555 |
| Tyrosine | 76.14 ± 26.81 | 91.19 ± 14.14 | 89.76 ± 32.79 | 82.07 ± 19.26 | 0.6074 | 0.5448 |
| Valine | 54.40 ± 19.08 | 40.16 ± 5.70 | 60.33 ± 7.25 | 51.91 ± 20.82 | 0.6410 | 0.3992 |
| Acetylornithine | 2.44 ± 2.34 | 1.18 ± 1.91 | 3.27 ± 1.82 | 2.57 ± 3.02 | 0.6537 | 0.5368 |
| ADMA | 2.17 ± 1.48 | 1.57 ± 0.75 | 1.31 ± 0.38 | 0.74 ± 0.69 | 0.3849 | 0.2322 |
| α-AAA | 7.09 ± 2.44 | 6.27 ± 2.80 | 5.35 ± 5.40 | 4.62 ± 0.51 | 0.6387 | 0.3711 |
| Carnosine | 0.84 ± 0.55 | 0.51 ± 0.19 | 0.38 ± 0.17 | 0.21 ± 0.09 | 0.2395 | 0.0732 |
| Creatinine | 16.70 ± 6.86 | 15.20 ± 2.31 | 10.38 ± 2.29 | 11.00 ± 1.91 | 0.2047 | 0.0722 |
| Histamine | 0.32 ± 0.29 | 0.23 ± 0.40 | 0.00 ± 0.00 | 0.25 ± 0.44 | 0.1252 | 0.9480 |
| Kynurenine | 0.38 ± 0.17 | 0.43 ± 0.09 | 0.39 ± 0.17 | 0.45 ± 0.16 | 0.9685 | 0.9024 |
| Met-SO | 1.88 ± 0.58 | 1.98 ± 0.95 | 2.81 ± 1.47 | 2.79 ± 1.40 | 0.3630 | 0.4562 |
| Putrescine | 2.96 ± 1.32 | 3.59 ± 1.84 | 3.00 ± 0.89 | 3.22 ± 0.51 | 0.9657 | 0.7542 |
| Spermidine | 45.35 ± 16.23 | 36.51 ± 5.33 | 23.81 ± 9.00 | 26.50 ± 7.43 | 0.1148 | 0.1306 |
| Spermine | 20.86 ± 11.70 | 14.61 ± 2.37 | 13.77 ± 7.30 | 9.65 ± 4.51 | 0.4232 | 0.1667 |
| T4-OH-Pro | 221.31 ± 22.58 | 202.10 ± 40.27 | 130.55 ± 4.57 | 130.59 ± 58.46 | 0.0024 | 0.1559 |
| Taurine | 469.08 ± 82.46 | 464.12 ± 361.99 | 196.01 ± 188.34 | 219.15 ± 200.75 | 0.0829 | 0.3633 |
| Total DMA | 0.12 ± 0.12 | 0.05 ± 0.05 | 0.12 ± 0.12 | 0.20 ± 0.16 | 0.9458 | 0.2015 |
References
- Comín-Anduix, B.; Boren, J.; Martinez, S.; Moro, C.; Centelles, J.J.; Trebukhina, R.; Petushok, N.; Lee, W.N.P.; Boros, L.G.; Cascante, M. The effect of thiamine supplementation on tumour roliferation: A metabolic control analysis study. Eur. J. Biochem. 2001, 268, 4177–4182. [Google Scholar] [CrossRef] [PubMed]
- Berthon, H.A.; Kuchel, P.W.; Nixon, P.F. High Control Coefficient of Transketolase in the Nonoxidative Pentose Phosphate Pathway of Human Erythrocytes: NMR, Antibody, and Computer Simulation Studies. Biochemistry 1992, 31, 12792–12798. [Google Scholar] [CrossRef] [PubMed]
- Deshpande, G.P.; Patterton, H.-G.; Faadiel Essop, M. The human transketolase-like proteins TKTL1 and TKTL2 are bona fide transketolases. BMC Struct. Biol. 2019, 19, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fritz, P.; Coy, J.F.; Mürdter, T.E.; Ott, G.; Alscher, M.D.; Friedel, G. TKTL-1 expression in lung cancer. Pathol. Res. Pract. 2012, 208, 203–209. [Google Scholar] [CrossRef]
- Chen, H.; Yue, J.X.; Yang, S.H.; Ding, H.; Zhao, R.W.; Zhang, S. Overexpression of transketolase-like gene 1 is associated with cell proliferation in uterine cervix cancer. J. Exp. Clin. Cancer Res. 2009, 28, 2–9. [Google Scholar] [CrossRef] [Green Version]
- Da Costa, I.A.; Hennenlotter, J.; Stühler, V.; Kühs, U.; Scharpf, M.; Todenhöfer, T.; Stenzl, A.; Bedke, J. Transketolase like 1 (TKTL1) expression alterations in prostate cancer tumorigenesis. Urol. Oncol. Semin. Orig. Investig. 2018, 36, 472.e21–472.e27. [Google Scholar] [CrossRef]
- Li, J.; Zhu, S.C.; Li, S.G.; Zhao, Y.; Xu, J.R.; Song, C.Y. TKTL1 promotes cell proliferation and metastasis in esophageal squamous cell carcinoma. Biomed. Pharmacother. 2015, 74, 71–76. [Google Scholar] [CrossRef]
- Diaz-Moralli, S.; Aguilar, E.; Marin, S.; Coy, J.F.; Dewerchin, M.; Antoniewicz, M.R.; Meca-Cortés, O.; Notebaert, L.; Ghesquière, B.; Eelen, G.; et al. A key role for transketolase-like 1 in tumor metabolic reprogramming. Oncotarget 2016, 7, 51875–51897. [Google Scholar] [CrossRef]
- Ahopelto, K.; Saukkonen, K.; Hagström, J.; Kauhanen, S.; Seppänen, H.; Böckelman, C.; Haglund, C. TKTL1 as a Prognostic Marker in Pancreatic Ductal Adenocarcinoma and Its Correlation with FDG-PET-CT. Oncology 2021, 99, 177–185. [Google Scholar] [CrossRef]
- Peltonen, R.; Ahopelto, K.; Hagström, J.; Böckelman, C.; Haglund, C.; Isoniemi, H. High TKTL1 expression as a sign of poor prognosis in colorectal cancer with synchronous rather than metachronous liver metastases. Cancer Biol. Ther. 2020, 21, 826–831. [Google Scholar] [CrossRef]
- Shi, Z.; Tang, Y.; Li, K.; Fan, Q. TKTL1 expression and its downregulation is implicated in cell proliferation inhibition and cell cycle arrest in esophageal squamous cell carcinoma. Tumor Biol. 2015, 36, 8519–8529. [Google Scholar] [CrossRef] [PubMed]
- Philipp, M.; Schwaab, J.; Dietz, C.T.; Hanfstein, B.; Kalmanti, L.; Munjal, U.; Mossner, M.; Nowak, D.; Seifarth, W.; Hofmann, W.K.; et al. Expression of transketolase-like gene 1 (TKTL1) depends on disease phase in patients with chronic myeloid leukaemia (CML). J. Cancer Res. Clin. Oncol. 2014, 140, 411–417. [Google Scholar] [CrossRef] [PubMed]
- Heller, S.; Maurer, G.D.; Wanka, C.; Hofmann, U.; Luger, A.L.; Bruns, I.; Steinbach, J.P.; Rieger, J. Gene suppression of transketolase-like protein 1 (TKTl1) sensitizes glioma cells to hypoxia and ionizing radiation. Int. J. Mol. Sci. 2018, 19, 2168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kämmerer, U.; Gires, O.; Pfetzer, N.; Wiegering, A.; Klement, R.J.; Otto, C. TKTL1 expression in human malign and benign cell lines. BMC Cancer 2015, 15, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lodi, A.; Tiziani, S.; Khanim, F.L.; Drayson, M.T.; Günther, U.L.; Bunce, C.M.; Viant, M.R. Hypoxia triggers major metabolic changes in AML cells without altering indomethacin-induced TCA cycle deregulation. ACS Chem. Biol. 2011, 6, 169–175. [Google Scholar] [CrossRef] [PubMed]
- Klco, J.M.; Spencer, D.H.; Lamprecht, T.L.; Sarkaria, S.M.; Wylie, T.; Magrini, V.; Hundal, J.; Walker, J.; Varghese, N.; Erdmann-Gilmore, P.; et al. Genomic impact of transient low-dose decitabine treatment on primary AMLcells. Blood 2013, 121, 1633–1643. [Google Scholar] [CrossRef] [Green Version]
- Contreras Mostazo, M.G.; Kurrle, N.; Casado, M.; Fuhrmann, D.; Alshamleh, I.; Häupl, B.; Martín-Sanz, P.; Brüne, B.; Serve, H.; Schwalbe, H.; et al. Metabolic plasticity is an essential requirement of acquired tyrosine kinase inhibitor resistance in chronic myeloid leukemia. Cancers 2020, 12, 3443. [Google Scholar] [CrossRef]
- Tsuchiya, S.; Yamabe, M.; Yamaguchi, Y.; Kobayashi, Y.; Konno, T.; Tada, K. Establishment and characterization of a human acute monocytic leukemia cell line (THP-1). Int. J. Cancer 1980, 26, 171–176. [Google Scholar] [CrossRef]
- Zhou, L.; Wang, F.; Sun, R.; Chen, X.; Zhang, M.; Xu, Q.; Wang, Y.; Wang, S.; Xiong, Y.; Guan, K.; et al. SIRT 5 promotes IDH 2 desuccinylation and G6 PD deglutarylation to enhance cellular antioxidant defense. EMBO Rep. 2016, 17, 811–822. [Google Scholar] [CrossRef]
- Draoui, N.; Feron, O. Lactate shuttles at a glance: From physiological paradigms to anti-cancer treatments. Dis. Model. Mech. 2011, 4, 727–732. [Google Scholar] [CrossRef] [Green Version]
- Mobasheri, A.; Richardson, S.; Mobasheri, R.; Shakibaei, M.; Hoyland, J.A. Hypoxia inducible factor-1 and facilitative glucose transporters GLUT1 and GLUT3: Putative molecular components of the oxygen and glucose sensing apparatus in articular chondrocytes. Histol. Histopathol. 2005, 20, 1327–1338. [Google Scholar] [CrossRef] [PubMed]
- Xia, H.; Chen, J.; Gao, H.; Kong, S.N.; Deivasigamani, A.; Shi, M.; Xie, T.; Hui, K.M. Hypoxia-induced modulation of glucose transporter expression impacts 18F-fluorodeoxyglucose PET-CT imaging in hepatocellular carcinoma. Eur. J. Nucl. Med. Mol. Imaging 2020, 47, 787–797. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Kim, E.-J.; Kim, D.-K.; Lee, J.-M.; Park, S.B.; Lee, I.-K.; Harris, R.A.; Lee, M.-O.; Choi, H.-S. Hypoxia Induces PDK4 Gene Expression through Induction of the Orphan Nuclear Receptor ERRγ. PLoS ONE 2012, 7, e46324. [Google Scholar] [CrossRef] [PubMed]
- Su, Z.; Liu, Y.; Zhang, H. Adaptive Cardiac Metabolism Under Chronic Hypoxia: Mechanism and Clinical Implications. Front. Cell Dev. Biol. 2021, 9, 625524. [Google Scholar] [CrossRef]
- Nowak, N.; Kulma, A.; Gutowicz, J. Up-regulation of key glycolysis proteins in cancer development. Open Life Sci. 2018, 13, 569–581. [Google Scholar] [CrossRef]
- Cunningham, K.F.; Beeson, G.C.; Beeson, C.C.; McDermott, P.J. Increased expression of estrogen-related receptor β during adaptation of adult cardiomyocytes to sustained hypoxia. Am. J. Cardiovasc. Dis. 2016, 6, 46–54. [Google Scholar]
- Leclerc, D.; Pham, D.N.T.; Lévesque, N.; Truongcao, M.; Foulkes, W.D.; Sapienza, C.; Rozen, R. Oncogenic role of PDK4 in human colon cancer cells. Br. J. Cancer 2017, 116, 930–936. [Google Scholar] [CrossRef]
- Riddle, S.R.; Ahmad, A.; Ahmad, S.; Deeb, S.S.; Malkki, M.; Schneider, B.K.; Allen, C.B.; White, C.W. Hypoxia induces hexokinase II gene expression in human lung cell line A549. Am. J. Physiol. Cell. Mol. Physiol. 2000, 278, L407–L416. [Google Scholar] [CrossRef] [Green Version]
- Lu, C.W.; Lin, S.C.; Chen, K.F.; Lai, Y.Y.; Tsai, S.J. Induction of pyruvate dehydrogenase kinase-3 by hypoxia-inducible factor-1 promotes metabolic switch and drug resistance. J. Biol. Chem. 2008, 283, 28106–28114. [Google Scholar] [CrossRef] [Green Version]
- Wyatt, E.; Wu, R.; Rabeh, W.; Park, H.-W.; Ghanefar, M.; Ardehali, H. Regulation and Cytoprotective Role of Hexokinase III. PLoS ONE 2010, 5, e13823. [Google Scholar] [CrossRef] [Green Version]
- Wilson, J.E. Isozymes of mammalian hexokinase: Structure, subcellular localization and metabolic function. J. Exp. Biol. 2003, 206, 2049–2057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bommer, G.T.; Van Schaftingen, E.; Veiga-da-Cunha, M. Metabolite Repair Enzymes Control Metabolic Damage in Glycolysis. Trends Biochem. Sci. 2020, 45, 228–243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Ma, K.; Sadana, P.; Chowdhury, F.; Gaillard, S.; Wang, F.; McDonnell, D.P.; Unterman, T.G.; Elam, M.B.; Park, E.A. Estrogen-related receptors stimulate pyruvate dehydrogenase kinase isoform 4 gene expression. J. Biol. Chem. 2006, 281, 39897–39906. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoo, H.C.; Yu, Y.C.; Sung, Y.; Han, J.M. Glutamine reliance in cell metabolism. Exp. Mol. Med. 2020, 52, 1496–1516. [Google Scholar] [CrossRef]
- Jones, M.E. Conversion of Glutamate to Ornithine and Proline: Pyrroline-5-Carboxylate, a Possible Modulator of Arginine Requirements. J. Nutr. 1985, 115, 509–515. [Google Scholar] [CrossRef]
- Simandi, Z.; Pajer, K.; Karolyi, K.; Sieler, T.; Jiang, L.L.; Kolostyak, Z.; Sari, Z.; Fekecs, Z.; Pap, A.; Patsalos, A.; et al. Arginine methyltransferase PRMT8 provides cellular stress tolerance in aging motoneurons. J. Neurosci. 2018, 38, 7683–7700. [Google Scholar] [CrossRef]
- Bhutia, Y.D.; Ganapathy, V. Glutamine transporters in mammalian cells and their functions in physiology and cancer. Biochim. Biophys. Acta-Mol. Cell Res. 2016, 1863, 2531–2539. [Google Scholar] [CrossRef]
- Garcia-Bermudez, J.; Baudrier, L.; La, K.; Zhu, X.G.; Fidelin, J.; Sviderskiy, V.O.; Papagiannakopoulos, T.; Molina, H.; Snuderl, M.; Lewis, C.A.; et al. Aspartate is a limiting metabolite for cancer cell proliferation under hypoxia and in tumours. Nat. Cell Biol. 2018, 20, 775–781. [Google Scholar] [CrossRef]
- Reid, M.A.; Allen, A.E.; Liu, S.; Liberti, M.V.; Liu, P.; Liu, X.; Dai, Z.; Gao, X.; Wang, Q.; Liu, Y.; et al. Serine synthesis through PHGDH coordinates nucleotide levels by maintaining central carbon metabolism. Nat. Commun. 2018, 9, 5442. [Google Scholar] [CrossRef]
- Yamaguchi, N.; Weinberg, E.M.; Nguyen, A.; Liberti, M.V.; Goodarzi, H.; Janjigian, Y.Y.; Paty, P.B.; Saltz, L.B.; Kingham, T.P.; Loo, J.; et al. PCK1 and DHODH drive colorectal cancer liver metastatic colonization and hypoxic growth by promoting nucleotide synthesis. Elife 2019, 8, e52135. [Google Scholar] [CrossRef]
- Arber, D.A.; Orazi, A.; Hasserjian, R.; Thiele, J.; Borowitz, M.J.; Le Beau, M.M.; Bloomfield, C.D.; Cazzola, M.; Vardiman, J.W. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 2016, 127, 2391–2405. [Google Scholar] [CrossRef] [PubMed]
- Bill, M.; Mrózek, K.; Kohlschmidt, J.; Eisfeld, A.K.; Walker, C.J.; Nicolet, D.; Papaioannou, D.; Blachly, J.S.; Orwick, S.; Carroll, A.J.; et al. Mutational landscape and clinical outcome of patients with de novo acute myeloid leukemia and rearrangements involving 11q23/KMT2A. Proc. Natl. Acad. Sci. USA 2020, 117, 26340–26346. [Google Scholar] [CrossRef] [PubMed]
- Adnan-Awad, S.; Kankainen, M.; Mustjoki, S. Mutational landscape of chronic myeloid leukemia: More than a single oncogene leukemia. Leuk. Lymphoma 2021, 62, 2064–2078. [Google Scholar] [CrossRef] [PubMed]
- DiNardo, C.D.; Cortes, J.E. Mutations in AML: Prognostic and therapeutic implications. Hematol. Am. Soc. Hematol. Educ. Progr. 2016, 2016, 348–355. [Google Scholar] [CrossRef] [Green Version]
- Estey, E.; Döhner, H. Acute myeloid leukaemia. Lancet 2006, 368, 1894–1907. [Google Scholar] [CrossRef]
- Lindsley, R.C.; Mar, B.G.; Mazzola, E.; Grauman, P.V.; Shareef, S.; Allen, S.L.; Pigneux, A.; Wetzler, M.; Stuart, R.K.; Erba, H.P.; et al. Acute myeloid leukemia ontogeny is defined by distinct somatic mutations. Blood 2015, 125, 1367–1376. [Google Scholar] [CrossRef] [Green Version]
- Boulais, P.E.; Frenette, P.S. Making sense of hematopoietic stem cell niches. Blood 2015, 125, 2621–2629. [Google Scholar] [CrossRef] [Green Version]
- Morikawa, T.; Takubo, K. Hypoxia regulates the hematopoietic stem cell niche. Pflugers Arch. 2016, 468, 13–22. [Google Scholar] [CrossRef]
- Klco, J.M.; Mullighan, C.G. Advances in germline predisposition to acute leukaemias and myeloid neoplasms. Nat. Rev. Cancer 2021, 21, 122–137. [Google Scholar] [CrossRef]
- Jones, C.L.; Stevens, B.M.; D’Alessandro, A.; Reisz, J.A.; Culp-Hill, R.; Nemkov, T.; Pei, S.; Khan, N.; Adane, B.; Ye, H.; et al. Inhibition of Amino Acid Metabolism Selectively Targets Human Leukemia Stem Cells. Cancer Cell 2018, 34, 724–740.e4. [Google Scholar] [CrossRef] [Green Version]
- Lagunas-Rangel, F.A.; Chávez-Valencia, V.; Gómez-Guijosa, M.Á.; Cortes-Penagos, C. Acute myeloid leukemia—genetic alterations and their clinical prognosis. Int. J. Hematol. Stem Cell Res. 2017, 11, 329–339. [Google Scholar]
- Xu, X.; Zur Hausen, A.; Coy, J.F.; Löchelt, M. Transketolase-like protein 1 (TKTL1) is required for rapid cell growth and full viability of human tumor cells. Int. J. Cancer 2009, 124, 1330–1337. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yao, C.-F.; Xu, F.-J.; Qu, Y.-Y.; Li, J.-T.; Lin, Y.; Cao, Z.-L.; Lin, P.-C.; Xu, W.; Zhao, S.-M.; et al. APC/CCDH1 synchronizes ribose-5-phosphate levels and DNA synthesis to cell cycle progression. Nat. Commun. 2019, 10, 2502. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Zhou, Y. Crucial role of the pentose phosphate pathway in malignant tumors. Oncol. Lett. 2019, 17, 4213–4221. [Google Scholar] [CrossRef] [Green Version]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- The Human Protein Atlas TKTL1 Protein Expression Summary—The Human Protein Atlas. Available online: https://www.proteinatlas.org/ENSG00000007350-TKTL1 (accessed on 24 February 2022).
- Krockenberger, M.; Engel, J.B.; Schmidt, M.; Kohrenhagen, N.; Häusler, S.F.M.; Dombrowski, Y.; Kapp, M.; Dietl, J.; Honig, A. Expression of transketolase-like 1 protein (TKTL1) in human endometrial cancer. Anticancer Res. 2010, 30, 1653–1659. [Google Scholar]
- Hutchinson, D.; Ho, V.; Dodd, M.; Dawson, H.N.; Zumwalt, A.C.; Colton, C.A. TKTL1 is activated by promoter hypomethylation and contributes to head and neck squamous cell carcinoma carcinogenesis via increased aerobic glycolysis and HIF1α stabilization Wenyue. NIH Public Access 2008, 148, 825–832. [Google Scholar] [CrossRef] [Green Version]
- Avhandling, A.; Lindberg, M.L. Acute Myeloid Leukemia—Apoptotic Signalling and Gene Expression Associated with Treatment Response; Karolinska Institutet: Stockholm, Sweden, 2013; ISBN 9789175493459. [Google Scholar]
- Sirover, M.A. GAPDH and Hypoxia. In Glyceraldehyde-3-phosphate Dehydrogenase (GAPDH); Elsevier: Amsterdam, The Netherlands, 2017; pp. 155–165. ISBN 9780128098523. [Google Scholar]
- Yang, H.C.; Wu, Y.H.; Yen, W.C.; Liu, H.Y.; Hwang, T.L.; Stern, A.; Chiu, D.T.Y. The Redox Role of G6PD in Cell Growth, Cell Death, and Cancer. Cells 2019, 8, 1055. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Y.; Cabassa-Hourton, C.; Planchais, S.; Lebreton, S.; Savouré, A. The proline cycle as an eukaryotic redox valve. J. Exp. Bot. 2021, 72, 6856–6866. [Google Scholar] [CrossRef]
- Suzuki, M.; Sasabe, J.; Miyoshi, Y.; Kuwasako, K.; Muto, Y.; Hamase, K.; Matsuoka, M.; Imanishi, N.; Aiso, S. Glycolytic flux controls D-serine synthesis through glyceraldehyde-3-phosphate dehydrogenase in astrocytes. Proc. Natl. Acad. Sci. USA 2015, 112, E2217–E2224. [Google Scholar] [CrossRef] [Green Version]
- de Atauri, P.; Benito, A.; Vizán, P.; Zanuy, M.; Mangues, R.; Marín, S.; Cascante, M. Carbon metabolism and the sign of control coefficients in metabolic adaptations underlying K-ras transformation. Biochim. Biophys. Acta 2011, 1807, 746–754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tarragó-Celada, J.; Foguet, C.; Tarrado-Castellarnau, M.; Marin, S.; Hernández-Alias, X.; Perarnau, J.; Morrish, F.; Hockenbery, D.; Gomis, R.R.; Ruppin, E.; et al. Cysteine and folate metabolism are targetable vulnerabilities of metastatic colorectal cancer. Cancers 2021, 13, 425. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing. Available online: https://www.r-project.org (accessed on 1 July 2021).
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Duplication Time (hours) | % Increment Versus WT | |
|---|---|---|
| THP-1WT Normoxia | 38.4 ± 2.24 | -- |
| THP-1KD Normoxia | 49.4 ± 2.48 ** | 25.1 |
| THP-1WT Hypoxia | 113.5 ± 41.81 | -- |
| THP-1KD Hypoxia | 151.3 ± 19.38 | 28.6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baptista, I.; Karakitsou, E.; Cazier, J.-B.; Günther, U.L.; Marin, S.; Cascante, M. TKTL1 Knockdown Impairs Hypoxia-Induced Glucose-6-phosphate Dehydrogenase and Glyceraldehyde-3-phosphate Dehydrogenase Overexpression. Int. J. Mol. Sci. 2022, 23, 3574. https://doi.org/10.3390/ijms23073574
Baptista I, Karakitsou E, Cazier J-B, Günther UL, Marin S, Cascante M. TKTL1 Knockdown Impairs Hypoxia-Induced Glucose-6-phosphate Dehydrogenase and Glyceraldehyde-3-phosphate Dehydrogenase Overexpression. International Journal of Molecular Sciences. 2022; 23(7):3574. https://doi.org/10.3390/ijms23073574
Chicago/Turabian StyleBaptista, Inês, Effrosyni Karakitsou, Jean-Baptiste Cazier, Ulrich L. Günther, Silvia Marin, and Marta Cascante. 2022. "TKTL1 Knockdown Impairs Hypoxia-Induced Glucose-6-phosphate Dehydrogenase and Glyceraldehyde-3-phosphate Dehydrogenase Overexpression" International Journal of Molecular Sciences 23, no. 7: 3574. https://doi.org/10.3390/ijms23073574
APA StyleBaptista, I., Karakitsou, E., Cazier, J. -B., Günther, U. L., Marin, S., & Cascante, M. (2022). TKTL1 Knockdown Impairs Hypoxia-Induced Glucose-6-phosphate Dehydrogenase and Glyceraldehyde-3-phosphate Dehydrogenase Overexpression. International Journal of Molecular Sciences, 23(7), 3574. https://doi.org/10.3390/ijms23073574







