Trans-Reduction of Cerebral Small Vessel Disease Proteins by Notch-Derived EGF-like Sequences
Abstract
:1. Introduction
2. Results
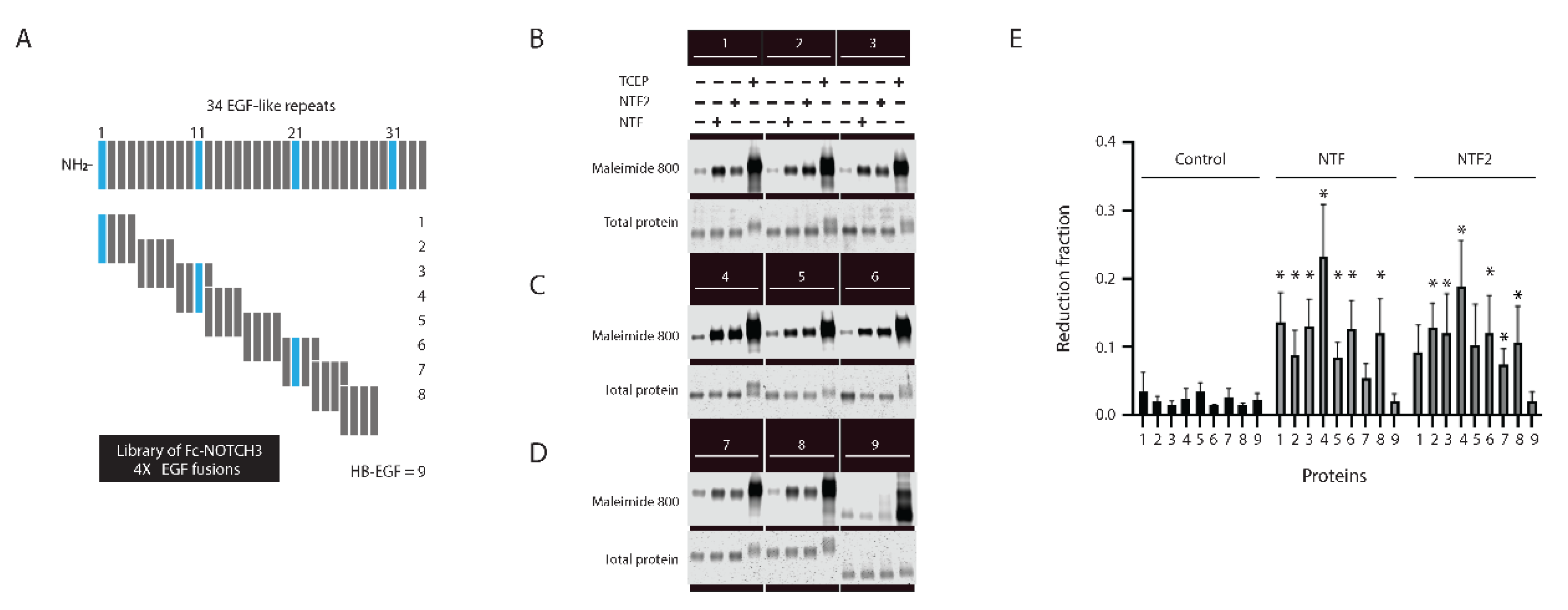
2.1. Trans-Reduction of NOTCH3 EGF-like Domains 1–3 by NTF2
2.2. Sequence Preference of NTF2 for NOTCH3 Trans-Reduction
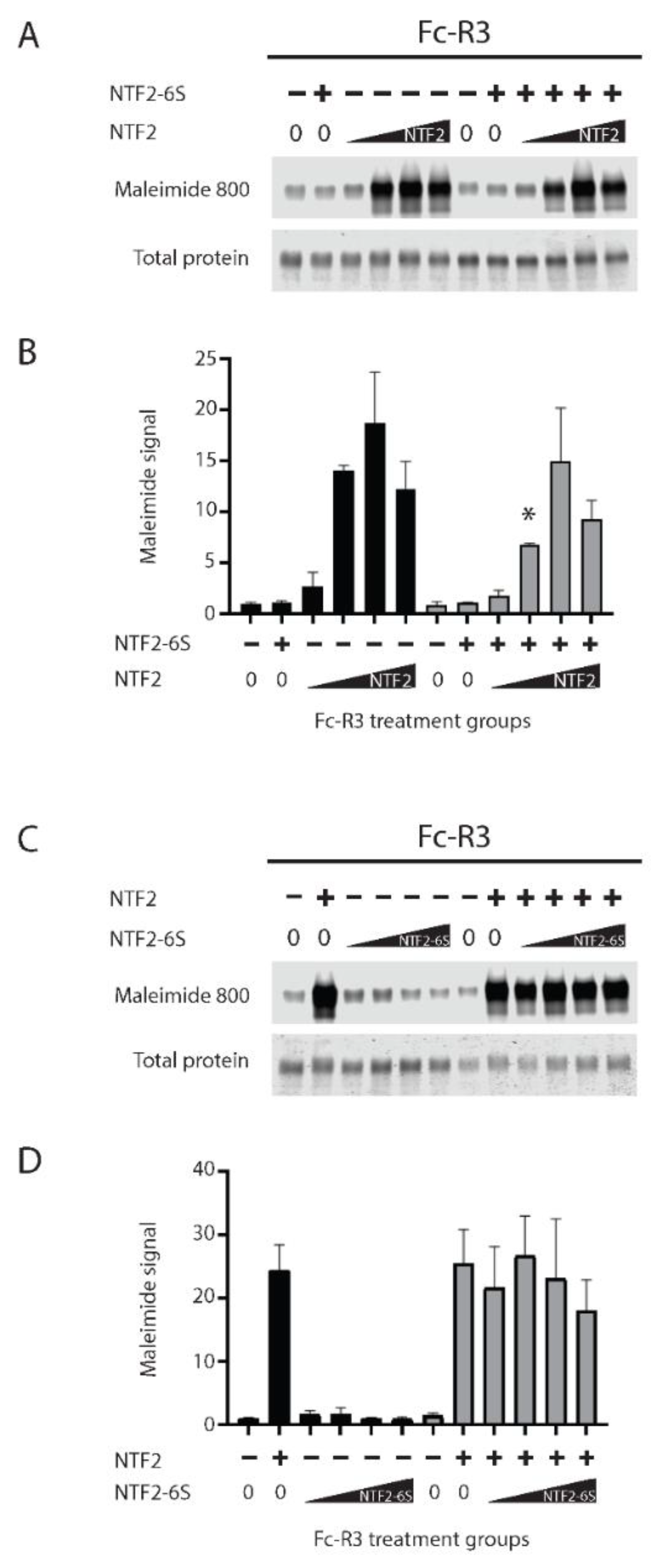
2.3. The Role of Cysteines of NTF2 in Trans-Reduction
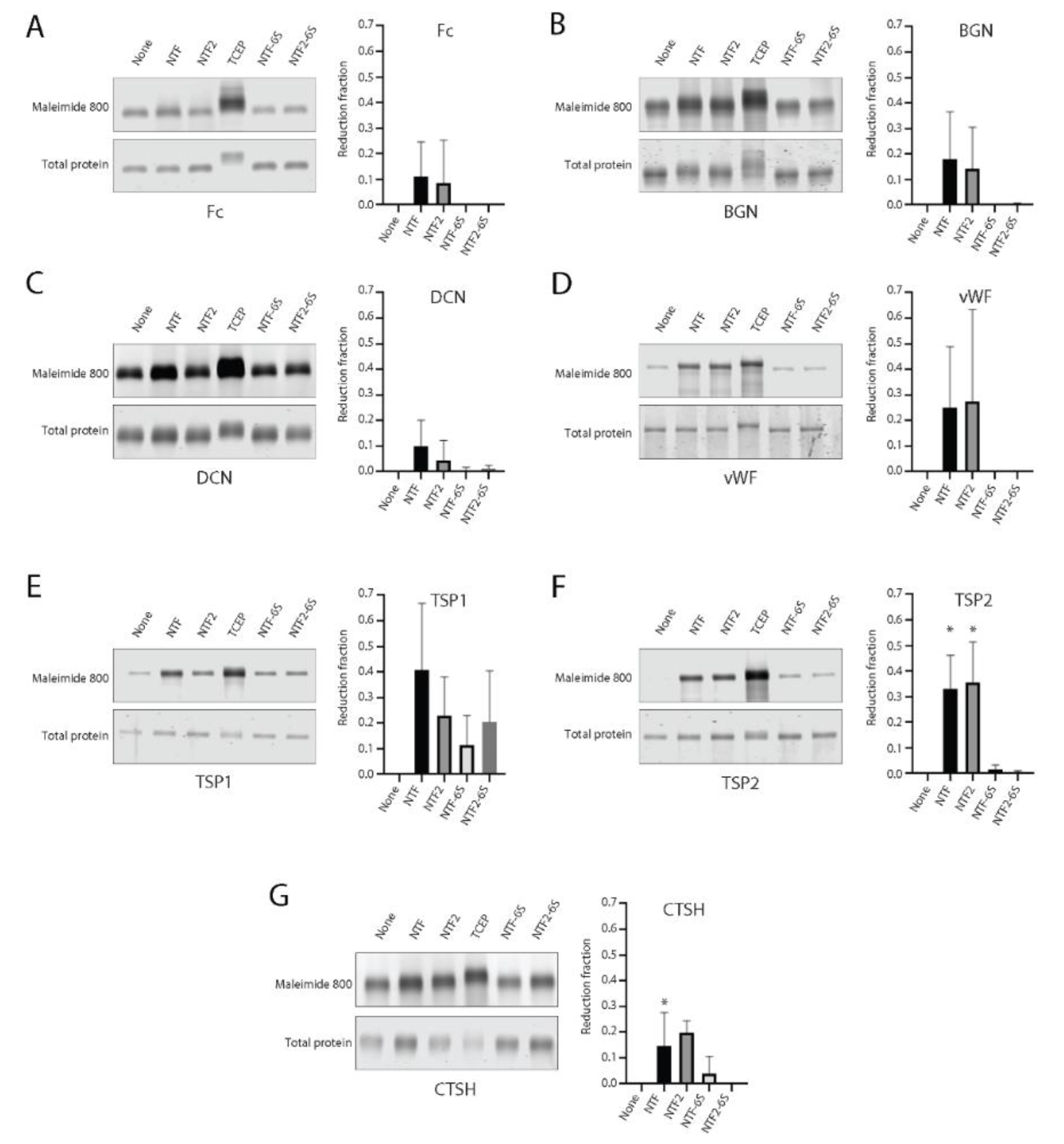
2.4. Comparison of Targeting of Additional Vascular Matrix Proteins by NTF2
2.5. Susceptibility of CADASIL Mutants of NOTCH3 to NTF2 Trans-Reduction
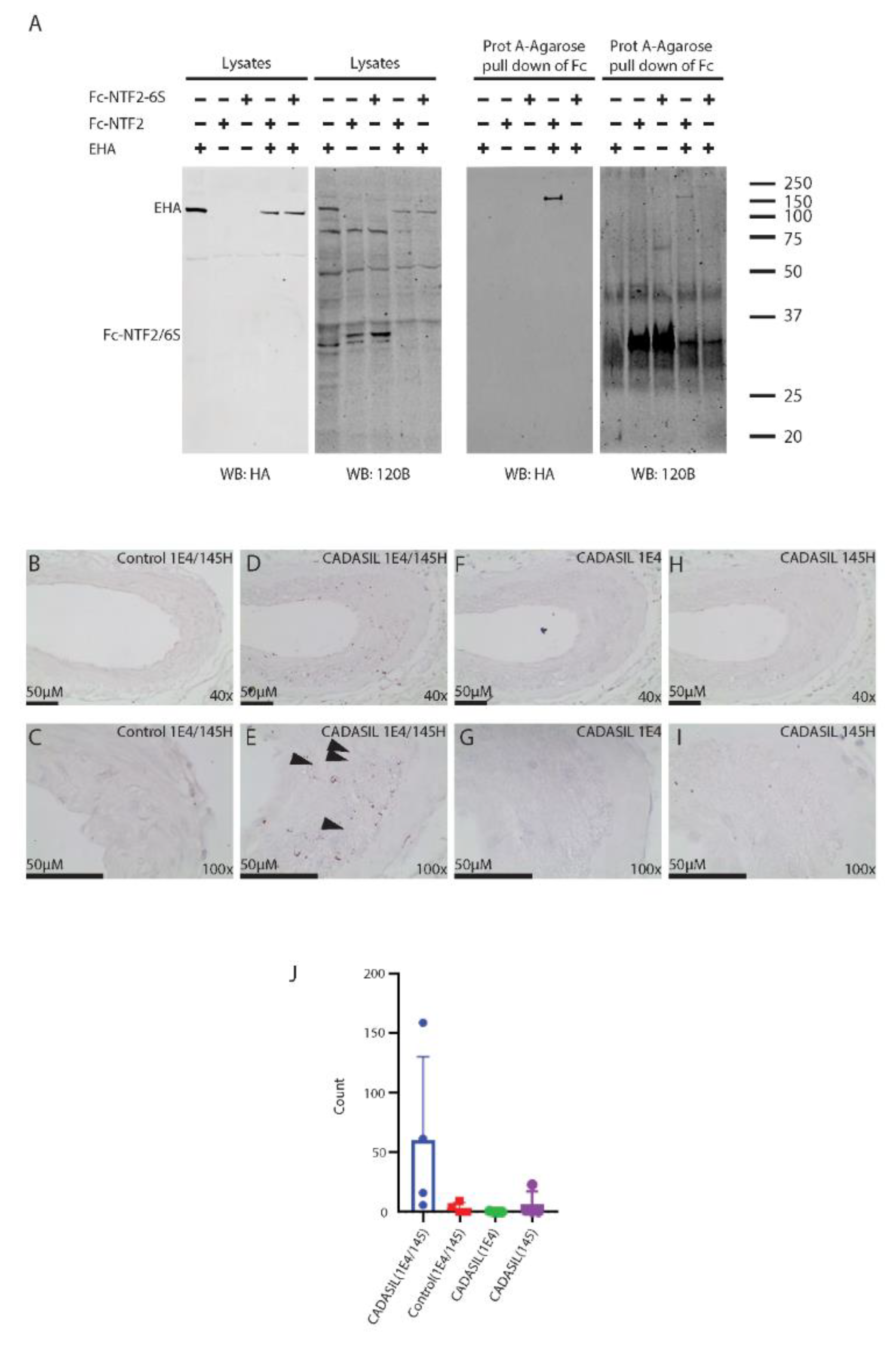
2.6. Trans-Reduction of NOTCH3 N-Terminal Protein by NOTCH3 Ectodomain
2.7. In Tissue Localization of NTF2 and NOTCH3 Complexes
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. DNA Constructs and Recombinant NOTCH3 Protein Generation
4.3. Cell Culture, Transfections, and Protein Co-Precipitation
4.4. Protein Analysis and Immunoblotting
4.5. Proximity Ligation Assay
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kim, H.J.; Ha, S.; Lee, H.Y.; Lee, K.J. ROSics: Chemistry and proteomics of cysteine modifications in redox biology. Mass Spectrom. Rev. 2015, 34, 184–208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reddie, K.G.; Carroll, K.S. Expanding the functional diversity of proteins through cysteine oxidation. Curr. Opin. Chem. Biol. 2008, 12, 746–754. [Google Scholar] [CrossRef] [PubMed]
- Barford, D. The role of cysteine residues as redox-sensitive regulatory switches. Curr. Opin. Struct. Biol. 2004, 14, 679–686. [Google Scholar] [CrossRef] [PubMed]
- Hidaka, Y.; Shimamoto, S. Folding of peptides and proteins: Role of disulfide bonds, recent developments. Biomol. Concepts 2013, 4, 597–604. [Google Scholar] [CrossRef]
- Lu, S.; Fan, S.B.; Yang, B.; Li, Y.X.; Meng, J.M.; Wu, L.; Li, P.; Zhang, K.; Zhang, M.J.; Fu, Y.; et al. Mapping native disulfide bonds at a proteome scale. Nat. Methods 2015, 12, 329–331. [Google Scholar] [CrossRef]
- Fra, A.; Yoboue, E.D.; Sitia, R. Cysteines as Redox Molecular Switches and Targets of Disease. Front. Mol. Neurosci. 2017, 10, 167. [Google Scholar] [CrossRef] [Green Version]
- Hogg, P.J. Disulfide bonds as switches for protein function. Trends Biochem. Sci. 2003, 28, 210–214. [Google Scholar] [CrossRef]
- Chiu, J.; Hogg, P.J. Allosteric disulfides: Sophisticated molecular structures enabling flexible protein regulation. J. Biol. Chem. 2019, 294, 2949–2960. [Google Scholar] [CrossRef] [Green Version]
- Kozlov, G.; Määttänen, P.; Thomas, D.Y.; Gehring, K. A structural overview of the PDI family of proteins. FEBS J. 2010, 277, 3924–3936. [Google Scholar] [CrossRef]
- Holmgren, A. Thioredoxin and glutaredoxin systems. J. Biol. Chem. 1989, 264, 13963–13966. [Google Scholar] [CrossRef]
- Chiu, J.; Wong, J.W.; Gerometta, M.; Hogg, P.J. Mechanism of dimerization of a recombinant mature vascular endothelial growth factor C. Biochemistry 2014, 53, 7–9. [Google Scholar] [CrossRef] [PubMed]
- Glushchenko, A.V.; Jacobsen, D.W. Molecular targeting of proteins by L-homocysteine: Mechanistic implications for vascular disease. Antioxid Redox Signal. 2007, 9, 1883–1898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saxena, V.P.; Wetlaufer, D.B. Formation of three-dimensional structure in proteins. I. Rapid nonenzymic reactivation of reduced lysozyme. Biochemistry 1970, 9, 5015–5023. [Google Scholar] [CrossRef] [PubMed]
- Okumura, M.; Saiki, M.; Yamaguchi, H.; Hidaka, Y. Acceleration of disulfide-coupled protein folding using glutathione derivatives. FEBS J. 2011, 278, 1137–1144. [Google Scholar] [CrossRef]
- Young, K.Z.; Lee, S.J.; Zhang, X.; Cartee, N.M.P.; Torres, M.; Keep, S.G.; Gabbireddy, S.R.; Fontana, J.L.; Qi, L.; Wang, M.M. NOTCH3 is non-enzymatically fragmented in inherited cerebral small-vessel disease. J. Biol. Chem. 2020, 295, 1960–1972. [Google Scholar] [CrossRef]
- Young, K.Z.; Cartee, N.M.P.; Ivanova, M.I.; Wang, M.M. Thiol-mediated and catecholamine-enhanced multimerization of a cerebrovascular disease enriched fragment of NOTCH3. Exp. Neurol. 2020, 328, 113261. [Google Scholar] [CrossRef]
- Joutel, A.; Corpechot, C.; Ducros, A.; Vahedi, K.; Chabriat, H.; Mouton, P.; Alamowitch, S.; Domenga, V.; Cecillion, M.; Marechal, E.; et al. Notch3 mutations in CADASIL, a hereditary adult-onset condition causing stroke and dementia. Nature 1996, 383, 707–710. [Google Scholar] [CrossRef]
- Joutel, A.; Vahedi, K.; Corpechot, C.; Troesch, A.; Chabriat, H.; Vayssiere, C.; Cruaud, C.; Maciazek, J.; Weissenbach, J.; Bousser, M.G.; et al. Strong clustering and stereotyped nature of Notch3 mutations in CADASIL patients. Lancet 1997, 350, 1511–1515. [Google Scholar] [CrossRef]
- Chabriat, H.; Joutel, A.; Dichgans, M.; Tournier-Lasserve, E.; Bousser, M.G. Cadasil. Lancet Neurol. 2009, 8, 643–653. [Google Scholar] [CrossRef]
- Wang, M.M. Cadasil. Handb. Clin. Neurol. 2018, 148, 733–743. [Google Scholar] [CrossRef]
- Zhang, X.; Lee, S.J.; Young, K.Z.; Josephson, D.A.; Geschwind, M.D.; Wang, M.M. Latent NOTCH3 epitopes unmasked in CADASIL and regulated by protein redox state. Brain Res. 2014, 1583, 230–236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Lee, S.J.; Wang, M.M. Hydrolysis of a second Asp-Pro site at the N-terminus of NOTCH3 in inherited vascular dementia. Sci. Rep. 2021, 11, 17246. [Google Scholar] [CrossRef] [PubMed]
- Holbrook, L.M.; Kwong, L.S.; Metcalfe, C.L.; Fenouillet, E.; Jones, I.M.; Barclay, A.N. OX133, a monoclonal antibody recognizing protein-bound N-ethylmaleimide for the identification of reduced disulfide bonds in proteins. MAbs 2016, 8, 672–677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cartee, N.M.P.; Lee, S.J.; Keep, S.G.; Wang, M.M. Context-dependent monoclonal antibodies against protein carbamidomethyl-cysteine. PLoS ONE 2020, 15, e0242376. [Google Scholar] [CrossRef]
- Wouters, M.A.; Rigoutsos, I.; Chu, C.K.; Feng, L.L.; Sparrow, D.B.; Dunwoodie, S.L. Evolution of distinct EGF domains with specific functions. Protein. Sci. 2005, 14, 1091–1103. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Lee, S.J.; Young, M.F.; Wang, M.M. The small leucine-rich proteoglycan BGN accumulates in CADASIL and binds to NOTCH3. Transl Stroke Res. 2015, 6, 148–155. [Google Scholar] [CrossRef] [Green Version]
- Joutel, A.; Andreux, F.; Gaulis, S.; Domenga, V.; Cecillon, M.; Battail, N.; Piga, N.; Chapon, F.; Godfrain, C.; Tournier-Lasserve, E. The ectodomain of the Notch3 receptor accumulates within the cerebrovasculature of CADASIL patients. J. Clin. Investig. 2000, 105, 597–605. [Google Scholar] [CrossRef]
- Zhang, X.; Meng, H.; Blaivas, M.; Rushing, E.J.; Moore, B.E.; Schwartz, J.; Lopes, M.B.; Worrall, B.B.; Wang, M.M. Von Willebrand Factor permeates small vessels in CADASIL and inhibits smooth muscle gene expression. Transl Stroke Res. 2012, 3, 138–145. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.J.; Zhang, X.; Wang, M.M. Vascular accumulation of the small leucine-rich proteoglycan decorin in CADASIL. Neuroreport 2014, 25, 1059. [Google Scholar] [CrossRef] [Green Version]
- Dong, H.; Blaivas, M.; Wang, M.M. Bidirectional encroachment of collagen into the tunica media in cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy. Brain Res. 2012, 1456, 64–71. [Google Scholar] [CrossRef] [Green Version]
- Dong, H.; Ding, H.; Young, K.; Blaivas, M.; Christensen, P.J.; Wang, M.M. Advanced intimal hyperplasia without luminal narrowing of leptomeningeal arteries in CADASIL. Stroke 2013, 44, 1456–1458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rutten, J.W.; Van Eijsden, B.J.; Duering, M.; Jouvent, E.; Opherk, C.; Pantoni, L.; Federico, A.; Dichgans, M.; Markus, H.S.; Chabriat, H.; et al. The effect of NOTCH3 pathogenic variant position on CADASIL disease severity: NOTCH3 EGFr 1-6 pathogenic variant are associated with a more severe phenotype and lower survival compared with EGFr 7-34 pathogenic variant. Genet. Med. 2019, 21, 676–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, H.; Zhang, X.; Yu, G.; Lee, S.J.; Chen, Y.E.; Prudovsky, I.; Wang, M.M. Biochemical characterization and cellular effects of CADASIL mutants of NOTCH3. PLoS ONE 2012, 7, e44964. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cartee, N.M.P.; Lee, S.J.; Young, K.Z.; Zhang, X.; Wang, M.M. Trans-Reduction of Cerebral Small Vessel Disease Proteins by Notch-Derived EGF-like Sequences. Int. J. Mol. Sci. 2022, 23, 3671. https://doi.org/10.3390/ijms23073671
Cartee NMP, Lee SJ, Young KZ, Zhang X, Wang MM. Trans-Reduction of Cerebral Small Vessel Disease Proteins by Notch-Derived EGF-like Sequences. International Journal of Molecular Sciences. 2022; 23(7):3671. https://doi.org/10.3390/ijms23073671
Chicago/Turabian StyleCartee, Naw May Pearl, Soo Jung Lee, Kelly Z. Young, Xiaojie Zhang, and Michael M. Wang. 2022. "Trans-Reduction of Cerebral Small Vessel Disease Proteins by Notch-Derived EGF-like Sequences" International Journal of Molecular Sciences 23, no. 7: 3671. https://doi.org/10.3390/ijms23073671
APA StyleCartee, N. M. P., Lee, S. J., Young, K. Z., Zhang, X., & Wang, M. M. (2022). Trans-Reduction of Cerebral Small Vessel Disease Proteins by Notch-Derived EGF-like Sequences. International Journal of Molecular Sciences, 23(7), 3671. https://doi.org/10.3390/ijms23073671





