Abstract
Plant transcriptomes encompass a large number of functional non-coding RNAs (ncRNAs), only some of which have protein-coding capacity. Since their initial discovery, ncRNAs have been classified into two broad categories based on their biogenesis and mechanisms of action, housekeeping ncRNAs and regulatory ncRNAs. With advances in RNA sequencing technology and computational methods, bioinformatics resources continue to emerge and update rapidly, including workflow for in silico ncRNA analysis, up-to-date platforms, databases, and tools dedicated to ncRNA identification and functional annotation. In this review, we aim to describe the biogenesis, biological functions, and interactions with DNA, RNA, protein, and microorganism of five major regulatory ncRNAs (miRNA, siRNA, tsRNA, circRNA, lncRNA) in plants. Then, we systematically summarize tools for analysis and prediction of plant ncRNAs, as well as databases. Furthermore, we discuss the silico analysis process of these ncRNAs and present a protocol for step-by-step computational analysis of ncRNAs. In general, this review will help researchers better understand the world of ncRNAs at multiple levels.
1. Introduction
In the last few years, a number of non-coding RNAs (ncRNAs) have been described in plants involved in several processes, ranging from RNA maturation, splicing, regulation of transcription, post-transcriptional RNA modifications, and nucleosome remodeling. Therefore, it is unquestionable that ncRNAs play a significant role in gene regulatory network [1,2,3,4]. With extensive transcriptome analysis, up to 90% of the eukaryotic genome is transcribed into RNA, of which only 1–2% corresponds to protein-coding mRNA [5,6]. Although the remaining transcripts lack minimal protein-coding capacity and poorly conserved sequences [2,5,7], the emergence of ncRNAs as novel ribose regulators of gene expression sheds light on the so-called “dark matter” of the genome.
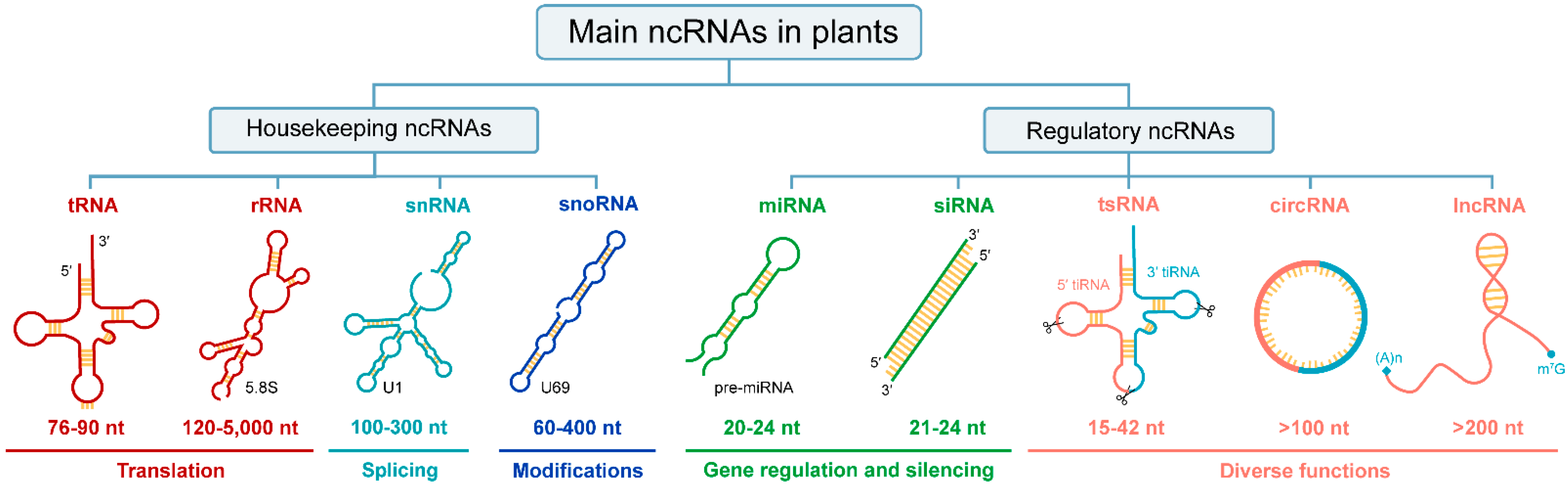
Current studies have revealed that ncRNAs can be transcribed from DNA sequences in protein-coding genes, intergenic or intronic regions [8]. In terms of their regulatory roles, ncRNAs could be divided into two major categories in plants (Figure 1). Among them, housekeeping ncRNAs are necessary for fundamental biological processes of life, so the content is relatively constant. Regulatory ncRNAs vary in size, shape, and accumulation patterns, so their expression is temporal and spatially specific [9]. Notably, any ncRNAs classification system is defined as an intelligent construct that is unlikely to perfectly reflect nature [10]. To date, many ncRNAs have not been described in plants, such as PIWI-interacting RNA (piRNA), an animal-specific small silencing RNA [11,12], enhancer RNAs (eRNAs), which play critical role in transcriptional activation in mammalian cells and are transcribed from enhancers [13,14,15], and Y RNAs, which are necessary for DNA replication in humans [16,17].
Figure 1.
ncRNAs category in plants. From top to bottom, there are primary classification, secondary classification, abbreviations, secondary structures, size, and functions of ncRNAs. Since some ncRNAs contain multiple types, one is selected and annotated with text in the lower right corner of the secondary structure. The sizes of ncRNAs are approximate. Diverse functions include gene expression regulation, translation inhibition, plant immunity, stress response, etc. Abbreviations: tRNA, transfer RNA; rRNA, ribosomal RNA; snRNA, small nuclear RNA; snoRNA, small nucleolar RNA; miRNA, micro RNA; siRNA, small interfering RNA; tsRNA, tRNA-derived small RNA; circRNA, circular RNA; lncRNA, long non-coding RNA; tiRNA, stress-induced tRNA or tRNA halves; nt, nucleotides.
So far, the biogenesis of some regulatory ncRNAs has been clearly described [18,19,20]; however, siRNA and tsRNA are poorly defined in plants. As further studies have shown, ncRNAs participate in the maintenance of homeostasis in plants by ncRNA-associated interaction with other biomolecules and microorganisms, which is of great significance to growth, development, differentiation, and reproduction of plants.
With the advancement of high-throughput RNA-seq technologies, the diversity of ncRNAs world has been unveiled. To date, numerous studies have applied RNA-seq technology to discover known and novel classes of ncRNAs in diverse tissues and developmental stages [21]. These precious data have been mined and stored in public databases.
In this review, we mainly focus on five regulatory ncRNAs, including microRNA (miRNA), small interfering RNA (siRNA), tRNA-derived small RNA (tsRNA), circular RNA (circRNA), and long ncRNA (lncRNA) in plants. Here, we will show a regulator ncRNAs biogenesis landscape and give a hypothesis of tsRNAs biogenesis in plants based on previous studies. Besides, we will discuss the ncRNAs interaction with DNA, RNA, protein, and microorganism for helping our understanding of the dynamic inter-molecular networks within plant cells. Furthermore, we will summarize the algorithms, databases, and RNA-seq-based analysis pipelines of the regulatory ncRNAs in the field of plants to incentivize researchers to make greater use of RNA-seq technologies and bioinformatics approaches to discover the individual, as well diverse, regulatory ncRNAs in the exceedingly complicated landscape of ncRNAs.
2. Biogenesis and Functions of ncRNAs in Plants
Currently, massive endogenous ncRNAs with various regulatory potentials have been discovered in various plant species [22,23]. Based on their average size, regulatory ncRNAs could be further categorized into small RNAs (18–30 nt), medium-sized ncRNAs (31–200 nt), and lncRNAs (>200 nt). In addition, it can be classified into linear or circular according to its morphology (Figure 1). In general, 200 nt is regarded as the dividing line in the regulatory ncRNAs world, but this size consideration is arbitrary because circRNAs, eRNAs, and promoter-associated transcripts (PATs) have displayed variable lengths [8]. Recently, regulatory small RNAs (sRNAs), namely, miRNA and siRNA, are considered to have tiny sizes but play important roles in response to stress or environmental changes by regulating the expression of target genes [24,25,26,27]. Likewise, lncRNAs were contemplated as transcriptional noise but later gained importance as one of the wide-ranging and heterogeneous groups of ncRNAs [28]. Notably, unlike other linear regulatory ncRNAs, circRNAs are a novel class of ncRNAs that lack free 5′ and 3′ terminus, which have been extensively explored in the past few years [29]. Besides, many small ncRNAs derived from tRNAs, called tsRNAs, have also been identified in plants with a broad size range of 15–42 nt [30,31,32]. Here, we classified tsRNAs as regulatory ncRNA according to their diverse functions (Figure 1). In general, the functions of some regulatory ncRNAs are similar, while a few are distinct, nevertheless overlapping in silencing pathways [33]. Next, we will introduce their biogenesis and functions in plants in detail.
2.1. miRNA
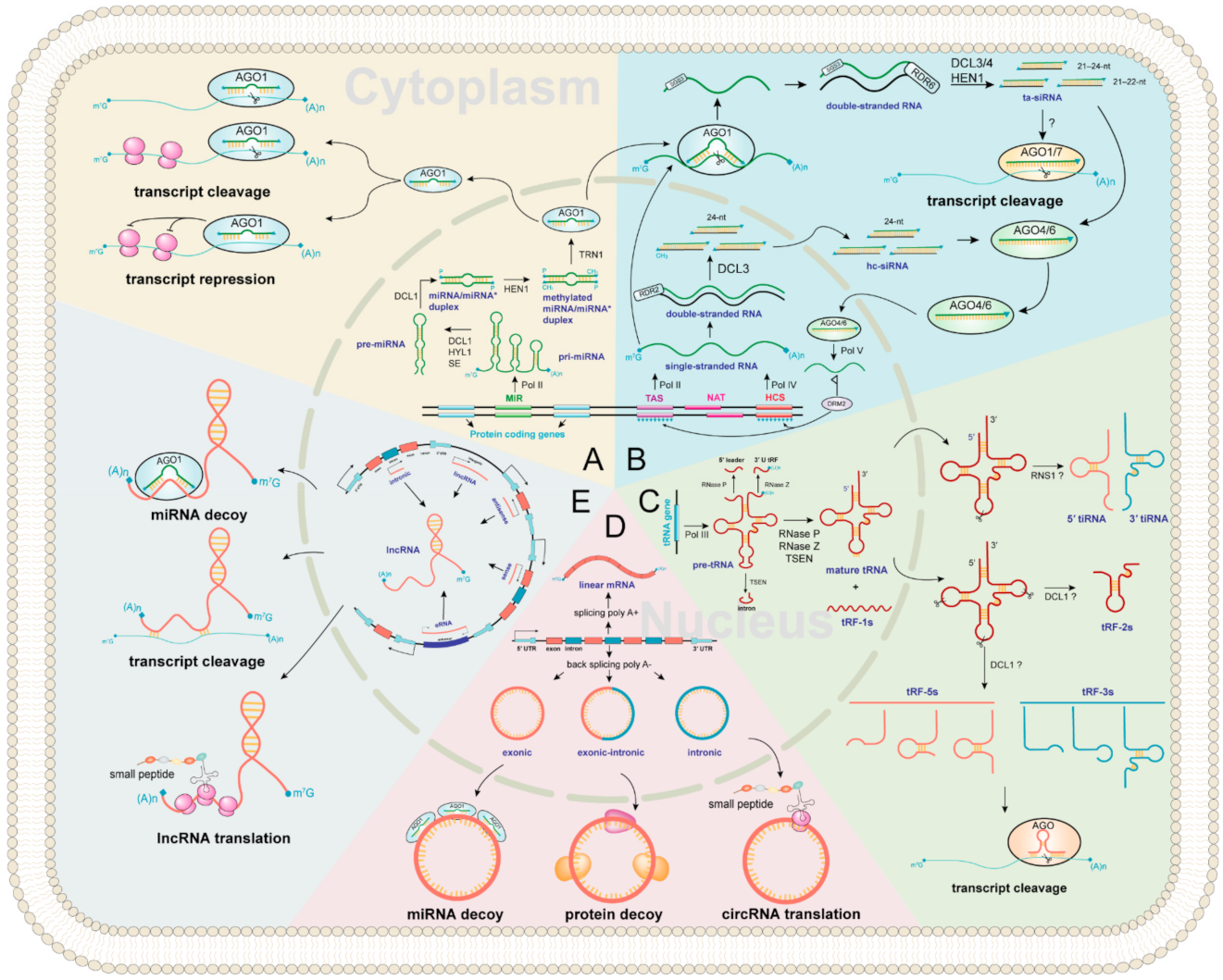
miRNA biogenesis is a multistep process involving transcription, processing, modification, and assembly of the RNA-induced silencing complex (RISC) (Figure 2A) [34,35,36]. First, primary miRNAs (pri-miRNAs) are transcribed by RNA POLYMERASE II (Pol II) containing hairpin RNA secondary structures. Then, an RNase III family DICER-LIKE (DCL) enzyme, usually DCL1 [37], assisted by HYPONASTIC LEAVES 1 (HYL1) and SERRATE (SE), cleaves from the base of the pri-miRNA hairpin to yield a precursor-miRNA (pre-miRNA) hairpin and cleaves again to release a miRNA/miRNA* duplex [38]. Next, the 3′-most nucleotides of the initial miRNA/miRNA* duplex are then 2′-O-methylated by the nuclear HUA ENHANCER 1 (HEN1) protein for stabilizing miRNA [39]. Finally, most mature miRNA strands are incorporated into ARGONAUTE 1 (AGO1) in the nucleus (unlike in animals, where it occurs in the cytoplasm [40]), with the removal of the miRNA* strand and the transport of the miRNA-AGO1 complex to the cytoplasm, where miRNAs induce post-transcriptional gene silencing by transcript cleavage and translation repression [24,35,41,42].
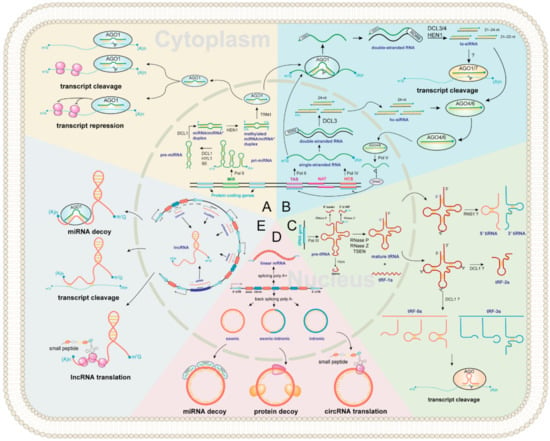
Figure 2.
Regulator ncRNAs biogenesis landscape and functions in plants. The inside of the circle represents the nucleus, and the outside represents cytoplasm. All ncRNAs types are marked in purple. The background of each color represents biogenesis and functions of (A) miRNA; (B) siRNA; (C) tsRNA; (D) circRNA; (E) lncRNA, respectively.
As post-transcriptional gene regulators, miRNAs are up- or down-regulated for improving plant productivity and stress tolerance in numerous species [43,44]. Therefore, studying the expression patterns of miRNAs can help us better understand the regulatory networks of stress response and environmental adaptation. In general, miRNAs have several features in regulatory pathways. (1) Evolutionarily conserved RNAs tend to have conserved targets in related plant species. For example, miR159 targets MYB genes and is down-regulated in salt-stress responses among Arabidopsis [45], tobacco [46], and kidney bean [47]. miR172 targets AP2 genes for regulating floral development in Arabidopsis, rice, soybean, barley, and maize [27]. A list of conserved miRNAs suggests a common regulatory mechanism across different species. (2) A single miRNA can participate in a variety of stress responses and developmental processes. For instance, cold-inducible miR393 targets TIR1/AFB genes and is up-regulated for enhancing cold tolerance [48]. miR393 is also induced by PAMP flagellin (flg22) to down-regulate the levels of TIR1/AFB genes for antibacterial defense [49]. Besides, miR393 is also involved in regulating arbuscule formation [50], inhibiting root elongation, and promoting lateral root initiation [51]. (3) Multiple miRNAs can participate in one biological process. For example, in rice, miR156, miR396, and miR397 cooperate in the regulation of grain size. miR156, miR393, and miR444 participate in tillering together [27]. (4) The expression patterns of miRNAs rely on the specific condition. As mentioned above, miRNAs are up- or down-regulated according to specific stress or specific tissue. Another example can also be supported. miR1425 will influence the number of fertile pollen grains by regulating a pentatricopeptide repeat (PPR)-containing protein under cold stress [52].
2.2. siRNA
According to the mode of action, siRNA can be simply divided into three secondary categories, namely trans-acting siRNAs (ta-siRNA), heterochromatic siRNAs (hc-siRNA), and natural antisense siRNAs (nat-siRNAs). However, in fact, ta-siRNAs belong to the so-called “secondary siRNAs” category, including ta-siRNA and phased siRNAs. Here, since many of the known ta-siRNAs are also phased [10], we use ta-siRNA instead of “secondary siRNAs” for discussion. Generally, they both play a role in transcriptional gene silencing by complementary target mRNAs or directing DNA and histone methylation through RNA-directed DNA methylation (RdDM) process [53,54].
ta-siRNAs are generated from TAS genes (Figure 2B) [24,55]. Firstly, TAS genes are transcribed into single-stranded RNAs by RNA Pol II, and then they loose the cap and mostly also the poly-A end upon miRNA-AGO1 complex guided cleavage. Secondly, the 5′ or 3′ cleavage fragments are protected by SUPPRESSOR OF GENE SILENCING 3 (SGS3) and converted to double-stranded RNA (dsRNA) by RNA-dependent RNA polymerases 6 (RDR6) [56]. Finally, they are methylated and processed into 21–24 nt ta-siRNAs by HEN1 and various DCL activities. The 21–22 nt size class are loaded onto AGO1 or AGO7 to induce post-transcriptional gene silencing of complementary target mRNAs in the cytoplasm, while some ta-siRNAs are incorporated into AGO4/6 to guide RNA Pol V-mediated de novo DNA methylation of TAS genes [54]. In Arabidopsis, miR173 targets TAS1 and TAS2 genes to generate ta-siRNAs [55,57]. The TAS1 ta-siRNAs target the heat stress transcription factor genes, HEAT-INDUCED TAS1 TARGET 1 (HTT1) and HTT2, to regulate plant thermotolerance [58].
nat-siRNAs can be divided into two categories, cis-NAT-siRNAs and trans-NAT-siRNAs. However, only cis-NAT-siRNAs have been described in plants. trans-NAT-siRNAs remain only a hypothetical possibility. Therefore, in this review, cis-NAT-siRNAs are collectively referred to as nat-siRNAs. Previously, nat-siRNAs were thought to be generated by the hybridization of separately transcribed complementary RNAs. However, to date, many of the nat-siRNAs investigated depend on RDR for their accumulation [10,59,60,61,62]. This RDR dependency suggests that the precursor dsRNA did not derive from the hybridization of two separately transcribed, complementary mRNAs. Thus, the biogenesis of nat-siRNAs is not well defined and appears to be very complex with some important unanswered questions. Based on available data, Zhang et al. speculated that there are at least five possible mechanisms to generate nat-siRNAs [63]. However, it is clear that nat-siRNAs can be induced by salt [59], pathogen [63], and control sperm function during double fertilization in Arabidopsis [61].
The biogenesis of hc-siRNA begins with the transcription of RNA Pol IV from the intergenic or repetitive genomic regions to generate single-stranded siRNA precursors [64,65,66], which are converted into dsRNA and processed into 24 nt siRNA duplexes. Methylated hc-siRNAs are loaded into AGO4 in the cytoplasm and are transported to the nucleus [67], followed by the recruitment of these hc-siRNA-AGO4 complexes to RNA Pol V transcripts. The subsequent recruitment of DOMAINS REARRANGED METHYLASE 2 (DRM2) catalyzes DNA methylation at RdDM target loci [53,67].
2.3. tsRNA
With a broad size range of 15–42 nt, tsRNAs are a new category of regulatory ncRNAs, which are classified into five categories according to the cleavage sites, namely tRF-1s, tRF-2s, tRF-3s, tRF-5s, and tiRNA (Figure 2C). However, the study in plants has just started, and many questions remain to be answered. For example, the biogenesis pathway of tsRNAs in plants is still unclear, and the physiological function of certain tsRNA in plants is currently very limited [68]. In this review, we propose a hypothesis of tsRNAs biogenesis in plants based on previous studies. First, RNA poly III transcribes tRNA gene as precursor tRNA (pre-tRNA) [69], which includes a 5′ leader, a mature tRNA backbone, a 3′ U trailer, and sometimes an intron [70]. Then, the 5′ leader, 3′ U trailer, and intronic sequences are cleaved by RNase P, RNase Z, and tRNA-splicing endonucleases (TSEN) to produce mature tRNA and tRF-1s (tRF-1s could be derived from 3′-end of pre-tRNA) [71,72,73]. The mature tRNA (73–90 nt) forms a secondary cloverleaf structure with a D-loop (left), a T-loop (right), anticodon loop (bottom), a variable loop, and an acceptor stem (Figure 2C). Finally, the mature tRNAs could be cleavaged by Arabidopsis S-like Ribonuclease 1 (RNS1) and/or DCL1 to form tRF-2s, tRF-3s, tRF-5s, and tiRNA (Figure 2C) [30,31]. In mammals, tsRNAs incorporate into silencing AGO and trigger RNA interference [74]. Likewise, AGO-associated tsRNAs have been predicted in Arabidopsis and rice [30,75]. An in vitro assay has shown that certain tsRNAs regulate gene expression by translation inhibition, and tsRNA-AGO1 complex tends to target transposable element transcripts and probably maintains genome stability [31,76,77].
2.4. circRNA
CircRNAs were first discovered in plant viruses by Sanger’s group in 1976. Studies have shown that circRNAs are circular, single-stranded, and covalently closed RNA biomolecules [78]. The composition of circRNAs can be divided into three categories (Figure 2D). (1) Exonic circRNAs are formed by lariat-driven circularization and intron pairing-driven circularization [79]. (2) Intronic circRNAs are the source of introns generated by the partial degradation of introns after the formation of the lasso structure. (3) Exonic-intronic circRNAs, which are composed of exons and introns, are cyclized during splicing. In 2013, Jeck et al., proposed that exon skipping and intron pairing reduced the distance between splicing sites and promoted the reverse splicing of pre-mRNA [80]. This leads to the deletion of the 3′ and 5′ ends of circRNAs [81]. Several distinct functional mechanisms for animal circRNAs have been identified, suggesting that plant circRNAs may exhibit similar conserved functions. These include miRNA decoys [82], transcriptional modulation [83], translation of circRNAs into small peptides [84]. Besides, circRNAs can play an important role in plant development and stress responses. For example, Vv-circATS1 responds to cold stress by regulating the expression of stimulus-responsive genes in grape [85]. Under dehydration-stressed conditions, many differentially expressed circRNAs have been detected in wheat [86], pear [87], maize, and Arabidopsis [88]. These studies suggest that circRNAs have post-transcriptional roles. However, the mechanism of this remains to be elucidated.
2.5. lncRNA
The biogenesis of lncRNAs can be divided into five categories according to the transcribed site by Pol II: (1) sense lncRNAs are transcribed on the same strand as exons; (2) antisense lncRNAs are transcribed on the opposite strand of exons; (3) intronic lncRNAs are transcribed on introns; (4) intergenic lncRNAs are located between two distinct genes; (5) enhancer lncRNAs emerge from an enhancer region of protein-coding genes (Figure 2E) [89]. They can control target regulation by multiple ways, including chromatin remodeling [90,91,92], transcriptional repression, RNA splicing and transcriptional enhancer [93,94]. In addition, lncRNAs may encode small peptides (Figure 2E), which are required for various cellular processes [95]. Notably, numerous plant lncRNAs are regulated by abiotic stresses. For example, many differentially expressed lncRNAs have been identified in Arabidopsis under drought, cold, salinity, heat, and abscisic acid stresses [96]. Besides, biotic stress-responsive lncRNAs have also been identified in wheat [97], Arabidopsis [98], and tomato [99].
3. ncRNA-Associated Interaction
Recent studies have demonstrated that ncRNAs are involved in the maintenance of plant homeostasis through ncRNA-associated interaction with DNA, RNA, protein, and microorganism, respectively, and have important implications for plant growth, development, differentiation, and reproduction [100,101]. Typically, these ncRNAs interact with genes or gene products (such as proteins and various RNAs) in the nucleus and cytoplasm region, thereby affecting biological processes and altering their cell fate [102,103,104,105,106,107,108]. Therefore, further discussions of how ncRNAs interact with other biological macromolecules will help advance our understanding of the landscape of the dynamic inter-molecular networks within plant cells.
3.1. ncRNAs Interact with RNAs
A series of experimental methods, such as PAR-CLIP [109], HITS-CLIP [110], CLASH [111], and LIGR-seq [112], were developed to define ncRNAs function and how they interact with other RNAs. Notably, LIGR-seq is a novel technology that can be used to detect RNA duplexes at scale without prior knowledge. Besides, several computational studies have also predicted that snoRNAs can interact with other RNA types, thereby regulating biological functions and cell signaling pathways [113].
Furthermore, the interactions among congener and isotypic ncRNAs can influence several biological processes, including epigenetic modifications and translation. For example, miRNA response elements (MRE), such as circRNAs, lncRNAs, and eRNAs, act as competing endogenous RNAs (ceRNAs) with rich implications for gene regulation in various physiological and pathophysiological processes at the post-transcriptional level [114]. ncRNAs can also act as gene sponges to regulate gene expression. It is well known that circRNAs and lncRNAs can generate internal regulatory networks through circRNA/lncRNA–miRNA-gene [115]. To date, the mechanisms of ncRNAs–RNAs interaction have been well defined in mammals and Homo sapiens [116,117,118], but little has been studied in plants. Hence, it is emergent to calculate the interactions between ncRNAs and RNAs in plants.
3.2. ncRNAs Intact with DNAs
So far, numerous ncRNAs have been reported to be involved in the regulation of gene expression in the nucleus as direct regulators [119,120,121,122]. They may play roles in nucleosome positioning, chromatin marking, and transcriptional regulation. Currently, those ncRNAs that interact with the global genome can be detected through deep-sequencing technology, such as GRID-seq [123].
Furthermore, some lncRNAs have been demonstrated to be able to divide the nuclear region into distinct compartments and participate in the organization of multi-chromosomal regions [124,125,126]. These lncRNAs have a close affinity to chromosomes through nuclear matrix factors, and they also provide favorable advantage for lncRNAs to interact with functional DNA elements related to transcriptional regulation. Therefore, lncRNAs can not only interfere with the expression of protein-coding genes, which are close to lncRNA genes, but also spread throughout the nucleus, close to spatial affinity sites, and regulate the expression of genes on chromosomes [127,128].
3.3. ncRNAs Interact with Proteins
ncRNA–protein interactions play a crucial role in regulating cell metabolism. It is widely known that numerous RNA binding proteins (RBPs) can change the fate or function of the bound RNAs in the nucleus region. To date, the RNA–protein regulatory relationship can be detected by CLIP-seq [129]. Therefore, a series of approaches were also developed to reveal how and where ncRNAs interacted with corresponding proteins [110,130].
Generally, ncRNAs achieve and regulate various functions by forming various ribonucleoprotein (RNP) complexes with proteins. For example, snRNPs, which are composed of snRNAs and proteins, can direct both canonical splicing and alternative splicing. Besides, numerous nucleotides in pre-rRNA, pre-snRNA, and pre-tRNA undergo post-transcriptional modification by nucleolar RNP particles [131]. Small ncRNAs, such as miRNA and siRNA, can also influence the regulation of gene expression by interacting with AGO family proteins during RNA interference pathway [132,133,134,135,136].
As for circRNA, research has shown that they can serve as protein sponges to transport proteins into specific subcellular compartments. On the other hand, lncRNA can achieve their function via recruitment, inhibition, and acting indirectly through genome organization and transcription [137]. ALTERNATIVE SPLICING COMPETITOR (ASCO) lncRNAs have been reported to be regulators of alternative splicing in Arabidopsis through interactions with splicing factors [138]. Apparently, with more and more thorough studies of the interactions between ncRNAs and proteins, people may gradually recognize a far more complex regulation network of interactions in plants.
3.4. ncRNAs Interact with Microbe
Plants have an animal-like innate immune system [139,140]. When attacked by pathogenic microorganisms, plants recognize pathogen-associated molecules through plasma membrane-associated pattern recognition receptors (PRR) and trigger immunity (PTI) [141]. Some microorganisms inhibit the signal transduction of plant PTIs by secreting small ncRNAs as effector molecules, leading to the occurrence of diseases. In addition, plants also mainly utilize extracellular vesicles to transport small ncRNAs into pathogens to suppress virulence-related genes [60].
Currently known microorganisms regulate their pathogenic capacity based on two modes of small ncRNA regulation. (1) The first way is to regulate their own toxicity through sRNAs derived from microorganisms, such as the sRNA of the entomopathogenic fungi Metarhizium anisopliae and Sclerotinia sclerotiorum, respectively, in conidia formation, and sclerotia are differentially expressed [142,143]. (2) The second way is to inhibit the small ncRNAs and RNAi pathways of plants through microbial effector proteins to achieve pathogenic effects, e.g., two effectors from the oomycete plant pathogen Phytophthora sojae suppress RNA silencing in plants by inhibiting the biogenesis of small ncRNAs [144].
In addition to the innate immune system, plants have evolved two ways of regulating small ncRNAs in response to infection by pathogenic microorganisms. The first way is that plant endogenous sRNAs are involved in the regulation of immune responses. For example, Arabidopsis miR863-3p fine tunes plant immune responses during infection by sequentially silencing negative and positive regulators of plant immunity [145]. Another study reported that the disease resistance protein (R protein) SNC1 represses the transcription of miRNA and ta-siRNA loci, probably through the transcriptional corepressor TPR1. This study revealed an additional layer of regulation—a regulatory circuit formed by miRNAs, ta-siRNAs, and NLR proteins to modulate and fine tune the trade-off between plant growth and defense [146]. The second pathway is plant growth through RNAi mechanisms to regulate the infectivity of microorganisms. We know that small ncRNAs are produced by DCLs and act through AGOs to silence target genes [147,148,149]. AGO2 is the only Arabidopsis AGO that is highly induced by bacterial infection [150]. In rhizobia-legume symbiosis, AGO7 of Lotus japonicas is required for TAS3 ta-siRNA biogenesis, and it is important for the development of nitrogen-fixing nodules in plant roots [151,152].
4. Bioinformatics Resources for ncRNA Analysis
In the past two decades, substantial research efforts have been devoted to discovering non-coding regulatory RNAs and studying their functions, including miRNAs, lncRNAs, and circRNAs. With the development of next-generation sequencing (NGS), high-throughput sequencing has been widely used to characterize the ncRNA transcriptomes under various conditions, which provides an unprecedented opportunity to discover ncRNAs and identify differentially expressed ncRNA transcripts. However, rapidly growing sequencing data have created challenges for the identification, annotation, and storage of ncRNAs. Here, we systematically summarize the prediction tools, database resources, and integration workflows of various ncRNAs in plants.
4.1. ncRNA Prediction Tools in Plants
Next-generation sequencing offers unprecedented opportunities to discover and quantify various ncRNAs. To date, numerous computational methods have been developed for single ncRNA category, such as miRDeep-P2, miR-island, phasiRNAClassifier, NATpare, tsRFinder, RNAplonc, FEELnc, Circle-Map, and CircMarker [153,154,155,156,157,158,159,160,161]. Besides, integrated analysis of multiple ncRNAs has also been published, such as mirTools 2.0 and sRNAtools [154,162]. To select suitable tools and platforms for ncRNA prediction, we systematically summarize the tools and platforms for plant ncRNA prediction and analysis (Table 1).

Table 1.
List of ncRNA prediction tools in plants.
4.2. ncRNA Databases in Plants
With the decreasing cost of NGS sequencing and the advent of various tools for prediction and characterization of ncRNAs, the number of annotated ncRNAs has grown exponentially. Therefore, relevant databases of ncRNAs are emerging rapidly. There are lots of ncRNA-related databases in plants, most of which focus on a single-type ncRNA, such as PmiREN2.0 [212], MepmiRDB [212,213], GreeNC v2.0 [214,215], and AtCircDB v2.0 [216]. Due to the lack of a unified way to access ncRNA information, fragmented data make it challenging and incompatible for ncRNA search and comparison. Here, we comprehensively summarize high-quality databases of various ncRNAs in plants. Data sources and stored ncRNA information are listed in Table 2.

Table 2.
List of ncRNA repositories and ncRNA interaction repositories in plants.
4.3. RNA-Seq-Based Pipeline for ncRNA Analysis in Plants
Currently, the research on various ncRNA functions using RNA-Seq and sRNA-seq technology has achieved great success. However, only a few tools and platforms are available for the analysis and prediction of most ncRNAs in plants, such as mirTools 2.0 and sRNAtools [154,162]. Most tools only predict and interpret a single ncRNA, such as miRDeep-p, FEELnc, Circle-Map, CPAT, and many other pipelines [159,160,172,245]. Choosing appropriate tools for comprehensive analysis of plant ncRNAs is a great challenge. To overcome this challenge, we summarize a comprehensive workflow for ncRNA analysis (Figure 3), which provides a general analysis pipeline from traditional RNA-seq/sRNA-seq to specific ncRNA identification.
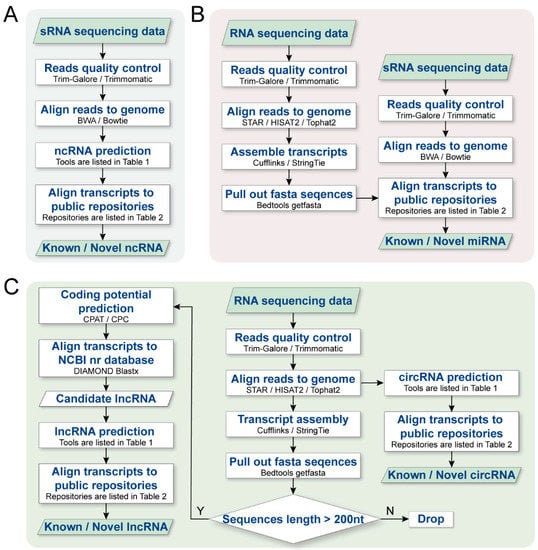
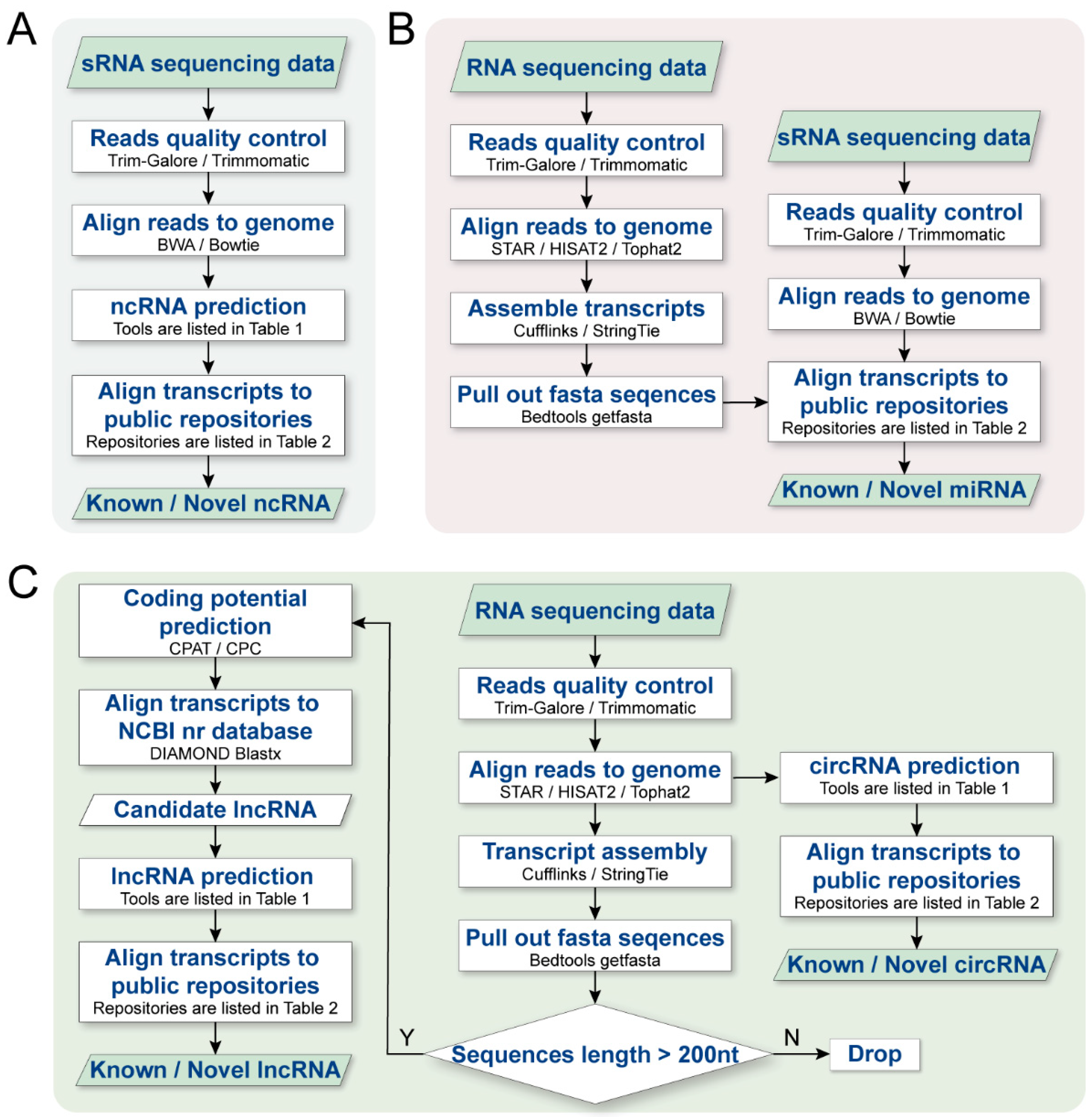
Figure 3.
The analysis workflow for differentiating between different classes of ncRNAs in sRNA-seq/RNA-seq datasets. (A) General analysis workflow from sRNA-seq data to ncRNA prediction (including data preprocessing, sequence alignment, ncRNA prediction, and related database alignment), (B) represents the general analysis process from RNA-seq to lncRNA/circRNA prediction (the lncRNA analysis process includes data preprocessing, sequence alignment, transcript assembly, sequence length filtering, transcript encoding potential estimation, database alignment, and lncRNA prediction. The circRNA analysis process includes data preprocessing, sequence alignment, circRNA prediction, and related database alignment), (C) represents the general process from sRNA-seq/RNA-seq to miRNA prediction (where analysis process from RNA-seq to miRNA prediction is like the general analysis process in A, while sRNA-seq to miRNA prediction includes data preprocessing, sequence alignment, miRNA prediction, and related database alignment).
In this workflow, we present a protocol for step-by-step computational analysis of ncRNAs, which mainly consists of three parts: (1) raw fastq data preprocessing, including read quality filtering and 3′ adapter trimming; (2) alignment and annotation; (3) sequence feature analysis, including ncRNA data alignment and novel ncRNA prediction.
5. Discussion
Although identification and characterization of biogenesis and fundamental functions of regulator ncRNAs are necessary, efforts to understand the depth and diversity of ncRNAs are the way forward. In the past decade, with the advancement of high-throughput sequencing technology, many bioinformatics methods and tools have emerged, which has enabled the discovery and research of many ncRNAs. Studies have found that ncRNAs mainly include housekeeping ncRNAs, such as tRNA, rRNA, etc., which are necessary to maintain plant life activities, and regulatory ncRNAs, such as miRNA, lncRNA, circRNA, etc., which play a role in regulating the special life process of plants. The interactions between these ncRNAs and the regulation of the expression of other encoded genes constitute a complex regulatory network of ncRNAs in plants.
Compared with numerous studies, such as miRNA and lncRNA, the functional characterization of circRNA is still in its infancy. Even in humans and animals, there are only a few reports on the functional study of circRNAs. Although many plants circRNAs have been identified from plant circRNA databases or sequencing analyses, the functional mechanism of only one circRNA from Arabidopsis has been revealed [83]. Due to the overlapping sequence characteristics of circRNAs, it is difficult to knock out circRNAs by traditional RNA interference methods, and the CRISPR-Cas system will enable new strategies for further circRNA function research.
At present, the research of these ncRNAs is more focused on their regulatory mechanisms during the growth and development of plant organs or tissues and in special environments. However, the expression and changes of ncRNAs in different cell types remain unclear [246]. In recent years, only a few reports have investigated the regulatory mechanisms of ncRNAs at the single-cell level in humans or animals [247,248,249], such as Luo et al. using scRNA-seq in three cancer-type data, where a total of 154 characteristic lncRNA genes related to effector, depletion, and regulatory T cell states were identified [249]. With the maturity of scRNA-seq technology, it is believed that the regulatory mechanisms of plant ncRNAs at the cellular level can be analyzed.
6. Conclusions
In this review, we systematically summarized the functions and regulatory relationships of major classes of ncRNAs in plants. Besides, we reviewed and summarized the computational methods, tools, and knowledge bases of ncRNA in plants in detail. Notably, we proposed a general protocol for step-by-step computational analysis of different types of ncRNAs to help researchers choose appropriate ncRNA analysis tools and platforms. We hope this work contributes to a better understanding of the complex ncRNA world.
Author Contributions
Conceptualization, M.C., P.Z.; investigation, Y.H.; writing—original draft preparation, H.C., Y.H., L.Z.; writing—review and editing, H.C., Y.H., L.Z., S.X., Q.N., P.Z., M.C.; Tables/Figures, H.C., Y.H. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Sciences Foundation of China (32070677); the 151 Talent Project of Zhejiang Province (first level); Jiangsu Collaborative Innovation Center for Modern Crop Production and Collaborative Innovation Center for Modern Crop Production cosponsored by province and ministry.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| ncRNA | non-coding RNA |
| piRNA | PIWI-interacting RNA |
| eRNA | enhancer RNA |
| lncRNA | long ncRNA |
| circRNA | circular RNA |
| PATs | promoter-associated transcripts |
| sRNA | small RNA |
| miRNA | microRNA |
| siRNA | small interfering RNA |
| tsRNA | tRNA-derived small RNA |
| RISC | RNA-induced silencing complex |
| pri-miRNA | primary miRNA |
| Pol II | RNA POLYMERASE II |
| DCL | DICER-LIKE |
| HYL1 | HYPONASTIC LEAVES 1 |
| SE | SERRATE |
| pre-miRNA | precursor-miRNA |
| HEN1 | HUA ENHANCER 1 |
| AGO1 | ARGONAUTE 1 |
| flg22 | PAMP flagellin |
| PPR | pentatricopeptide repeat |
| ta-siRNA | trans-acting siRNAs |
| hc-siRNA | heterochromatic siRNAs |
| nat-siRNAs | natural antisense siRNAs |
| RdDM | RNA-directed DNA methylation |
| SGS3 | SUPPRESSOR OF GENE SILENCING 3 |
| dsRNA | double-stranded RNA |
| RDR6 | RNA-dependent RNA polymerases 6 |
| HTT1 | HEAT-INDUCED TAS1 TARGET 1 |
| DRM2 | DOMAINS REARRANGED METHYLASE 2 |
| pre-tRNA | precursor tRNA |
| TSEN | tRNA-splicing endonucleases |
| RNS1 | ribonuclease 1 |
| MRE | miRNA response element |
| ceRNA | competitive endogenous RNA |
| RBP | RNA binding protein |
| RNP | ribonucleoprotein |
| ASCO | ALTERNATIVE SPLICING COMPETITOR |
| PRR | pattern recognition receptors |
| PTI | pattern trigger immunity |
| R protein | resistance protein |
| NGS | next-generation sequencing |
References
- Rincon-Riveros, A.; Morales, D.; Rodriguez, J.A.; Villegas, V.E.; Lopez-Kleine, L. Bioinformatic Tools for the Analysis and Prediction of ncRNA Interactions. Int. J. Mol. Sci. 2021, 22, 11397. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Zhang, Y.; Chen, X.; Chen, Y. Plant Noncoding RNAs: Hidden Players in Development and Stress Responses. Annu. Rev. Cell Dev. Biol. 2019, 35, 407–431. [Google Scholar] [CrossRef] [PubMed]
- Jampala, P.; Garhewal, A.; Lodha, M. Functions of long non-coding RNA in Arabidopsis thaliana. Plant Signal. Behav. 2021, 16, 1925440. [Google Scholar] [CrossRef]
- Yao, R.W.; Wang, Y.; Chen, L.L. Cellular functions of long noncoding RNAs. Nat. Cell Biol. 2019, 21, 542–551. [Google Scholar] [CrossRef]
- Pauli, A.; Rinn, J.L.; Schier, A.F. Non-coding RNAs as regulators of embryogenesis. Nat. Rev. Genet. 2011, 12, 136–149. [Google Scholar] [CrossRef]
- Rai, M.I.; Alam, M.; Lightfoot, D.A.; Gurha, P.; Afzal, A.J. Classification and experimental identification of plant long non-coding RNAs. Genomics 2019, 111, 997–1005. [Google Scholar] [CrossRef]
- Ariel, F.; Romero-Barrios, N.; Jegu, T.; Benhamed, M.; Crespi, M. Battles and hijacks: Noncoding transcription in plants. Trends Plant Sci. 2015, 20, 362–371. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Wu, W.; Chen, Q.; Chen, M. Non-Coding RNAs and their Integrated Networks. J. Integr. Bioinform. 2019, 16, 20190027. [Google Scholar] [CrossRef]
- Vivek, A.T.; Kumar, S. Computational methods for annotation of plant regulatory non-coding RNAs using RNA-seq. Brief. Bioinform. 2021, 22, bbaa322. [Google Scholar] [CrossRef]
- Axtell, M.J. Classification and comparison of small RNAs from plants. Annu. Rev. Plant Biol. 2013, 64, 137–159. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.K.; Gabel, H.W.; Kamath, R.S.; Tewari, M.; Pasquinelli, A.; Rual, J.F.; Kennedy, S.; Dybbs, M.; Bertin, N.; Kaplan, J.M.; et al. Functional genomic analysis of RNA interference in C. elegans. Science 2005, 308, 1164–1167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ozata, D.M.; Gainetdinov, I.; Zoch, A.; O’Carroll, D.; Zamore, P.D. PIWI-interacting RNAs: Small RNAs with big functions. Nat. Rev. Genet. 2019, 20, 89–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, R.; Cao, C.; Xue, Y. Enhancer RNA: Biogenesis, function, and regulation. Essays Biochem. 2020, 64, 883–894. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.K.; Hemberg, M.; Gray, J.M.; Costa, A.M.; Bear, D.M.; Wu, J.; Harmin, D.A.; Laptewicz, M.; Barbara-Haley, K.; Kuersten, S.; et al. Widespread transcription at neuronal activity-regulated enhancers. Nature 2010, 465, 182–187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, T.; Qian, J. EnhancerAtlas 2.0: An updated resource with enhancer annotation in 586 tissue/cell types across nine species. Nucleic Acids Res. 2020, 48, D58–D64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christov, C.P.; Gardiner, T.J.; Szuts, D.; Krude, T. Functional requirement of noncoding Y RNAs for human chromosomal DNA replication. Mol. Cell. Biol. 2006, 26, 6993–7004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, A.T.; Langley, A.R.; Christov, C.P.; Kheir, E.; Shafee, T.; Gardiner, T.J.; Krude, T. Dynamic interaction of Y RNAs with chromatin and initiation proteins during human DNA replication. J. Cell Sci. 2011, 124, 2058–2069. [Google Scholar] [CrossRef] [Green Version]
- Achkar, N.P.; Cambiagno, D.A.; Manavella, P.A. miRNA Biogenesis: A Dynamic Pathway. Trends Plant Sci. 2016, 21, 1034–1044. [Google Scholar] [CrossRef]
- Wang, H.V.; Chekanova, J.A. Long Noncoding RNAs in Plants. Adv Exp Med Biol 2017, 1008, 133–154. [Google Scholar] [CrossRef]
- Ashwal-Fluss, R.; Meyer, M.; Pamudurti, N.R.; Ivanov, A.; Bartok, O.; Hanan, M.; Evantal, N.; Memczak, S.; Rajewsky, N.; Kadener, S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell 2014, 56, 55–66. [Google Scholar] [CrossRef] [Green Version]
- Parvathaneni, R.K.; Bertolini, E.; Shamimuzzaman, M.; Vera, D.L.; Lung, P.Y.; Rice, B.R.; Zhang, J.; Brown, P.J.; Lipka, A.E.; Bass, H.W.; et al. The regulatory landscape of early maize inflorescence development. Genome Biol. 2020, 21, 165. [Google Scholar] [CrossRef] [PubMed]
- Bonnet, E.; Van de Peer, Y.; Rouze, P. The small RNA world of plants. New Phytol. 2006, 171, 451–468. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Hao, L.; Li, D.; Zhu, L.; Hu, S. Long non-coding RNAs and their biological roles in plants. Genom. Proteom. Bioinform. 2015, 13, 137–147. [Google Scholar] [CrossRef] [Green Version]
- Chen, X. Small RNAs and their roles in plant development. Annu. Rev. Cell Dev. Biol. 2009, 25, 21–44. [Google Scholar] [CrossRef] [Green Version]
- D’Ario, M.; Griffiths-Jones, S.; Kim, M. Small RNAs: Big Impact on Plant Development. Trends Plant Sci. 2017, 22, 1056–1068. [Google Scholar] [CrossRef] [Green Version]
- Martinez, G.; Kohler, C. Role of small RNAs in epigenetic reprogramming during plant sexual reproduction. Curr. Opin. Plant Biol. 2017, 36, 22–28. [Google Scholar] [CrossRef]
- Tang, J.; Chu, C. MicroRNAs in crop improvement: Fine-tuners for complex traits. Nat. Plants 2017, 3, 17077. [Google Scholar] [CrossRef]
- Chekanova, J.A. Long non-coding RNAs and their functions in plants. Curr. Opin. Plant Biol. 2015, 27, 207–216. [Google Scholar] [CrossRef] [Green Version]
- Sablok, G.; Zhao, H.; Sun, X. Plant Circular RNAs (circRNAs): Transcriptional Regulation Beyond miRNAs in Plants. Mol. Plant 2016, 9, 192–194. [Google Scholar] [CrossRef] [Green Version]
- Alves, C.S.; Vicentini, R.; Duarte, G.T.; Pinoti, V.F.; Vincentz, M.; Nogueira, F.T. Genome-wide identification and characterization of tRNA-derived RNA fragments in land plants. Plant Mol. Biol. 2017, 93, 35–48. [Google Scholar] [CrossRef] [Green Version]
- Cognat, V.; Morelle, G.; Megel, C.; Lalande, S.; Molinier, J.; Vincent, T.; Small, I.; Duchene, A.M.; Marechal-Drouard, L. The nuclear and organellar tRNA-derived RNA fragment population in Arabidopsis thaliana is highly dynamic. Nucleic Acids Res. 2017, 45, 3460–3472. [Google Scholar] [CrossRef]
- Megel, C.; Morelle, G.; Lalande, S.; Duchene, A.M.; Small, I.; Marechal-Drouard, L. Surveillance and cleavage of eukaryotic tRNAs. Int. J. Mol. Sci. 2015, 16, 1873–1893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Axtell, M.J.; Meyers, B.C. Revisiting Criteria for Plant MicroRNA Annotation in the Era of Big Data. Plant Cell 2018, 30, 272–284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rogers, K.; Chen, X. Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell 2013, 25, 2383–2399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, Y.; Jia, T.; Chen, X. The ‘how’ and ‘where’ of plant microRNAs. New Phytol. 2017, 216, 1002–1017. [Google Scholar] [CrossRef] [Green Version]
- Park, M.Y.; Wu, G.; Gonzalez-Sulser, A.; Vaucheret, H.; Poethig, R.S. Nuclear processing and export of microRNAs in Arabidopsis. Proc. Natl. Acad. Sci. USA 2005, 102, 3691–3696. [Google Scholar] [CrossRef] [Green Version]
- Fahlgren, N.; Howell, M.D.; Kasschau, K.D.; Chapman, E.J.; Sullivan, C.M.; Cumbie, J.S.; Givan, S.A.; Law, T.F.; Grant, S.R.; Dangl, J.L.; et al. High-throughput sequencing of Arabidopsis microRNAs: Evidence for frequent birth and death of MIRNA genes. PLoS ONE 2007, 2, e219. [Google Scholar] [CrossRef]
- Kurihara, Y.; Watanabe, Y. Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc. Natl. Acad. Sci. USA 2004, 101, 12753–12758. [Google Scholar] [CrossRef] [Green Version]
- Yu, B.; Yang, Z.; Li, J.; Minakhina, S.; Yang, M.; Padgett, R.W.; Steward, R.; Chen, X. Methylation as a crucial step in plant microRNA biogenesis. Science 2005, 307, 932–935. [Google Scholar] [CrossRef] [Green Version]
- Axtell, M.J.; Westholm, J.O.; Lai, E.C. Vive la difference: Biogenesis and evolution of microRNAs in plants and animals. Genome Biol. 2011, 12, 221. [Google Scholar] [CrossRef] [Green Version]
- Baumberger, N.; Baulcombe, D.C. Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc. Natl. Acad. Sci. USA 2005, 102, 11928–11933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bologna, N.G.; Iselin, R.; Abriata, L.A.; Sarazin, A.; Pumplin, N.; Jay, F.; Grentzinger, T.; Dal Peraro, M.; Voinnet, O. Nucleo-cytosolic Shuttling of ARGONAUTE1 Prompts a Revised Model of the Plant MicroRNA Pathway. Mol. Cell 2018, 69, 709–719.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shriram, V.; Kumar, V.; Devarumath, R.M.; Khare, T.S.; Wani, S.H. MicroRNAs as Potential Targets for Abiotic Stress Tolerance in Plants. Front. Plant Sci. 2016, 7, 817. [Google Scholar] [CrossRef]
- Koroban, N.V.; Kudryavtseva, A.V.; Krasnov, G.S.; Sadritdinova, A.F.; Fedorova, M.S.; Snezhkina, A.V.; Bolsheva, N.L.; Muravenko, O.V.; Dmitriev, A.A.; Melnikova, N.V. The role of microRNA in abiotic stress response in plants. Mol. Biol. 2016, 50, 387–394. [Google Scholar] [CrossRef]
- Chen, L.; Wang, T.; Zhao, M.; Tian, Q.; Zhang, W.H. Identification of aluminum-responsive microRNAs in Medicago truncatula by genome-wide high-throughput sequencing. Planta 2012, 235, 375–386. [Google Scholar] [CrossRef] [PubMed]
- Frazier, T.P.; Sun, G.; Burklew, C.E.; Zhang, B. Salt and drought stresses induce the aberrant expression of microRNA genes in tobacco. Mol. Biotechnol. 2011, 49, 159–165. [Google Scholar] [CrossRef]
- Jones-Rhoades, M.W.; Bartel, D.P. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell 2004, 14, 787–799. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, K.; Li, D.; Yan, J.; Zhang, W. Enhanced Cold Tolerance and Tillering in Switchgrass (Panicum virgatum L.) by Heterologous Expression of Osa-miR393a. Plant Cell Physiol. 2017, 58, 2226–2240. [Google Scholar] [CrossRef] [Green Version]
- Navarro, L.; Dunoyer, P.; Jay, F.; Arnold, B.; Dharmasiri, N.; Estelle, M.; Voinnet, O.; Jones, J.D. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 2006, 312, 436–439. [Google Scholar] [CrossRef] [Green Version]
- Etemadi, M.; Gutjahr, C.; Couzigou, J.M.; Zouine, M.; Lauressergues, D.; Timmers, A.; Audran, C.; Bouzayen, M.; Becard, G.; Combier, J.P. Auxin perception is required for arbuscule development in arbuscular mycorrhizal symbiosis. Plant Physiol. 2014, 166, 281–292. [Google Scholar] [CrossRef] [Green Version]
- Gifford, M.L.; Dean, A.; Gutierrez, R.A.; Coruzzi, G.M.; Birnbaum, K.D. Cell-specific nitrogen responses mediate developmental plasticity. Proc. Natl. Acad. Sci. USA 2008, 105, 803–808. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeong, D.H.; Park, S.; Zhai, J.; Gurazada, S.G.; De Paoli, E.; Meyers, B.C.; Green, P.J. Massive analysis of rice small RNAs: Mechanistic implications of regulated microRNAs and variants for differential target RNA cleavage. Plant Cell 2011, 23, 4185–4207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, J.; Johnson, L.M.; Jacobsen, S.E.; Patel, D.J. DNA methylation pathways and their crosstalk with histone methylation. Nat. Rev. Mol. Cell. Biol. 2015, 16, 519–532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matzke, M.A.; Kanno, T.; Matzke, A.J. RNA-Directed DNA Methylation: The Evolution of a Complex Epigenetic Pathway in Flowering Plants. Annu. Rev. Plant Biol. 2015, 66, 243–267. [Google Scholar] [CrossRef]
- Fei, Q.; Xia, R.; Meyers, B.C. Phased, secondary, small interfering RNAs in posttranscriptional regulatory networks. Plant Cell 2013, 25, 2400–2415. [Google Scholar] [CrossRef] [Green Version]
- Peragine, A.; Yoshikawa, M.; Wu, G.; Albrecht, H.L.; Poethig, R.S. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 2004, 18, 2368–2379. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.M.; Chen, L.T.; Patel, K.; Li, Y.H.; Baulcombe, D.C.; Wu, S.H. 22-Nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proc. Natl. Acad. Sci. USA 2010, 107, 15269–15274. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Liu, J.; Liu, Z.; Li, X.; Wu, F.; He, Y. HEAT-INDUCED TAS1 TARGET1 Mediates Thermotolerance via HEAT STRESS TRANSCRIPTION FACTOR A1a-Directed Pathways in Arabidopsis. Plant Cell 2014, 26, 1764–1780. [Google Scholar] [CrossRef] [Green Version]
- Borsani, O.; Zhu, J.; Verslues, P.E.; Sunkar, R.; Zhu, J.K. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 2005, 123, 1279–1291. [Google Scholar] [CrossRef] [Green Version]
- Katiyar-Agarwal, S.; Morgan, R.; Dahlbeck, D.; Borsani, O.; Villegas, A.J.; Zhu, J.K.; Staskawicz, B.J.; Jin, H. A pathogen-inducible endogenous siRNA in plant immunity. Proc. Natl. Acad. Sci. USA 2006, 103, 18002–18007. [Google Scholar] [CrossRef] [Green Version]
- Ron, M.; Alandete, S.M.; Eshed, W.L.; Fletcher, J.C.; McCormick, S. Proper regulation of a sperm-specific cis-nat-siRNA is essential for double fertilization in Arabidopsis. Genes Dev. 2010, 24, 1010–1021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Xia, J.; Lii, Y.E.; Barrera-Figueroa, B.E.; Zhou, X.; Gao, S.; Lu, L.; Niu, D.; Chen, Z.; Leung, C.; et al. Genome-wide analysis of plant nat-siRNAs reveals insights into their distribution, biogenesis and function. Genome Biol. 2012, 13, R20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Lii, Y.; Wu, Z.; Polishko, A.; Zhang, H.; Chinnusamy, V.; Lonardi, S.; Zhu, J.K.; Liu, R.; Jin, H. Mechanisms of small RNA generation from cis-NATs in response to environmental and developmental cues. Mol. Plant 2013, 6, 704–715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blevins, T.; Podicheti, R.; Mishra, V.; Marasco, M.; Wang, J.; Rusch, D.; Tang, H.; Pikaard, C.S. Identification of Pol IV and RDR2-dependent precursors of 24 nt siRNAs guiding de novo DNA methylation in Arabidopsis. eLife 2015, 4, e09591. [Google Scholar] [CrossRef]
- Zhai, J.; Bischof, S.; Wang, H.; Feng, S.; Lee, T.F.; Teng, C.; Chen, X.; Park, S.Y.; Liu, L.; Gallego-Bartolome, J.; et al. A One Precursor One siRNA Model for Pol IV-Dependent siRNA Biogenesis. Cell 2015, 163, 445–455. [Google Scholar] [CrossRef] [Green Version]
- Zhou, M.; Palanca, A.; Law, J.A. Locus-specific control of the de novo DNA methylation pathway in Arabidopsis by the CLASSY family. Nat. Genet. 2018, 50, 865–873. [Google Scholar] [CrossRef]
- Law, J.A.; Jacobsen, S.E. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 2010, 11, 204–220. [Google Scholar] [CrossRef]
- Zhu, L.; Ow, D.W.; Dong, Z. Transfer RNA-derived small RNAs in plants. Sci. China Life Sci. 2018, 61, 155–161. [Google Scholar] [CrossRef]
- Schramm, L.; Hernandez, N. Recruitment of RNA polymerase III to its target promoters. Genes Dev. 2002, 16, 2593–2620. [Google Scholar] [CrossRef] [Green Version]
- Tocchini-Valentini, G.D.; Fruscoloni, P.; Tocchini-Valentini, G.P. Processing of multiple-intron-containing pretRNA. Proc. Natl. Acad. Sci. USA 2009, 106, 20246–20251. [Google Scholar] [CrossRef] [Green Version]
- Ceballos, M.; Vioque, A. tRNase Z. Protein Pept. Lett. 2007, 14, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Frank, D.N.; Pace, N.R. Ribonuclease P: Unity and diversity in a tRNA processing ribozyme. Annu. Rev. Biochem. 1998, 67, 153–180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abelson, J.; Trotta, C.R.; Li, H. tRNA splicing. J. Biol. Chem. 1998, 273, 12685–12688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maute, R.L.; Schneider, C.; Sumazin, P.; Holmes, A.; Califano, A.; Basso, K.; Dalla-Favera, R. tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc. Natl. Acad. Sci. USA 2013, 110, 1404–1409. [Google Scholar] [CrossRef] [Green Version]
- Loss-Morais, G.; Waterhouse, P.M.; Margis, R. Description of plant tRNA-derived RNA fragments (tRFs) associated with argonaute and identification of their putative targets. Biol. Direct 2013, 8, 6. [Google Scholar] [CrossRef] [Green Version]
- Ivanov, P.; Emara, M.M.; Villen, J.; Gygi, S.P.; Anderson, P. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol. Cell 2011, 43, 613–623. [Google Scholar] [CrossRef] [Green Version]
- Nowacka, M.; Strozycki, P.M.; Jackowiak, P.; Hojka-Osinska, A.; Szymanski, M.; Figlerowicz, M. Identification of stable, high copy number, medium-sized RNA degradation intermediates that accumulate in plants under non-stress conditions. Plant Mol. Biol. 2013, 83, 191–204. [Google Scholar] [CrossRef] [Green Version]
- Kolakofsky, D. Isolation and characterization of Sendai virus DI-RNAs. Cell 1976, 8, 547–555. [Google Scholar] [CrossRef]
- Liu, C.X.; Li, X.; Nan, F.; Jiang, S.; Gao, X.; Guo, S.K.; Xue, W.; Cui, Y.; Dong, K.; Ding, H.; et al. Structure and Degradation of Circular RNAs Regulate PKR Activation in Innate Immunity. Cell 2019, 177, 865–880.e21. [Google Scholar] [CrossRef]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Liu, S.; Zhang, L.; Issaian, A.; Hill, R.C.; Espinosa, S.; Shi, S.; Cui, Y.; Kappel, K.; Das, R.; et al. A unified mechanism for intron and exon definition and back-splicing. Nature 2019, 573, 375–380. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Conn, V.M.; Hugouvieux, V.; Nayak, A.; Conos, S.A.; Capovilla, G.; Cildir, G.; Jourdain, A.; Tergaonkar, V.; Schmid, M.; Zubieta, C.; et al. A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat. Plants 2017, 3, 17053. [Google Scholar] [CrossRef] [PubMed]
- Pamudurti, N.R.; Bartok, O.; Jens, M.; Ashwal-Fluss, R.; Stottmeister, C.; Ruhe, L.; Hanan, M.; Wyler, E.; Perez-Hernandez, D.; Ramberger, E.; et al. Translation of CircRNAs. Mol. Cell 2017, 66, 9–21.e7. [Google Scholar] [CrossRef] [Green Version]
- Gao, Z.; Li, J.; Luo, M.; Li, H.; Chen, Q.; Wang, L.; Song, S.; Zhao, L.; Xu, W.; Zhang, C.; et al. Characterization and Cloning of Grape Circular RNAs Identified the Cold Resistance-Related Vv-circATS1. Plant Physiol. 2019, 180, 966–985. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, M.; Wei, S.; Qin, F.; Zhao, H.; Suo, B. Identification of Circular RNAs and Their Targets in Leaves of Triticum aestivum L. under Dehydration Stress. Front. Plant Sci. 2016, 7, 2024. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Lin, J.; Wang, H.; Li, X.; Yang, Q.; Li, H.; Chang, Y. Identification and characterization of circRNAs in Pyrus betulifolia Bunge under drought stress. PLoS ONE 2018, 13, e0200692. [Google Scholar] [CrossRef]
- Zhang, P.; Fan, Y.; Sun, X.; Chen, L.; Terzaghi, W.; Bucher, E.; Li, L.; Dai, M. A large-scale circular RNA profiling reveals universal molecular mechanisms responsive to drought stress in maize and Arabidopsis. Plant J. 2019, 98, 697–713. [Google Scholar] [CrossRef]
- Waititu, J.K.; Zhang, C.; Liu, J.; Wang, H. Plant Non-Coding RNAs: Origin, Biogenesis, Mode of Action and Their Roles in Abiotic Stress. Int. J. Mol. Sci. 2020, 21, 8401. [Google Scholar] [CrossRef]
- Chen, M.; Penfield, S. Feedback regulation of COOLAIR expression controls seed dormancy and flowering time. Science 2018, 360, 1014–1017. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.H.; Sung, S. Coordination of the vernalization response through a VIN3 and FLC gene family regulatory network in Arabidopsis. Plant Cell 2013, 25, 454–469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marquardt, S.; Raitskin, O.; Wu, Z.; Liu, F.; Sun, Q.; Dean, C. Functional consequences of splicing of the antisense transcript COOLAIR on FLC transcription. Mol. Cell 2014, 54, 156–165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, Y.; Xuan, A.; Bu, C.; Ci, D.; Tian, M.; Zhang, D. Osmotic stress-responsive promoter upstream transcripts (PROMPTs) act as carriers of MYB transcription factors to induce the expression of target genes in Populus simonii. Plant Biotechnol. J. 2019, 17, 164–177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bardou, F.; Ariel, F.; Simpson, C.G.; Romero-Barrios, N.; Laporte, P.; Balzergue, S.; Brown, J.W.; Crespi, M. Long noncoding RNA modulates alternative splicing regulators in Arabidopsis. Dev. Cell 2014, 30, 166–176. [Google Scholar] [CrossRef] [Green Version]
- Matsumoto, A.; Nakayama, K.I. Hidden Peptides Encoded by Putative Noncoding RNAs. Cell Struct. Funct. 2018, 43, 75–83. [Google Scholar] [CrossRef] [Green Version]
- Di, C.; Yuan, J.; Wu, Y.; Li, J.; Lin, H.; Hu, L.; Zhang, T.; Qi, Y.; Gerstein, M.B.; Guo, Y.; et al. Characterization of stress-responsive lncRNAs in Arabidopsis thaliana by integrating expression, epigenetic and structural features. Plant J. 2014, 80, 848–861. [Google Scholar] [CrossRef]
- Xin, M.; Wang, Y.; Yao, Y.; Song, N.; Hu, Z.; Qin, D.; Xie, C.; Peng, H.; Ni, Z.; Sun, Q. Identification and characterization of wheat long non-protein coding RNAs responsive to powdery mildew infection and heat stress by using microarray analysis and SBS sequencing. BMC Plant Biol. 2011, 11, 61. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Q.H.; Stephen, S.; Taylor, J.; Helliwell, C.A.; Wang, M.B. Long noncoding RNAs responsive to Fusarium oxysporum infection in Arabidopsis thaliana. New Phytol. 2014, 201, 574–584. [Google Scholar] [CrossRef]
- Wang, J.; Yu, W.; Yang, Y.; Li, X.; Chen, T.; Liu, T.; Ma, N.; Yang, X.; Liu, R.; Zhang, B. Genome-wide analysis of tomato long non-coding RNAs and identification as endogenous target mimic for microRNA in response to TYLCV infection. Sci. Rep. 2015, 5, 16946. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Liu, C. Coding or Noncoding, the Converging Concepts of RNAs. Front. Genet. 2019, 10, 496. [Google Scholar] [CrossRef]
- Fernandes, J.; Acuna, S.M.; Aoki, J.I.; Floeter-Winter, L.M.; Muxel, S.M. Long Non-Coding RNAs in the Regulation of Gene Expression: Physiology and Disease. Non-Coding RNA 2019, 5, 17. [Google Scholar] [CrossRef] [Green Version]
- Shi, T.; Guo, D.; Xu, H.; Su, G.; Chen, J.; Zhao, Z.; Shi, J.; Wedemeyer, M.; Attenello, F.; Zhang, L.; et al. HOTAIRM1, an enhancer lncRNA, promotes glioma proliferation by regulating long-range chromatin interactions within HOXA cluster genes. Mol. Biol. Rep. 2020, 47, 2723–2733. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Lei, C.; He, Q.; Pan, Z.; Xiao, D.; Tao, Y. Nuclear functions of mammalian MicroRNAs in gene regulation, immunity and cancer. Mol. Cancer 2018, 17, 64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamamura, S.; Imai-Sumida, M.; Tanaka, Y.; Dahiya, R. Interaction and cross-talk between non-coding RNAs. Cell. Mol. Life Sci. 2018, 75, 467–484. [Google Scholar] [CrossRef] [PubMed]
- Kazimierczyk, M.; Kasprowicz, M.K.; Kasprzyk, M.E.; Wrzesinski, J. Human Long Noncoding RNA Interactome: Detection, Characterization and Function. Int. J. Mol. Sci. 2020, 21, 1027. [Google Scholar] [CrossRef] [Green Version]
- Grillone, K.; Riillo, C.; Scionti, F.; Rocca, R.; Tradigo, G.; Guzzi, P.H.; Alcaro, S.; Di Martino, M.T.; Tagliaferri, P.; Tassone, P. Non-coding RNAs in cancer: Platforms and strategies for investigating the genomic “dark matter”. J. Exp. Clin. Cancer Res. 2020, 39, 117. [Google Scholar] [CrossRef]
- Liao, W.; Liu, Y.; Huang, H.; Xie, H.; Gong, W.; Liu, D.; Tian, F.; Huang, R.; Yi, F.; Zhou, J. Intersectional analysis of chronic mild stress-induced lncRNA-mRNA interaction networks in rat hippocampus reveals potential anti-depression/anxiety drug targets. Neurobiol. Stress 2021, 15, 100347. [Google Scholar] [CrossRef]
- Dragomir, M.P.; Knutsen, E.; Calin, G.A. SnapShot: Unconventional miRNA Functions. Cell 2018, 174, 1038.e1. [Google Scholar] [CrossRef]
- Hafner, M.; Landthaler, M.; Burger, L.; Khorshid, M.; Hausser, J.; Berninger, P.; Rothballer, A.; Ascano, M.J.; Jungkamp, A.C.; Munschauer, M.; et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 2010, 141, 129–141. [Google Scholar] [CrossRef] [Green Version]
- Chi, S.W.; Zang, J.B.; Mele, A.; Darnell, R.B. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature 2009, 460, 479–486. [Google Scholar] [CrossRef] [Green Version]
- Helwak, A.; Kudla, G.; Dudnakova, T.; Tollervey, D. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell 2013, 153, 654–665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, E.; Sterne-Weiler, T.; O’Hanlon, D.; Blencowe, B.J. Global Mapping of Human RNA-RNA Interactions. Mol. Cell 2016, 62, 618–626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bergeron, D.; Fafard-Couture, E.; Scott, M.S. Small nucleolar RNAs: Continuing identification of novel members and increasing diversity of their molecular mechanisms of action. Biochem. Soc. Trans. 2020, 48, 645–656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef] [Green Version]
- Zhang, P.; Meng, X.; Chen, H.; Liu, Y.; Xue, J.; Zhou, Y.; Chen, M. PlantCircNet: A database for plant circRNA-miRNA-mRNA regulatory networks. Database (Oxford) 2017, 2017, bax089. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, X.; Lin, C.; Jia, X.; Zhu, H.; Song, J.; Zhang, Y. Noncoding RNAs regulate alternative splicing in Cancer. J. Exp. Clin. Cancer Res. 2021, 40, 11. [Google Scholar] [CrossRef]
- Wong, C.M.; Tsang, F.H.; Ng, I.O. Non-coding RNAs in hepatocellular carcinoma: Molecular functions and pathological implications. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 137–151. [Google Scholar] [CrossRef]
- Wang, J.; Zhu, S.; Meng, N.; He, Y.; Lu, R.; Yan, G.R. ncRNA-Encoded Peptides or Proteins and Cancer. Mol. Ther. 2019, 27, 1718–1725. [Google Scholar] [CrossRef]
- Seila, A.C.; Calabrese, J.M.; Levine, S.S.; Yeo, G.W.; Rahl, P.B.; Flynn, R.A.; Young, R.A.; Sharp, P.A. Divergent transcription from active promoters. Science 2008, 322, 1849–1851. [Google Scholar] [CrossRef] [Green Version]
- Post-transcriptional processing generates a diversity of 5’-modified long and short RNAs. Nature 2009, 457, 1028–1032. [CrossRef]
- Taft, R.J.; Simons, C.; Nahkuri, S.; Oey, H.; Korbie, D.J.; Mercer, T.R.; Holst, J.; Ritchie, W.; Wong, J.J.; Rasko, J.E.; et al. Nuclear-localized tiny RNAs are associated with transcription initiation and splice sites in metazoans. Nat. Struct. Mol. Biol. 2010, 17, 1030–1034. [Google Scholar] [CrossRef] [PubMed]
- Taft, R.J.; Glazov, E.A.; Cloonan, N.; Simons, C.; Stephen, S.; Faulkner, G.J.; Lassmann, T.; Forrest, A.R.; Grimmond, S.M.; Schroder, K.; et al. Tiny RNAs associated with transcription start sites in animals. Nat. Genet. 2009, 41, 572–578. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhou, B.; Chen, L.; Gou, L.T.; Li, H.; Fu, X.D. GRID-seq reveals the global RNA-chromatin interactome. Nat. Biotechnol. 2017, 35, 940–950. [Google Scholar] [CrossRef] [PubMed]
- Engreitz, J.M.; Ollikainen, N.; Guttman, M. Long non-coding RNAs: Spatial amplifiers that control nuclear structure and gene expression. Nat. Rev. Mol. Cell. Biol. 2016, 17, 756–770. [Google Scholar] [CrossRef] [Green Version]
- Hasegawa, Y.; Brockdorff, N.; Kawano, S.; Tsutui, K.; Tsutui, K.; Nakagawa, S. The matrix protein hnRNP U is required for chromosomal localization of Xist RNA. Dev. Cell 2010, 19, 469–476. [Google Scholar] [CrossRef] [Green Version]
- Chu, C.; Zhang, Q.C.; Da, R.S.; Flynn, R.A.; Bharadwaj, M.; Calabrese, J.M.; Magnuson, T.; Heard, E.; Chang, H.Y. Systematic discovery of Xist RNA binding proteins. Cell 2015, 161, 404–416. [Google Scholar] [CrossRef] [Green Version]
- West, J.A.; Davis, C.P.; Sunwoo, H.; Simon, M.D.; Sadreyev, R.I.; Wang, P.I.; Tolstorukov, M.Y.; Kingston, R.E. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol. Cell 2014, 55, 791–802. [Google Scholar] [CrossRef] [Green Version]
- Engreitz, J.M.; Pandya-Jones, A.; McDonel, P.; Shishkin, A.; Sirokman, K.; Surka, C.; Kadri, S.; Xing, J.; Goren, A.; Lander, E.S.; et al. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science 2013, 341, 1237973. [Google Scholar] [CrossRef] [Green Version]
- Li, J.H.; Liu, S.; Zhou, H.; Qu, L.H.; Yang, J.H. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014, 42, D92–D97. [Google Scholar] [CrossRef] [Green Version]
- Licatalosi, D.D.; Mele, A.; Fak, J.J.; Ule, J.; Kayikci, M.; Chi, S.W.; Clark, T.A.; Schweitzer, A.C.; Blume, J.E.; Wang, X.; et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature 2008, 456, 464–469. [Google Scholar] [CrossRef] [Green Version]
- Lafontaine, D.L. Noncoding RNAs in eukaryotic ribosome biogenesis and function. Nat. Struct. Mol. Biol. 2015, 22, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Hutvagner, G.; Simard, M.J. Argonaute proteins: Key players in RNA silencing. Nat. Rev. Mol. Cell. Biol. 2008, 9, 22–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyoshi, T.; Ito, K.; Murakami, R.; Uchiumi, T. Structural basis for the recognition of guide RNA and target DNA heteroduplex by Argonaute. Nat. Commun. 2016, 7, 11846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schraivogel, D.; Meister, G. Import routes and nuclear functions of Argonaute and other small RNA-silencing proteins. Trends Biochem. Sci. 2014, 39, 420–431. [Google Scholar] [CrossRef] [PubMed]
- Hegge, J.W.; Swarts, D.C.; Chandradoss, S.D.; Cui, T.J.; Kneppers, J.; Jinek, M.; Joo, C.; van der Oost, J. DNA-guided DNA cleavage at moderate temperatures by Clostridium butyricum Argonaute. Nucleic Acids Res. 2019, 47, 5809–5821. [Google Scholar] [CrossRef] [Green Version]
- Kuhn, C.D.; Joshua-Tor, L. Eukaryotic Argonautes come into focus. Trends Biochem. Sci. 2013, 38, 263–271. [Google Scholar] [CrossRef]
- Long, Y.; Wang, X.; Youmans, D.T.; Cech, T.R. How do lncRNAs regulate transcription? Sci. Adv. 2017, 3, eaao2110. [Google Scholar] [CrossRef] [Green Version]
- Rigo, R.; Bazin, J.; Romero-Barrios, N.; Moison, M.; Lucero, L.; Christ, A.; Benhamed, M.; Blein, T.; Huguet, S.; Charon, C.; et al. The Arabidopsis lncRNA ASCO modulates the transcriptome through interaction with splicing factors. Embo Rep. 2020, 21, e48977. [Google Scholar] [CrossRef]
- Cook, D.E.; Mesarich, C.H.; Thomma, B.P. Understanding plant immunity as a surveillance system to detect invasion. Annu. Rev. Phytopathol. 2015, 53, 541–563. [Google Scholar] [CrossRef]
- Jones, J.D.; Vance, R.E.; Dangl, J.L. Intracellular innate immune surveillance devices in plants and animals. Science 2016, 354, aaf6395. [Google Scholar] [CrossRef] [Green Version]
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Q.; Wang, Z.; Zhang, J.; Meng, H.; Huang, B. Genome-wide identification and profiling of microRNA-like RNAs from Metarhizium anisopliae during development. Fungal Biol. 2012, 116, 1156–1162. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Fu, Y.; Xie, J.; Li, B.; Jiang, D.; Li, G.; Cheng, J. Identification of microRNA-like RNAs in a plant pathogenic fungus Sclerotinia sclerotiorum by high-throughput sequencing. Mol. Genet. Genom. 2012, 287, 275–282. [Google Scholar] [CrossRef]
- Qiao, Y.; Liu, L.; Xiong, Q.; Flores, C.; Wong, J.; Shi, J.; Wang, X.; Liu, X.; Xiang, Q.; Jiang, S.; et al. Oomycete pathogens encode RNA silencing suppressors. Nat. Genet. 2013, 45, 330–333. [Google Scholar] [CrossRef] [Green Version]
- Niu, D.; Lii, Y.E.; Chellappan, P.; Lei, L.; Peralta, K.; Jiang, C.; Guo, J.; Coaker, G.; Jin, H. miRNA863-3p sequentially targets negative immune regulator ARLPKs and positive regulator SERRATE upon bacterial infection. Nat. Commun. 2016, 7, 11324. [Google Scholar] [CrossRef] [Green Version]
- Cai, Q.; Liang, C.; Wang, S.; Hou, Y.; Gao, L.; Liu, L.; He, W.; Ma, W.; Mo, B.; Chen, X. The disease resistance protein SNC1 represses the biogenesis of microRNAs and phased siRNAs. Nat. Commun. 2018, 9, 5080. [Google Scholar] [CrossRef]
- Weiberg, A.; Wang, M.; Bellinger, M.; Jin, H. Small RNAs: A new paradigm in plant-microbe interactions. Annu. Rev. Phytopathol. 2014, 52, 495–516. [Google Scholar] [CrossRef]
- Zhang, H.; Tao, Z.; Hong, H.; Chen, Z.; Wu, C.; Li, X.; Xiao, J.; Wang, S. Transposon-derived small RNA is responsible for modified function of WRKY45 locus. Nat. Plants 2016, 2, 16016. [Google Scholar] [CrossRef]
- Deng, Y.; Zhai, K.; Xie, Z.; Yang, D.; Zhu, X.; Liu, J.; Wang, X.; Qin, P.; Yang, Y.; Zhang, G.; et al. Epigenetic regulation of antagonistic receptors confers rice blast resistance with yield balance. Science 2017, 355, 962–965. [Google Scholar] [CrossRef]
- Zhang, X.; Zhao, H.; Gao, S.; Wang, W.C.; Katiyar-Agarwal, S.; Huang, H.D.; Raikhel, N.; Jin, H. Arabidopsis Argonaute 2 regulates innate immunity via miRNA393(*)-mediated silencing of a Golgi-localized SNARE gene, MEMB12. Mol. Cell 2011, 42, 356–366. [Google Scholar] [CrossRef] [Green Version]
- Yan, J.; Cai, X.; Luo, J.; Sato, S.; Jiang, Q.; Yang, J.; Cao, X.; Hu, X.; Tabata, S.; Gresshoff, P.M.; et al. The REDUCED LEAFLET genes encode key components of the trans-acting small interfering RNA pathway and regulate compound leaf and flower development in Lotus japonicus. Plant Physiol. 2010, 152, 797–807. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Lei, M.; Yan, Z.; Wang, Q.; Chen, A.; Sun, J.; Luo, D.; Wang, Y. The REL3-mediated TAS3 ta-siRNA pathway integrates auxin and ethylene signaling to regulate nodulation in Lotus japonicus. New Phytol. 2014, 201, 531–544. [Google Scholar] [CrossRef] [PubMed]
- Kuang, Z.; Wang, Y.; Li, L.; Yang, X. miRDeep-P2: Accurate and fast analysis of the microRNA transcriptome in plants. Bioinformatics 2019, 35, 2521–2522. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Ding, C.; Lang, X.; Guo, G.; Chen, J.; Su, X. Small noncoding RNA discovery and profiling with sRNAtools based on high-throughput sequencing. Brief. Bioinform. 2021, 22, 463–473. [Google Scholar] [CrossRef] [Green Version]
- Patel, P.; Mathioni, S.; Kakrana, A.; Shatkay, H.; Meyers, B.C. Reproductive phasiRNAs in grasses are compositionally distinct from other classes of small RNAs. New Phytol. 2018, 220, 851–864. [Google Scholar] [CrossRef] [Green Version]
- Thody, J.; Folkes, L.; Moulton, V. NATpare: A pipeline for high-throughput prediction and functional analysis of nat-siRNAs. Nucleic Acids Res. 2020, 48, 6481–6490. [Google Scholar] [CrossRef]
- Wang, Q.; Li, T.; Xu, K.; Zhang, W.; Wang, X.; Quan, J.; Jin, W.; Zhang, M.; Fan, G.; Wang, M.B.; et al. The tRNA-Derived Small RNAs Regulate Gene Expression through Triggering Sequence-Specific Degradation of Target Transcripts in the Oomycete Pathogen Phytophthora sojae. Front. Plant Sci. 2016, 7, 1938. [Google Scholar] [CrossRef] [Green Version]
- Negri, T.; Alves, W.; Bugatti, P.H.; Saito, P.; Domingues, D.S.; Paschoal, A.R. Pattern recognition analysis on long noncoding RNAs: A tool for prediction in plants. Brief. Bioinform. 2019, 20, 682–689. [Google Scholar] [CrossRef]
- Wucher, V.; Legeai, F.; Hedan, B.; Rizk, G.; Lagoutte, L.; Leeb, T.; Jagannathan, V.; Cadieu, E.; David, A.; Lohi, H.; et al. FEELnc: A tool for long non-coding RNA annotation and its application to the dog transcriptome. Nucleic Acids Res. 2017, 45, e57. [Google Scholar] [CrossRef] [Green Version]
- Prada-Luengo, I.; Krogh, A.; Maretty, L.; Regenberg, B. Sensitive detection of circular DNAs at single-nucleotide resolution using guided realignment of partially aligned reads. BMC Bioinform. 2019, 20, 663. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Chu, C.; Pei, J.; Mandoiu, I.; Wu, Y. CircMarker: A fast and accurate algorithm for circular RNA detection. BMC Genom. 2018, 19, 572. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Liu, Q.; Wang, X.; Zheng, J.; Wang, T.; You, M.; Sheng, S.Z.; Shi, Q. mirTools 2.0 for non-coding RNA discovery, profiling, and functional annotation based on high-throughput sequencing. RNA Biol. 2013, 10, 1087–1092. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stocks, M.B.; Mohorianu, I.; Beckers, M.; Paicu, C.; Moxon, S.; Thody, J.; Dalmay, T.; Moulton, V. The UEA sRNA Workbench (version 4.4): A comprehensive suite of tools for analyzing miRNAs and sRNAs. Bioinformatics 2018, 34, 3382–3384. [Google Scholar] [CrossRef]
- Zhang, Y.; Leclercq, J.; Wu, S.; Ortega-Abboud, E.; Pointet, S.; Tang, C.; Hu, S.; Montoro, P. Genome-wide analysis in Hevea brasiliensis laticifers revealed species-specific post-transcriptional regulations of several redox-related genes. Sci. Rep. 2019, 9, 5701. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Wan, Y.; Ito, H.; Ma, X.; Xie, T.; Wang, T.; Shao, C.; Meng, Y. PmiRDiscVali: An integrated pipeline for plant microRNA discovery and validation. BMC Genom. 2019, 20, 133. [Google Scholar] [CrossRef]
- An, J.; Lai, J.; Sajjanhar, A.; Lehman, M.L.; Nelson, C.C. miRPlant: An integrated tool for identification of plant miRNA from RNA sequencing data. BMC Bioinform. 2014, 15, 275. [Google Scholar] [CrossRef] [Green Version]
- Vitsios, D.M.; Enright, A.J. Chimira: Analysis of small RNA sequencing data and microRNA modifications. Bioinformatics 2015, 31, 3365–3367. [Google Scholar] [CrossRef] [Green Version]
- Sun, Z.; Evans, J.; Bhagwate, A.; Middha, S.; Bockol, M.; Yan, H.; Kocher, J.P. CAP-miRSeq: A comprehensive analysis pipeline for microRNA sequencing data. BMC Genom. 2014, 15, 423. [Google Scholar] [CrossRef] [Green Version]
- Vitsios, D.M.; Kentepozidou, E.; Quintais, L.; Benito-Gutierrez, E.; van Dongen, S.; Davis, M.P.; Enright, A.J. Mirnovo: Genome-free prediction of microRNAs from small RNA sequencing data and single-cells using decision forests. Nucleic Acids Res. 2017, 45, e177. [Google Scholar] [CrossRef]
- Tseng, K.C.; Chiang-Hsieh, Y.F.; Pai, H.; Chow, C.N.; Lee, S.C.; Zheng, H.Q.; Kuo, P.L.; Li, G.Z.; Hung, Y.C.; Lin, N.S.; et al. microRPM: A microRNA prediction model based only on plant small RNA sequencing data. Bioinformatics 2018, 34, 1108–1115. [Google Scholar] [CrossRef]
- Paicu, C.; Mohorianu, I.; Stocks, M.; Xu, P.; Coince, A.; Billmeier, M.; Dalmay, T.; Moulton, V.; Moxon, S. miRCat2: Accurate prediction of plant and animal microRNAs from next-generation sequencing datasets. Bioinformatics 2017, 33, 2446–2454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, X.; Li, L. miRDeep-P: A computational tool for analyzing the microRNA transcriptome in plants. Bioinformatics 2011, 27, 2614–2615. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Kim, T.K.; Baxter, D.; Scherler, K.; Gordon, A.; Fong, O.; Etheridge, A.; Galas, D.J.; Wang, K. sRNAnalyzer-a flexible and customizable small RNA sequencing data analysis pipeline. Nucleic Acids Res. 2017, 45, 12140–12151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Vieira, R.E.S.B.; Cui, J. miRDis: A Web tool for endogenous and exogenous microRNA discovery based on deep-sequencing data analysis. Brief. Bioinform. 2018, 19, 415–424. [Google Scholar] [CrossRef] [Green Version]
- Evers, M.; Huttner, M.; Dueck, A.; Meister, G.; Engelmann, J.C. miRA: Adaptable novel miRNA identification in plants using small RNA sequencing data. BMC Bioinform. 2015, 16, 370. [Google Scholar] [CrossRef] [Green Version]
- Yu, L.; Shao, C.; Ye, X.; Meng, Y.; Zhou, Y.; Chen, M. miRNA Digger: A comprehensive pipeline for genome-wide novel miRNA mining. Sci. Rep. 2016, 6, 18901. [Google Scholar] [CrossRef] [Green Version]
- Lei, J.; Sun, Y. miR-PREFeR: An accurate, fast and easy-to-use plant miRNA prediction tool using small RNA-Seq data. Bioinformatics 2014, 30, 2837–2839. [Google Scholar] [CrossRef] [Green Version]
- Douglass, S.; Hsu, S.W.; Cokus, S.; Goldberg, R.B.; Harada, J.J.; Pellegrini, M. A naive Bayesian classifier for identifying plant microRNAs. Plant J. 2016, 86, 481–492. [Google Scholar] [CrossRef] [Green Version]
- Higashi, S.; Fournier, C.; Gautier, C.; Gaspin, C.; Sagot, M.F. Mirinho: An efficient and general plant and animal pre-miRNA predictor for genomic and deep sequencing data. BMC Bioinform. 2015, 16, 179. [Google Scholar] [CrossRef]
- Kuenne, C.; Preussner, J.; Herzog, M.; Braun, T.; Looso, M. MIRPIPE: Quantification of microRNAs in niche model organisms. Bioinformatics 2014, 30, 3412–3413. [Google Scholar] [CrossRef] [Green Version]
- Chae, H.; Rhee, S.; Nephew, K.P.; Kim, S. BioVLAB-MMIA-NGS: microRNA-mRNA integrated analysis using high-throughput sequencing data. Bioinformatics 2015, 31, 265–267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Jiang, L.; Wang, J.; Gu, P.; Chen, M. MTide: An integrated tool for the identification of miRNA-target interaction in plants. Bioinformatics 2015, 31, 290–291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qian, K.; Auvinen, E.; Greco, D.; Auvinen, P. miRSeqNovel: An R based workflow for analyzing miRNA sequencing data. Mol. Cell. Probes 2012, 26, 208–211. [Google Scholar] [CrossRef]
- Jha, A.; Shankar, R. miReader: Discovering Novel miRNAs in Species without Sequenced Genome. PLoS ONE 2013, 8, e66857. [Google Scholar] [CrossRef] [Green Version]
- Yuan, T.; Huang, X.; Dittmar, R.L.; Du, M.; Kohli, M.; Boardman, L.; Thibodeau, S.N.; Wang, L. eRNA: A graphic user interface-based tool optimized for large data analysis from high-throughput RNA sequencing. BMC Genom. 2014, 15, 176. [Google Scholar] [CrossRef] [Green Version]
- Patra, D.; Fasold, M.; Langenberger, D.; Steger, G.; Grosse, I.; Stadler, P.F. plantDARIO: Web based quantitative and qualitative analysis of small RNA-seq data in plants. Front. Plant Sci. 2014, 5, 708. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Oliveira, L.F.; Christoff, A.P.; Margis, R. isomiRID: A framework to identify microRNA isoforms. Bioinformatics 2013, 29, 2521–2523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sablok, G.; Milev, I.; Minkov, G.; Minkov, I.; Varotto, C.; Yahubyan, G.; Baev, V. isomiRex: Web-based identification of microRNAs, isomiR variations and differential expression using next-generation sequencing datasets. FEBS Lett. 2013, 587, 2629–2634. [Google Scholar] [CrossRef] [Green Version]
- Mathelier, A.; Carbone, A. MIReNA: Finding microRNAs with high accuracy and no learning at genome scale and from deep sequencing data. Bioinformatics 2010, 26, 2226–2234. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Kim, D.I.; Park, J.H.; Choi, I.Y.; Shin, C. MiRAuto: An automated user-friendly microRNA prediction tool utilizing plant small RNA sequencing data. Mol. Cells 2013, 35, 342–347. [Google Scholar] [CrossRef] [Green Version]
- Mapleson, D.; Moxon, S.; Dalmay, T.; Moulton, V. MirPlex: A tool for identifying miRNAs in high-throughput sRNA datasets without a genome. J. Exp. Zool. B Mol. Dev. Evol. 2013, 320, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Breakfield, N.W.; Corcoran, D.L.; Petricka, J.J.; Shen, J.; Sae-Seaw, J.; Rubio-Somoza, I.; Weigel, D.; Ohler, U.; Benfey, P.N. High-resolution experimental and computational profiling of tissue-specific known and novel miRNAs in Arabidopsis. Genome Res. 2012, 22, 163–176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, F.; Xiao, P.; Chen, D.; Xu, L.; Zhang, B. miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 2012, 80, 75–84. [Google Scholar] [CrossRef]
- Hunt, M.; Banerjee, S.; Surana, P.; Liu, M.; Fuerst, G.; Mathioni, S.; Meyers, B.C.; Nettleton, D.; Wise, R.P. Small RNA discovery in the interaction between barley and the powdery mildew pathogen. BMC Genom. 2019, 20, 610. [Google Scholar] [CrossRef]
- Guo, Q.; Qu, X.; Jin, W. PhaseTank: Genome-wide computational identification of phasiRNAs and their regulatory cascades. Bioinformatics 2015, 31, 284–286. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Liberman, L.M.; Mukherjee, N.; Benfey, P.N.; Ohler, U. Integrated detection of natural antisense transcripts using strand-specific RNA sequencing data. Genome Res. 2013, 23, 1730–1739. [Google Scholar] [CrossRef] [Green Version]
- Yu, D.; Meng, Y.; Zuo, Z.; Xue, J.; Wang, H. NATpipe: An integrative pipeline for systematical discovery of natural antisense transcripts (NATs) and phase-distributed nat-siRNAs from de novo assembled transcriptomes. Sci. Rep. 2016, 6, 21666. [Google Scholar] [CrossRef] [Green Version]
- Dai, X.; Zhao, P.X. pssRNAMiner: A plant short small RNA regulatory cascade analysis server. Nucleic Acids Res. 2008, 36, W114–W118. [Google Scholar] [CrossRef] [Green Version]
- Meng, J.; Kang, Q.; Chang, Z.; Luan, Y. PlncRNA-HDeep: Plant long noncoding RNA prediction using hybrid deep learning based on two encoding styles. BMC Bioinform. 2021, 22, 242. [Google Scholar] [CrossRef]
- Simopoulos, C.; Weretilnyk, E.A.; Golding, G.B. Prediction of plant lncRNA by ensemble machine learning classifiers. BMC Genom. 2018, 19, 316. [Google Scholar] [CrossRef] [Green Version]
- Singh, U.; Khemka, N.; Rajkumar, M.S.; Garg, R.; Jain, M. PLncPRO for prediction of long non-coding RNAs (lncRNAs) in plants and its application for discovery of abiotic stress-responsive lncRNAs in rice and chickpea. Nucleic Acids Res. 2017, 45, e183. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.C.; Fang, S.S.; Wu, Y.; Zhang, J.H.; Chen, Y.; Liu, J.; Wu, B.; Wu, J.R.; Li, E.M.; Xu, L.Y.; et al. CNIT: A fast and accurate web tool for identifying protein-coding and long non-coding transcripts based on intrinsic sequence composition. Nucleic Acids Res. 2019, 47, W516–W522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nelson, A.; Devisetty, U.K.; Palos, K.; Haug-Baltzell, A.K.; Lyons, E.; Beilstein, M.A. Evolinc: A Tool for the Identification and Evolutionary Comparison of Long Intergenic Non-coding RNAs. Front. Genet. 2017, 8, 52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deshpande, S.; Shuttleworth, J.; Yang, J.; Taramonli, S.; England, M. PLIT: An alignment-free computational tool for identification of long non-coding RNAs in plant transcriptomic datasets. Comput Biol Med 2019, 105, 169–181. [Google Scholar] [CrossRef] [Green Version]
- Gong, Y.; Huang, H.T.; Liang, Y.; Trimarchi, T.; Aifantis, I.; Tsirigos, A. lncRNA-screen: An interactive platform for computationally screening long non-coding RNAs in large genomics datasets. BMC Genom. 2017, 18, 434. [Google Scholar] [CrossRef] [Green Version]
- Sun, P.; Wang, H.; Li, G. Rcirc: An R Package for circRNA Analyses and Visualization. Front. Genet. 2020, 11, 548. [Google Scholar] [CrossRef]
- Humphreys, D.T.; Fossat, N.; Demuth, M.; Tam, P.; Ho, J. Ularcirc: Visualization and enhanced analysis of circular RNAs via back and canonical forward splicing. Nucleic Acids Res. 2019, 47, e123. [Google Scholar] [CrossRef]
- Gaffo, E.; Bonizzato, A.; Kronnie, G.T.; Bortoluzzi, S. CirComPara: A Multi-Method Comparative Bioinformatics Pipeline to Detect and Study circRNAs from RNA-seq Data. Non-Coding RNA 2017, 3, 8. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Zhang, X.O.; Chen, T.; Xiang, J.F.; Yin, Q.F.; Xing, Y.H.; Zhu, S.; Yang, L.; Chen, L.L. Circular intronic long noncoding RNAs. Mol. Cell 2013, 51, 792–806. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Yu, Y.; Zhang, X.; Liu, C.; Ye, C.; Fan, L. PcircRNA_finder: A software for circRNA prediction in plants. Bioinformatics 2016, 32, 3528–3529. [Google Scholar] [CrossRef] [Green Version]
- You, X.; Conrad, T.O. Acfs: Accurate circRNA identification and quantification from RNA-Seq data. Sci. Rep. 2016, 6, 38820. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Kuang, Z.; Wang, Y.; Zhao, Y.; Tao, Y.; Cheng, C.; Yang, J.; Lu, X.; Hao, C.; Wang, T.; et al. PmiREN: A comprehensive encyclopedia of plant miRNAs. Nucleic Acids Res. 2020, 48, D1114–D1121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, D.; Lu, J.; Shao, W.; Ma, X.; Xie, T.; Ito, H.; Wang, T.; Xu, M.; Wang, H.; Meng, Y. MepmiRDB: A medicinal plant microRNA database. Database (Oxford) 2019, 2019, baz070. [Google Scholar] [CrossRef] [PubMed]
- Di Marsico, M.; Paytuvi, G.A.; Sanseverino, W.; Aiese, C.R. GreeNC 2.0: A comprehensive database of plant long non-coding RNAs. Nucleic Acids Res. 2022, 50, D1442–D1447. [Google Scholar] [CrossRef] [PubMed]
- Paytuvi, G.A.; Hermoso, P.A.; Anzar, M.D.L.I.; Sanseverino, W.; Aiese, C.R. GREENC: A Wiki-based database of plant lncRNAs. Nucleic Acids Res. 2016, 44, D1161–D1166. [Google Scholar] [CrossRef]
- Ye, J.; Wang, L.; Li, S.; Zhang, Q.; Zhang, Q.; Tang, W.; Wang, K.; Song, K.; Sablok, G.; Sun, X.; et al. AtCircDB: A tissue-specific database for Arabidopsis circular RNAs. Brief. Bioinform. 2019, 20, 58–65. [Google Scholar] [CrossRef]
- Yang, K.; Wen, X.; Mudunuri, S.B.; Sablok, G. Plant IsomiR Atlas: Large Scale Detection, Profiling, and Target Repertoire of IsomiRs in Plants. Front. Plant Sci. 2018, 9, 1881. [Google Scholar] [CrossRef] [Green Version]
- Yang, K.; Wen, X.; Mudunuri, S.; Varma, G.; Sablok, G. Diff isomiRs: Large-scale detection of differential isomiRs for understanding non-coding regulated stress omics in plants. Sci. Rep. 2019, 9, 1406. [Google Scholar] [CrossRef] [Green Version]
- Griffiths-Jones, S. The microRNA Registry. Nucleic Acids Res. 2004, 32, D109–D111. [Google Scholar] [CrossRef]
- Griffiths-Jones, S.; Grocock, R.J.; van Dongen, S.; Bateman, A.; Enright, A.J. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006, 34, D140–D144. [Google Scholar] [CrossRef]
- Kozomara, A.; Griffiths-Jones, S. miRBase: Integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011, 39, D152–D157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kozomara, A.; Griffiths-Jones, S. miRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014, 42, D68–D73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef] [PubMed]
- Zielezinski, A.; Dolata, J.; Alaba, S.; Kruszka, K.; Pacak, A.; Swida-Barteczka, A.; Knop, K.; Stepien, A.; Bielewicz, D.; Pietrykowska, H.; et al. mirEX 2.0—An integrated environment for expression profiling of plant microRNAs. BMC Plant Biol. 2015, 15, 144. [Google Scholar] [CrossRef] [Green Version]
- Bielewicz, D.; Dolata, J.; Zielezinski, A.; Alaba, S.; Szarzynska, B.; Szczesniak, M.W.; Jarmolowski, A.; Szweykowska-Kulinska, Z.; Karlowski, W.M. mirEX: A platform for comparative exploration of plant pri-miRNA expression data. Nucleic Acids Res. 2012, 40, D191–D197. [Google Scholar] [CrossRef] [Green Version]
- Szczesniak, M.W.; Deorowicz, S.; Gapski, J.; Kaczynski, L.; Makalowska, I. miRNEST database: An integrative approach in microRNA search and annotation. Nucleic Acids Res. 2012, 40, D198–D204. [Google Scholar] [CrossRef]
- Szczesniak, M.W.; Makalowska, I. miRNEST 2.0: A database of plant and animal microRNAs. Nucleic Acids Res. 2014, 42, D74–D77. [Google Scholar] [CrossRef] [Green Version]
- Johnson, C.; Bowman, L.; Adai, A.T.; Vance, V.; Sundaresan, V. CSRDB: A small RNA integrated database and browser resource for cereals. Nucleic Acids Res. 2007, 35, D829–D833. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Li, J.; Feng, J.; Liu, B.; Feng, L.; Yu, X.; Li, G.; Zhai, J.; Meyers, B.C.; Xia, R. sRNAanno-a database repository of uniformly annotated small RNAs in plants. Hortic. Res. 2021, 8, 45. [Google Scholar] [CrossRef]
- Lunardon, A.; Johnson, N.R.; Hagerott, E.; Phifer, T.; Polydore, S.; Coruh, C.; Axtell, M.J. Integrated annotations and analyses of small RNA-producing loci from 47 diverse plants. Genome Res. 2020, 30, 497–513. [Google Scholar] [CrossRef]
- Yi, X.; Zhang, Z.; Ling, Y.; Xu, W.; Su, Z. PNRD: A plant non-coding RNA database. Nucleic Acids Res. 2015, 43, D982–D989. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.; Li, G.; Zhu, S.; Zhang, S.; Fang, J. tasiRNAdb: A database of ta-siRNA regulatory pathways. Bioinformatics 2014, 30, 1045–1046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, N.; Singh, A.; Zahra, S.; Kumar, S. PtRFdb: A database for plant transfer RNA-derived fragments. Database (Oxford) 2018, 2018, bay063. [Google Scholar] [CrossRef]
- Thompson, A.; Zielezinski, A.; Plewka, P.; Szymanski, M.; Nuc, P.; Szweykowska-Kulinska, Z.; Jarmolowski, A.; Karlowski, W.M. tRex: A Web Portal for Exploration of tRNA-Derived Fragments in Arabidopsis thaliana. Plant Cell Physiol. 2018, 59, e1. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Lu, P.; Xu, Y.; Li, Z.; Yu, S.; Liu, J.; Wang, H.; Chua, N.H.; Cao, P. PLncDB V2.0: A comprehensive encyclopedia of plant long noncoding RNAs. Nucleic Acids Res. 2021, 49, D1489–D1495. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Liu, J.; Wang, H.; Wong, L.; Chua, N.H. PLncDB: Plant long non-coding RNA database. Bioinformatics 2013, 29, 1068–1071. [Google Scholar] [CrossRef] [Green Version]
- Szczesniak, M.W.; Bryzghalov, O.; Ciomborowska-Basheer, J.; Makalowska, I. CANTATAdb 2.0: Expanding the Collection of Plant Long Noncoding RNAs. Methods Mol. Biol. 2019, 1933, 415–429. [Google Scholar] [CrossRef]
- Szczesniak, M.W.; Rosikiewicz, W.; Makalowska, I. CANTATAdb: A Collection of Plant Long Non-Coding RNAs. Plant Cell Physiol. 2016, 57, e8. [Google Scholar] [CrossRef] [Green Version]
- Shao, Y.; Wei, J.; Wu, F.; Zhang, H.; Yang, D.; Liang, Z.; Jin, W. DsTRD: Danshen Transcriptional Resource Database. PLoS ONE 2016, 11, e0149747. [Google Scholar] [CrossRef] [Green Version]
- Xuan, H.; Zhang, L.; Liu, X.; Han, G.; Li, J.; Li, X.; Liu, A.; Liao, M.; Zhang, S. PLNlncRbase: A resource for experimentally identified lncRNAs in plants. Gene 2015, 573, 328–332. [Google Scholar] [CrossRef]
- Meng, X.; Hu, D.; Zhang, P.; Chen, Q.; Chen, M. CircFunBase: A database for functional circular RNAs. Database 2019, 2019, baz003. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.; Wang, C.; Guo, B.; Song, K.; Shi, C.; Jiang, X.; Wang, K.; Tan, Y.; Wang, L.; Wang, L.; et al. CropCircDB: A comprehensive circular RNA resource for crops in response to abiotic stress. Database 2019, 2019, baz053. [Google Scholar] [CrossRef] [Green Version]
- Chu, Q.; Bai, P.; Zhu, X.; Zhang, X.; Mao, L.; Zhu, Q.H.; Fan, L.; Ye, C.Y. Characteristics of plant circular RNAs. Brief. Bioinform. 2018, 21, 135–143. [Google Scholar] [CrossRef]
- Chu, Q.; Zhang, X.; Zhu, X.; Liu, C.; Mao, L.; Ye, C.; Zhu, Q.H.; Fan, L. PlantcircBase: A Database for Plant Circular RNAs. Mol. Plant 2017, 10, 1126–1128. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Park, H.J.; Dasari, S.; Wang, S.; Kocher, J.P.; Li, W. CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013, 41, e74. [Google Scholar] [CrossRef] [PubMed]
- Gawronski, K.; Kim, J. Single cell transcriptomics of noncoding RNAs and their cell-specificity. Wiley Interdiscip. Rev. RNA 2017, 8, e1433. [Google Scholar] [CrossRef]
- Gao, Y.; Shang, S.; Guo, S.; Li, X.; Zhou, H.; Liu, H.; Sun, Y.; Wang, J.; Wang, P.; Zhi, H.; et al. Lnc2Cancer 3.0: An updated resource for experimentally supported lncRNA/circRNA cancer associations and web tools based on RNA-seq and scRNA-seq data. Nucleic Acids Res. 2021, 49, D1251–D1258. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.L.; Xiong, J.H.; Zheng, W.J.; Wang, J.H.; Huang, Z.L.; Chen, Z.R.; Sun, X.Y.; Zheng, Y.M.; Zhou, K.R.; Li, B.; et al. ColorCells: A database of expression, classification and functions of lncRNAs in single cells. Brief. Bioinform. 2021, 22, bbaa325. [Google Scholar] [CrossRef]
- Luo, H.; Bu, D.; Shao, L.; Li, Y.; Sun, L.; Wang, C.; Wang, J.; Yang, W.; Yang, X.; Dong, J.; et al. Single-cell Long Non-coding RNA Landscape of T Cells in Human Cancer Immunity. Genom. Proteom. Bioinform. 2021, 19, 377–393. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).