Behavioral Neuroscience in the Era of Genomics: Tools and Lessons for Analyzing High-Dimensional Datasets
Abstract
:1. Introduction
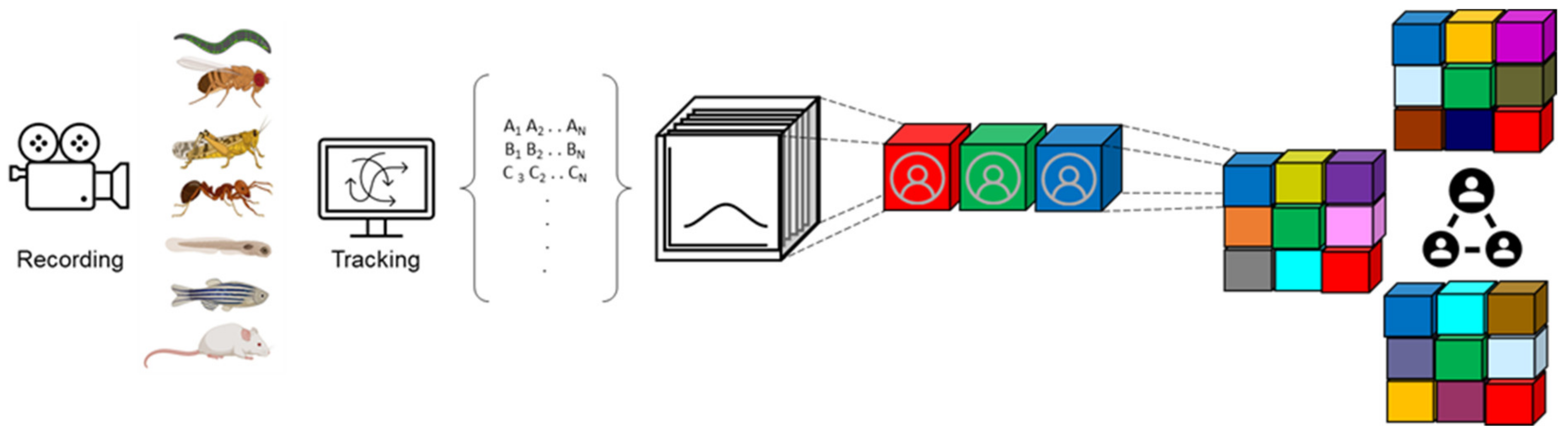
2. Social Behavior Generates High-Dimensional Datasets
3. Both Behavioral and Genomic Datasets Are High-Dimensional
4. Technological Advances in Other Fields Generate Large Datasets
5. Similarities and Differences between Genomics and Behavior Data
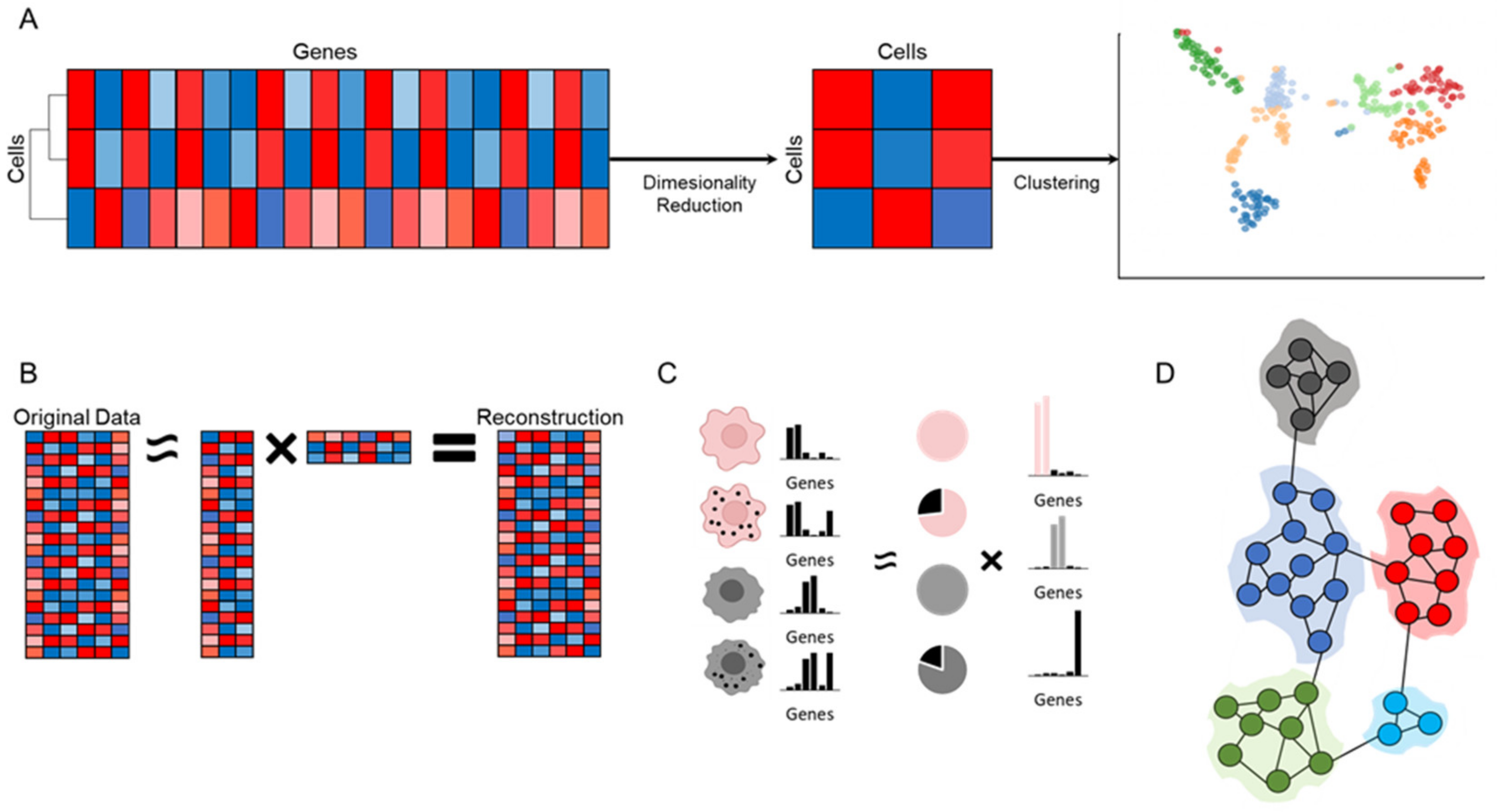
6. Dimensionality Reduction
7. Clustering Analysis
8. Variance as a Tool to Investigate Behavioral Phenomena
9. The Concept of Individuality and Group Identity
10. Variance and Individuality in Genomics
11. Future Perspective and Challenges
Funding
Conflicts of Interest
References
- Cande, J.; Namiki, S.; Qiu, J.; Korff, W.; Card, G.M.; Shaevitz, J.W.; Stern, D.L.; Berman, G.J. Optogenetic dissection of descending behavioral control in Drosophila. eLife 2018, 7, e34275. [Google Scholar] [CrossRef] [PubMed]
- Roemschied, F.A.; Pacheco, D.A.; Ireland, E.C.; Li, X.; Aragon, M.J.; Pang, R.; Murthy, M. Flexible Circuit Mechanisms for Context-Dependent Song Sequencing. bioRxiv 2021. [Google Scholar] [CrossRef]
- Dolensek, N.; Gehrlach, D.A.; Klein, A.S.; Gogolla, N. Facial expressions of emotion states and their neuronal correlates in mice. Science 2020, 368, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Graving, J.M.; Chae, D.; Naik, H.; Li, L.; Koger, B.; Costelloe, B.R.; Couzin, I.D. DeepPoseKit, a software toolkit for fast and robust animal pose estimation using deep learning. eLife 2019, 8, e47994. [Google Scholar] [CrossRef] [PubMed]
- Robie, A.A.; Hirokawa, J.; Edwards, A.W.; Umayam, L.A.; Lee, A.; Phillips, M.L.; Card, G.M.; Korff, W.; Rubin, G.M.; Simpson, J.H.; et al. Mapping the Neural Substrates of Behavior. Cell 2017, 170, 393–406.e28. [Google Scholar] [CrossRef] [Green Version]
- Ariel, G.; Ayali, A. Locust Collective Motion and Its Modeling. PLoS Comput. Biol. 2015, 11, e1004522. [Google Scholar] [CrossRef] [Green Version]
- Shemesh, Y.; Sztainberg, Y.; Forkosh, O.; Shlapobersky, T.; Chen, A.; Schneidman, E. High-order social interactions in groups of mice. eLife 2013, 2, e00759. [Google Scholar] [CrossRef]
- Karamihalev, S.; Brivio, E.; Flachskamm, C.; Stoffel, R.; Schmidt, M.V.; Chen, A. Social dominance mediates behavioral adaptation to chronic stress in a sex-specific manner. eLife 2020, 9, e58723. [Google Scholar] [CrossRef]
- Elliott, E.; Ezra-Nevo, G.; Regev, L.; Neufeld-Cohen, A.; Chen, A. Resilience to social stress coincides with functional DNA methylation of the Crf gene in adult mice. Nat. Neurosci. 2010, 13, 1351–1353. [Google Scholar] [CrossRef]
- Jolles, J.W.; Boogert, N.J.; Sridhar, V.H.; Couzin, I.D.; Manica, A. Consistent Individual Differences Drive Collective Behavior and Group Functioning of Schooling Fish. Curr. Biol. 2017, 27, 2862–2868.e7. [Google Scholar] [CrossRef] [Green Version]
- Rosenthal, S.B.; Twomey, C.R.; Hartnett, A.T.; Wu, H.S.; Couzin, I.D. Revealing the hidden networks of interaction in mobile animal groups allows prediction of complex behavioral contagion. Proc. Natl. Acad. Sci. USA 2015, 112, 4690–4695. [Google Scholar] [CrossRef] [Green Version]
- Versace, E.; Caffini, M.; Werkhoven, Z.; De Bivort, B.L. Individual, but not population asymmetries, are modulated by social environment and genotype in Drosophila melanogaster. Sci. Rep. 2020, 10, 4480. [Google Scholar] [CrossRef] [Green Version]
- Rooke, R.; Rasool, A.; Schneider, J.; Levine, J.D. Drosophila melanogaster behaviour changes in different social environments based on group size and density. Commun. Biol. 2020, 3, 304. [Google Scholar] [CrossRef]
- Anpilov, S.; Shemesh, Y.; Eren, N.; Harony-Nicolas, H.; Benjamin, A.; Dine, J.; Oliveira, V.E.M.; Forkosh, O.; Karamihalev, S.; Hüttl, R.; et al. Wireless Optogenetic Stimulation of Oxytocin Neurons in a Semi-natural Setup Dynamically Elevates Both Pro-social and Agonistic Behaviors. Neuron 2020, 107, 644–655.e7. [Google Scholar] [CrossRef]
- Marshall, J.D.; Aldarondo, D.E.; Dunn, T.W.; Wang, W.L.; Berman, G.J.; Ölveczky, B.P. Continuous Whole-Body 3D Kinematic Recordings across the Rodent Behavioral Repertoire. Neuron 2021, 109, 420–437.e8. [Google Scholar] [CrossRef]
- Netser, S.; Meyer, A.; Magalnik, H.; Zylbertal, A.; de la Zerda, S.H.; Briller, M.; Bizer, A.; Grinevich, V.; Wagner, S. Distinct dynamics of social motivation drive differential social behavior in laboratory rat and mouse strains. Nat. Commun. 2020, 11, 5908. [Google Scholar] [CrossRef]
- Davidson, J.D.; Sosna, M.M.G.; Twomey, C.R.; Sridhar, V.H.; Leblanc, S.P.; Couzin, I.D. Collective detection based on visual information in animal groups. J. R. Soc. Interface 2021, 18, 20210142. [Google Scholar] [CrossRef]
- Smith, J.E.; Pinter-Wollman, N. Observing the unwatchable: Integrating automated sensing, naturalistic observations and animal social network analysis in the age of big data. J. Anim. Ecol. 2021, 90, 62–75. [Google Scholar] [CrossRef]
- Von Ziegler, L.; Sturman, O.; Bohacek, J. Big behavior: Challenges and opportunities in a new era of deep behavior profiling. Neuropsychopharmacology 2021, 46, 33–44. [Google Scholar] [CrossRef]
- Hong, G.; Lieber, C.M. Novel electrode technologies for neural recordings. Nat. Rev. Neurosci. 2019, 20, 330–345. [Google Scholar] [CrossRef]
- Hedlund, E.; Deng, Q. Single-cell RNA sequencing: Technical advancements and biological applications. Mol. Asp. Med. 2018, 59, 36–46. [Google Scholar] [CrossRef]
- Halder, A.; Verma, A.; Biswas, D.; Srivastava, S. Recent advances in mass-spectrometry based proteomics software, tools and databases. Drug Discov. Today Technol. 2021, 39, 69–79. [Google Scholar] [CrossRef]
- Wills, G.D.; Wesley, A.L.; Moore, F.R.; Sisemore, D.A. Social interactions among rodent conspecifics: A review of experimental paradigms. Neurosci. Biobehav. Rev. 1983, 7, 315–323. [Google Scholar] [CrossRef]
- Modi, M.; Shuai, Y.; Turner, G.C. The Drosophila Mushroom Body: From Architecture to Algorithm in a Learning Circuit. Annu. Rev. Neurosci. 2020, 43, 465–484. [Google Scholar] [CrossRef] [Green Version]
- Bentzur, A.; Ben-Shaanan, S.; Benichou, J.I.C.; Costi, E.; Levi, M.; Ilany, A.; Shohat-Ophir, G. Early Life Experience Shapes Male Behavior and Social Networks in Drosophila. Curr. Biol. 2021, 31, 670. [Google Scholar] [CrossRef]
- Burmeister, S.S.; Jarvis, E.D.; Fernald, R.D. Rapid Behavioral and Genomic Responses to Social Opportunity. PLoS Biol. 2005, 3, e363. [Google Scholar] [CrossRef] [Green Version]
- Karvat, G.; Kimchi, T. Acetylcholine Elevation Relieves Cognitive Rigidity and Social Deficiency in a Mouse Model of Autism. Neuropsychopharmacology 2014, 39, 831–840. [Google Scholar] [CrossRef]
- Weissbrod, A.; Shapiro, A.; Vasserman, G.; Edry, L.; Dayan, M.; Yitzhaky, A.; Hertzberg, L.; Feinerman, O.; Kimchi, T. Automated long-term tracking and social behavioural phenotyping of animal colonies within a semi-natural environment. Nat. Commun. 2013, 4, 2018. [Google Scholar] [CrossRef] [Green Version]
- Forkosh, O.; Karamihalev, S.; Roeh, S.; Alon, U.; Anpilov, S.; Touma, C.; Nussbaumer, M.; Flachskamm, C.; Kaplick, P.M.; Shemesh, Y.; et al. Identity domains capture individual differences from across the behavioral repertoire. Nat. Neurosci. 2019, 22, 2023–2028. [Google Scholar] [CrossRef] [PubMed]
- Kabra, M.; Robie, A.; Rivera-Alba, M.; Branson, S.; Branson, K. JAABA: Interactive machine learning for automatic annotation of animal behavior. Nat. Methods 2013, 10, 64–67. [Google Scholar] [CrossRef] [PubMed]
- Shneiderman, B. The Eyes Have It: A Task by Data Type Taxonomy for Information Visualizations. In The Craft of Information Visualization; Bederson, B.B., Shneiderman, B., Eds.; Morgan Kaufmann: Burlington, MA, USA, 2003; pp. 364–371. [Google Scholar]
- Croft, D.P.; James, R.; Krause, J. Exploring Animal Social Networks; Princeton University Press: Princeton, NJ, USA, 2008. [Google Scholar]
- Whitehead, H. Analyzing Animal Societies; University of Chicago Press: Chicago, IL, USA, 2008. [Google Scholar]
- Goaillard, J.-M.; Marder, E. Ion Channel Degeneracy, Variability, and Covariation in Neuron and Circuit Resilience. Annu. Rev. Neurosci. 2021, 44, 335–357. [Google Scholar] [CrossRef] [PubMed]
- Keren, L.; Bosse, M.; Thompson, S.; Risom, T.; Vijayaragavan, K.; McCaffrey, E.; Marquez, D.; Angoshtari, R.; Greenwald, N.F.; Fienberg, H.; et al. MIBI-TOF: A multiplexed imaging platform relates cellular phenotypes and tissue structure. Sci. Adv. 2019, 5, eaax5851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moffitt, J.R.; Hao, J.; Wang, G.; Chen, K.H.; Babcock Hazen, P.; Zhuang, X. High-throughput single-cell gene-expression profiling with multiplexed error-robust fluorescence in situ hybridization. Proc. Natl. Acad. Sci. USA 2016, 113, 11046–11051. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Allen, W.E.; Wright, M.A.; Sylwestrak, E.L.; Samusik, N.; Vesuna, S.; Evans, K.; Liu, C.; Ramakrishnan, C.; Liu, J.; et al. Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science 2018, 361, eaat5691. [Google Scholar] [CrossRef] [Green Version]
- Alon, S.; Goodwin, D.R.; Sinha, A.; Wassie, A.T.; Chen, F.; Daugharthy, E.R.; Bando, Y.; Kajita, A.; Xue, A.G.; Marrett, K.; et al. Expansion sequencing: Spatially precise in situ transcriptomics in intact biological systems. Science 2021, 371, eaax2656. [Google Scholar] [CrossRef]
- Rodriques, S.G.; Stickels, R.R.; Goeva, A.; Martin, C.A.; Murray, E.; Vanderburg, C.R.; Welch, J.; Chen, L.M.; Chen, F.; Macosko, E.Z. Slide-seq: A scalable technology for measuring genome-wide expression at high spatial resolution. Science 2019, 363, 1463–1467. [Google Scholar] [CrossRef]
- Goltsev, Y.; Samusik, N.; Kennedy-Darling, J.; Bhate, S.; Hale, M.; Vazquez, G.; Black, S.; Nolan, G.P. Deep Profiling of Mouse Splenic Architecture with CODEX Multiplexed Imaging. Cell 2018, 174, 968–981.e15. [Google Scholar] [CrossRef] [Green Version]
- Ståhl, P.L.; Salmén, F.; Vickovic, S.; Lundmark, A.; Navarro, J.F.; Magnusson, J.; Giacomello, S.; Asp, M.; Westholm, J.O.; Huss, M.; et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 2016, 353, 78–82. [Google Scholar] [CrossRef] [Green Version]
- Driscoll, N.; Erickson, B.; Murphy, B.B.; Richardson, A.G.; Robbins, G.; Apollo, N.V.; Mentzelopoulos, G.; Mathis, T.; Hantanasirisakul, K.; Bagga, P.; et al. MXene-infused bioelectronic interfaces for multiscale electrophysiology and stimulation. Sci. Transl. Med. 2021, 13, eabf8629. [Google Scholar] [CrossRef]
- Liu, P.; Miller, E.W. Electrophysiology, Unplugged: Imaging Membrane Potential with Fluorescent Indicators. Acc. Chem. Res. 2020, 53, 11–19. [Google Scholar] [CrossRef]
- Kim, K.; Vöröslakos, M.; Seymour, J.P.; Wise, K.D.; Buzsáki, G.; Yoon, E. Artifact-free and high-temporal-resolution in vivo opto-electrophysiology with microLED optoelectrodes. Nat. Commun. 2020, 11, 2063. [Google Scholar] [CrossRef]
- Adam, Y. All-optical electrophysiology in behaving animals. J. Neurosci. Methods 2021, 353, 109101. [Google Scholar] [CrossRef]
- Tian, J.; Lin, Z.; Chen, Z.; Obaid, S.N.; Efimov, I.R.; Lu, L. Stretchable and Transparent Metal Nanowire Microelectrodes for Simultaneous Electrophysiology and Optogenetics Applications. Photonics 2021, 8, 220. [Google Scholar] [CrossRef]
- Steinmetz, N.; Koch, C.; Harris, K.; Carandini, M. Challenges and opportunities for large-scale electrophysiology with Neuropixels probes. Curr. Opin. Neurobiol. 2018, 50, 92–100. [Google Scholar] [CrossRef]
- Zhu, C.; Preissl, S.; Ren, B. Single-cell multimodal omics: The power of many. Nat. Methods 2020, 17, 11–14. [Google Scholar] [CrossRef]
- Tanay, A.; Regev, A. Scaling single-cell genomics from phenomenology to mechanism. Nature 2017, 541, 331–338. [Google Scholar] [CrossRef] [Green Version]
- Svensson, V.; Vento-Tormo, R.; Teichmann, S.A. Exponential scaling of single-cell RNA-seq in the past decade. Nat. Protoc. 2018, 13, 599–604. [Google Scholar] [CrossRef]
- Regev, A.; Teichmann, S.A.; Lander, E.S.; Amit, I.; Benoist, C.; Birney, E.; Bodenmiller, B.; Campbell, P.; Carninci, P.; Clatworthy, M.; et al. The Human Cell Atlas. eLife 2017, 6, e27041. [Google Scholar] [CrossRef]
- Svensson, V.; de Veiga Beltrame, E.; Pachter, L. A curated database reveals trends in single-cell transcriptomics. Database 2020, 2020, baaa073. [Google Scholar] [CrossRef]
- Tang, F.; Barbacioru, C.; Wang, Y.; Nordman, E.; Lee, C.; Xu, N.; Wang, X.; Bodeau, J.; Tuch, B.B.; Siddiqui, A.; et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods 2009, 6, 377–382. [Google Scholar] [CrossRef]
- Chen, X.; Teichmann, S.A.; Meyer, K.B. From Tissues to Cell Types and Back: Single-Cell Gene Expression Analysis of Tissue Architecture. Annu. Rev. Biomed. Data Sci. 2018, 1, 29–51. [Google Scholar] [CrossRef]
- Ding, J.; Adiconis, X.; Simmons, S.K.; Kowalczyk, M.S.; Hession, C.C.; Marjanovic, N.D.; Hughes, T.K.; Wadsworth, M.H.; Burks, T.; Nguyen, L.T.; et al. Systematic comparison of single-cell and single-nucleus RNA-sequencing methods. Nat. Biotechnol. 2020, 38, 737–746. [Google Scholar] [CrossRef]
- Mereu, E.; Lafzi, A.; Moutinho, C.; Ziegenhain, C.; McCarthy Davis, J.; Álvarez-Varela, A.; Batlle, E.; Sagar; Grün, D.; Lau, J.K.; et al. Benchmarking single-cell RNA-sequencing protocols for cell atlas projects. Nat. Biotechnol. 2020, 38, 747–755. [Google Scholar] [CrossRef]
- Zappia, L.; Phipson, B.; Oshlack, A. Exploring the single-cell RNA-seq analysis landscape with the scRNA-tools database. PLoS Comput. Biol. 2018, 14, e1006245. [Google Scholar] [CrossRef]
- Hie, B.; Peters, J.; Nyquist, S.K.; Shalek, A.K.; Berger, B.; Bryson, B.D. Computational methods for single-cell RNA sequencing. Annu. Rev. Biomed. Data Sci. 2020, 3, 339–364. [Google Scholar] [CrossRef]
- Van den Berge, K.; Hembach, K.M.; Soneson, C.; Tiberi, S.; Clement, L.; Love, M.I.; Patro, R.; Robinson, M.D. RNA sequencing data: Hitchhiker’s guide to expression analysis. Annu. Rev. Biomed. Data Sci. 2019, 2, 139–173. [Google Scholar] [CrossRef] [Green Version]
- Ilany, A.; Holekamp, K.E.; Akçay, E. Rank-dependent social inheritance determines social network structure in spotted hyenas. Science 2021, 373, 348–352. [Google Scholar] [CrossRef]
- Bendall, S.C.; Davis, K.L.; Amir, E.D.; Tadmor, M.D.; Simonds, E.F.; Chen, T.J.; Shenfeld, D.K.; Nolan Garry, P.; Peer, D. Single-cell trajectory detection uncovers progression and regulatory coordination in human B cell development. Cell 2014, 157, 714–725. [Google Scholar] [CrossRef] [Green Version]
- La Manno, G.; Soldatov, R.; Zeisel, A.; Braun, E.; Hochgerner, H.; Petukhov, V.; Lidschreiber, K.; Kastriti, M.E.; Lönnerberg, P.; Furlan, A.; et al. RNA velocity of single cells. Nature 2018, 560, 494–498. [Google Scholar] [CrossRef] [Green Version]
- Svensson, V.; Pachter, L. RNA Velocity: Molecular Kinetics from Single-Cell RNA-Seq. Mol. Cell. 2018, 72, 7–9. [Google Scholar] [CrossRef] [Green Version]
- Loh, P.-R.; Baym, M.; Berger, B. Compressive genomics. Nat. Biotechnol. 2012, 30, 627–630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, Y.W.; Daniels, N.M.; Danko, D.C.; Berger, B. Entropy-Scaling Search of Massive Biological Data. Cell Syst. 2015, 1, 130–140. [Google Scholar] [CrossRef] [Green Version]
- Cleary, B.; Cong, L.; Cheung, A.; Lander, E.S.; Regev, A. Efficient Generation of Transcriptomic Profiles by Random Composite Measurements. Cell 2017, 171, 1424–1436.e18. [Google Scholar] [CrossRef] [PubMed]
- Stein-O’Brien, G.L.; Arora, R.; Culhane, A.C.; Favorov, A.V.; Garmire, L.X.; Greene, C.S.; Goff, L.A.; Li, Y.; Ngom, A.; Ochs, M.F.; et al. Enter the Matrix: Factorization Uncovers Knowledge from Omics. Trends Genet. 2018, 34, 790–805. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van der Maaten, L.; Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res. 2008, 9, 2579–2605. [Google Scholar]
- Jacomy, M.; Venturini, T.; Heymann, S.; Bastian, M. ForceAtlas2, a Continuous Graph Layout Algorithm for Handy Network Visualization Designed for the Gephi Software. PLoS ONE 2014, 9, e98679. [Google Scholar] [CrossRef]
- Weinreb, C.; Wolock, S.; Klein, A.M. SPRING: A kinetic interface for visualizing high dimensional single-cell expression data. Bioinformatics 2018, 34, 1246–1248. [Google Scholar] [CrossRef]
- McInnes, L.; Healy, J.; Melville, J. UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. arXiv 2020, arXiv:1802.03426. [Google Scholar]
- Becht, E.; McInnes, L.; Healy, J.; Dutertre, C.; Kwok, I.W.H.; Ng, L.G.; Ginhoux, F.; Newell, E.W. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 2018, 37, 38–44. [Google Scholar] [CrossRef]
- Kotliar, D.; Veres, A.; Nagy, M.A.; Tabrizi, S.; Hodis, E.; Melton, D.A.; Sabeti, P.C. Identifying gene expression programs of cell-type identity and cellular activity with single-cell RNA-Seq. eLife 2019, 8, e43803. [Google Scholar] [CrossRef]
- Nelson, W.; Zitnik, M.; Wang, B.; Leskovec, J.; Goldenberg, A.; Sharan, R. To Embed or Not: Network Embedding as a Paradigm in Computational Biology. Front. Genet. 2019, 10, 381. [Google Scholar] [CrossRef] [Green Version]
- Eisen, M.B.; Spellman, P.T.; Brown, P.O.; Botstein, D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 1998, 95, 14863–14868. [Google Scholar] [CrossRef] [Green Version]
- Shafer, M.E.R. Cross-Species Analysis of Single-Cell Transcriptomic Data. Front. Cell Dev. Biol. 2019, 7, 175. [Google Scholar] [CrossRef] [Green Version]
- Chauvel, C.; Novoloaca, A.; Veyre, P.; Reynier, F.; Becker, J. Evaluation of integrative clustering methods for the analysis of multi-omics data. Brief. Bioinform. 2020, 21, 541–552. [Google Scholar] [CrossRef]
- Kiselev, V.Y.; Andrews, T.S.; Hemberg, M. Publisher Correction: Challenges in unsupervised clustering of single-cell RNA-seq data. Nat. Rev. Genet. 2019, 20, 310. [Google Scholar] [CrossRef] [Green Version]
- Zeng, W.; Chen, X.; Duren, Z.; Wang, Y.; Jiang, R.; Wong, W.H. DC3 is a method for deconvolution and coupled clustering from bulk and single-cell genomics data. Nat. Commun. 2019, 10, 4613. [Google Scholar] [CrossRef]
- Petegrosso, R.; Li, Z.; Kuang, R. Machine learning and statistical methods for clustering single-cell RNA-sequencing data. Brief. Bioinform. 2019, 21, 1209–1223. [Google Scholar] [CrossRef] [Green Version]
- Voelkl, B.; Altman, N.S.; Forsman, A.; Forstmeier, W.; Gurevitch, J.; Jaric, I.; Karp, N.A.; Kas, M.J.; Schielzeth, H.; van de Casteele, T.; et al. Reproducibility of animal research in light of biological variation. Nat. Rev. Neurosci. 2020, 21, 384–393. [Google Scholar] [CrossRef]
- O’Leary, T.; Sutton, A.C.; Marder, E. Computational models in the age of large datasets. Curr. Opin. Neurobiol. 2015, 32, 87–94. [Google Scholar] [CrossRef] [Green Version]
- Cohen, D. Optimizing reproduction in a randomly varying environment. J. Theor. Biol. 1966, 12, 119–129. [Google Scholar] [CrossRef]
- Miguel, M.C.; Parley, J.T.; Pastor-Satorras, R. Effects of Heterogeneous Social Interactions on Flocking Dynamics. Phys. Rev. Lett. 2018, 120, 068303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knebel, D.; Ayali, A.; Guershon, M.; Ariel, G. Intra- versus intergroup variance in collective behavior. Sci. Adv. 2019, 5, eaav0695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stern, S.; Kirst, C.; Bargmann, C.I. Neuromodulatory Control of Long-Term Behavioral Patterns and Individuality across Development. Cell 2017, 171, 1649–1662.e10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stevenson-Hinde, J.; Zunz, M. Subjective assessment of individual rhesus monkeys. Primates 1978, 19, 473–482. [Google Scholar] [CrossRef]
- Mather, J.A.; Anderson, R.C. Personalities of octopuses (Octopus rubescens). J. Comp. Psychol. 1993, 107, 336–340. [Google Scholar] [CrossRef]
- Boring, L.; Gosling, J.; Cleary, M.; Charo, I.F. Decreased lesion formation in CCR2−/− mice reveals a role for chemokines in the initiation of atherosclerosis. Nature 1998, 394, 894–897. [Google Scholar] [CrossRef]
- Forkosh, O. Animal behavior and animal personality from a non-human perspective: Getting help from the machine. Patterns 2021, 2, 100194. [Google Scholar] [CrossRef]
- Wice, E.W.; Saltz, J.B. Selection on heritable social network positions is context-dependent in Drosophila melanogaster. Nat. Commun. 2021, 12, 3357. [Google Scholar] [CrossRef]
- Bruijning, M.; Metcalf, C.J.E.; Jongejans, E.; Ayroles, J.F. The Evolution of Variance Control. Trends Ecol. Evol. 2020, 35, 22–33. [Google Scholar] [CrossRef]
- Hill, M.S.; Zande, P.V.; Wittkopp, P.J. Molecular and evolutionary processes generating variation in gene expression. Nat. Rev. Genet. 2021, 22, 203–215. [Google Scholar] [CrossRef]
- Dueck, H.; Eberwine, J.; Kim, J. Variation is function: Are single cell differences functionally important? Testing the hypothesis that single cell variation is required for aggregate function. BioEssays 2016, 38, 172–180. [Google Scholar] [CrossRef]
- Trapnell, C. Defining cell types and states with single-cell genomics. Genome Res. 2015, 25, 1491–1498. [Google Scholar] [CrossRef] [Green Version]
- Arnol, D.; Schapiro, D.; Bodenmiller, B.; Saez-Rodriguez, J.; Stegle, O. Modeling Cell-Cell Interactions from Spatial Molecular Data with Spatial Variance Component Analysis. Cell Rep. 2019, 29, 202–211.e6. [Google Scholar] [CrossRef] [Green Version]
- Gustafsson, J.; Held, F.; Robinson, J.L.; Björnson, E.; Jörnsten, R.; Nielsen, J. Sources of variation in cell-type RNA-Seq profiles. PLoS ONE 2020, 15, e0239495. [Google Scholar] [CrossRef]
- Foreman, R.; Wollman, R. Mammalian gene expression variability is explained by underlying cell state. Mol. Syst. Biol. 2020, 16, e9146. [Google Scholar] [CrossRef]
- Osorio, D.; Yu, X.; Zhong, Y.; Li, G.; Yu, P.; Serpedin, E.; Huang, J.Z.; Cai, J.J. Single-Cell Expression Variability Implies Cell Function. Cells 2019, 9, 14. [Google Scholar] [CrossRef] [Green Version]
- Shaffer, S.M.; Emert, B.L.; Reyes Hueros, R.A.; Cote, C.; Harmange, G.; Schaff, D.L.; Sizemore, A.E.; Gupte, R.; Torre, E.; Singh, A.; et al. Memory Sequencing Reveals Heritable Single-Cell Gene Expression Programs Associated with Distinct Cellular Behaviors. Cell 2020, 182, 947–959.e17. [Google Scholar] [CrossRef]
- Phillips, N.E.; Mandic, A.; Omidi, S.; Naef, F.; Suter, D.M. Memory and relatedness of transcriptional activity in mammalian cell lineages. Nat. Commun. 2019, 10, 1208. [Google Scholar] [CrossRef] [Green Version]
- Javer, A.; Currie, M.; Lee, C.W.; Hokanson, J.; Li, K.; Martineau, C.N.; Yemini, E.; Grundy, L.J.; Li, C.; Ch’ng, Q.; et al. An open-source platform for analyzing and sharing worm-behavior data. Nat. Methods 2018, 15, 645–646. [Google Scholar] [CrossRef]
- Butler, A.; Hoffman, P.; Smibert, P.; Papalexi, E.; Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018, 36, 411–420. [Google Scholar] [CrossRef]
- Stuart, T.; Butler, A.; Hoffman, P.; Hafemeister, C.; Papalexi, E.; Mauck William, M., 3rd; Hao, Y.; Stoeckius, M.; Smibert, P.; Satija, R. Comprehensive Integration of Single-Cell Data. Cell 2019, 177, 1888–1902.e21. [Google Scholar] [CrossRef]
- Wolf, F.A.; Angerer, P.; Theis, F.J. SCANPY: Large-scale single-cell gene expression data analysis. Genome Biol. 2018, 19, 15. [Google Scholar] [CrossRef] [Green Version]
- Branson, K.; Robie, A.; Bender, A.J.; Perona, P.; Dickinson, M.H. High-throughput ethomics in large groups of Drosophila. Nat. Methods 2009, 6, 451–457. [Google Scholar] [CrossRef]
- Mathis, A.; Mamidanna, P.; Cury, K.M.; Abe, T.; Murthy, V.N.; Mathis, M.W.; Bethge, M. DeepLabCut: Markerless pose estimation of user-defined body parts with deep learning. Nat. Neurosci. 2018, 21, 1281–1289. [Google Scholar] [CrossRef]
- Jezovit Jacob, A.; Alwash Nawar Levine Joel, D. Using Flies to Understand Social Networks. Front. Neural Circuits 2021, 15, 1662–5110. [Google Scholar] [CrossRef]
- Finn, K.R.; Silk, M.J.; Porter, M.A.; Pinter-Wollman, N. The use of multilayer network analysis in animal behaviour. Anim. Behav. 2019, 149, 7–22. [Google Scholar] [CrossRef] [Green Version]
- Rocha, L.E.C.; Ryckebusch, J.; Schoors, K.; Smith, M. The scaling of social interactions across animal species. Sci. Rep. 2021, 11, 12584. [Google Scholar] [CrossRef]
- Castles, M.; Heinsohn, R.; Marshall, H.H.; Lee Alexander, E.G.; Cowlishaw, G.; Carter Alecia, J. Social networks created with different techniques are not comparable. Anim. Behav. 2014, 96, 59–67. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bentzur, A.; Alon, S.; Shohat-Ophir, G. Behavioral Neuroscience in the Era of Genomics: Tools and Lessons for Analyzing High-Dimensional Datasets. Int. J. Mol. Sci. 2022, 23, 3811. https://doi.org/10.3390/ijms23073811
Bentzur A, Alon S, Shohat-Ophir G. Behavioral Neuroscience in the Era of Genomics: Tools and Lessons for Analyzing High-Dimensional Datasets. International Journal of Molecular Sciences. 2022; 23(7):3811. https://doi.org/10.3390/ijms23073811
Chicago/Turabian StyleBentzur, Assa, Shahar Alon, and Galit Shohat-Ophir. 2022. "Behavioral Neuroscience in the Era of Genomics: Tools and Lessons for Analyzing High-Dimensional Datasets" International Journal of Molecular Sciences 23, no. 7: 3811. https://doi.org/10.3390/ijms23073811
APA StyleBentzur, A., Alon, S., & Shohat-Ophir, G. (2022). Behavioral Neuroscience in the Era of Genomics: Tools and Lessons for Analyzing High-Dimensional Datasets. International Journal of Molecular Sciences, 23(7), 3811. https://doi.org/10.3390/ijms23073811





