Katanin-Dependent Microtubule Ordering in Association with ABA Is Important for Root Hydrotropism
Abstract
1. Introduction
2. Results and Discussion
2.1. Hydrotropic Bending of WT Col-0 Roots Is Reduced by Oryzalin Treatment
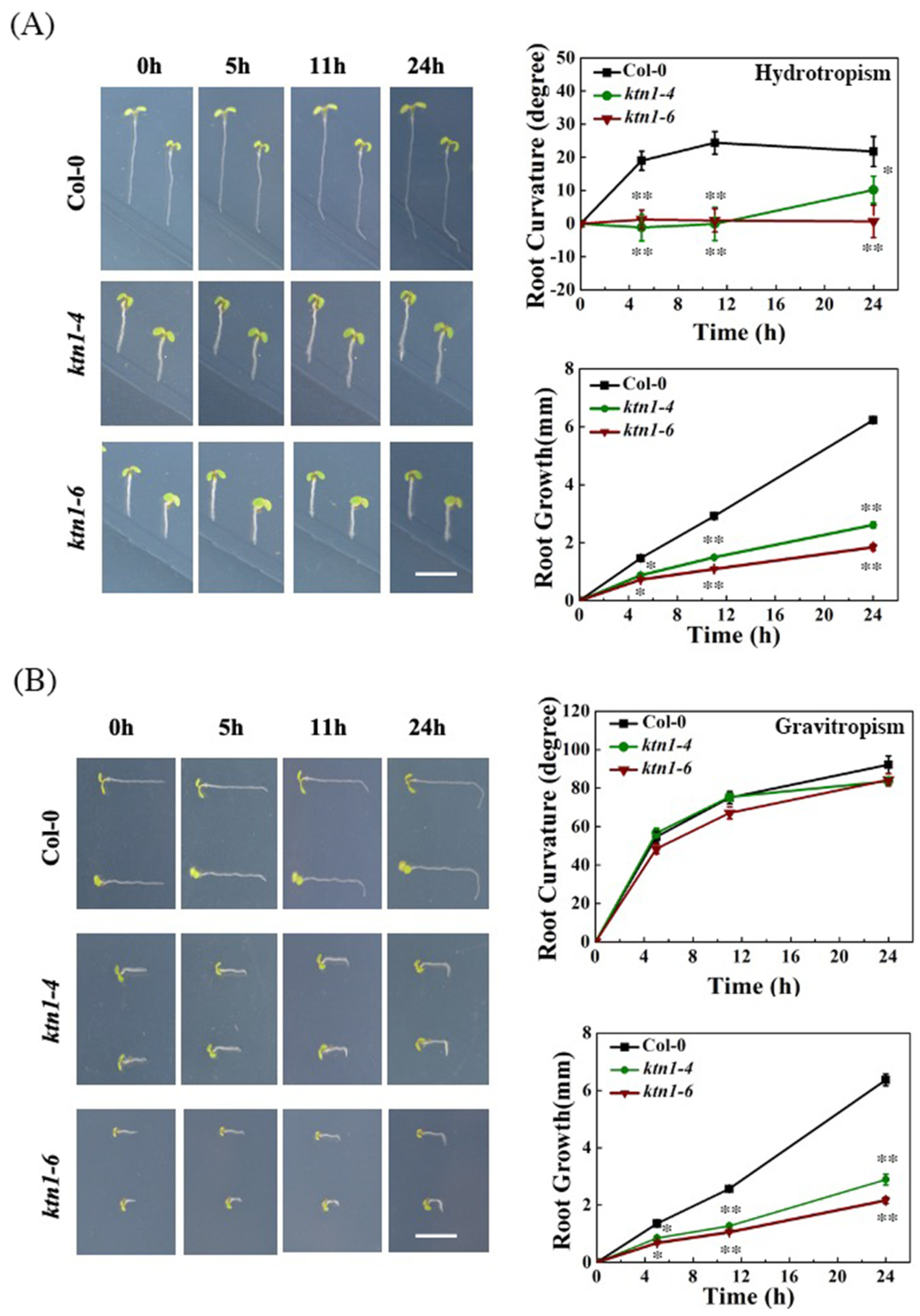
2.2. Katanin Is Required for Root Hydrotropism in Arabidopsis
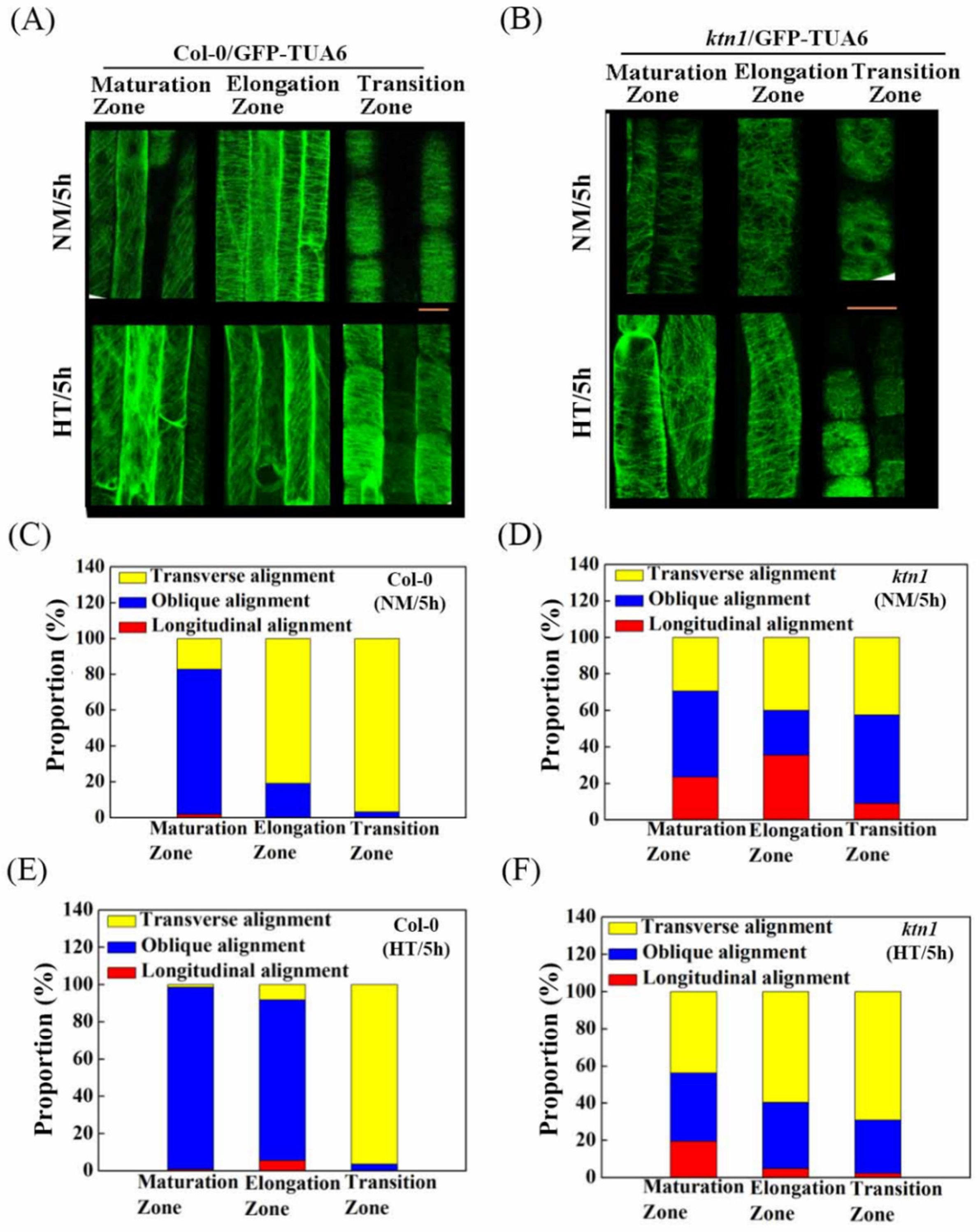
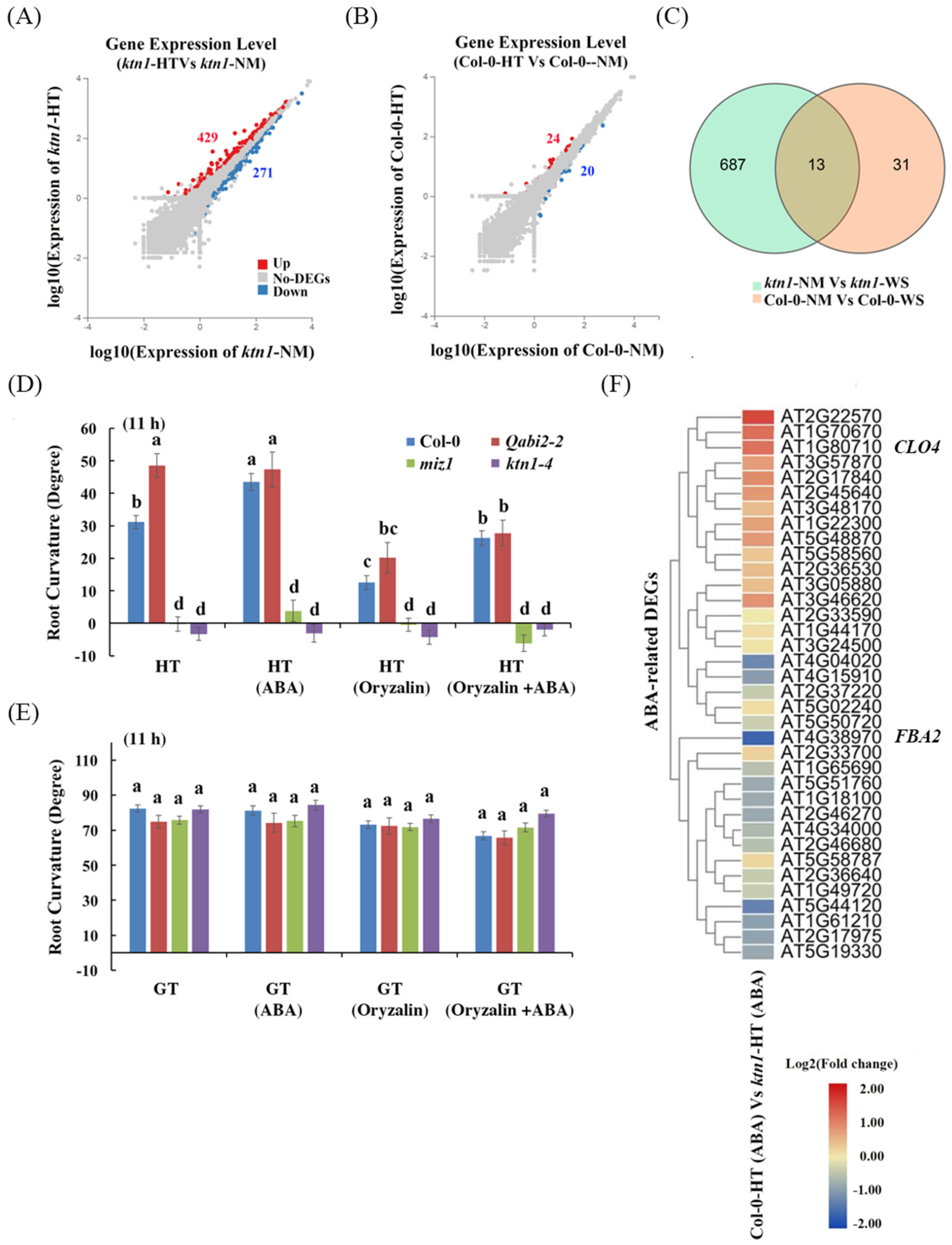
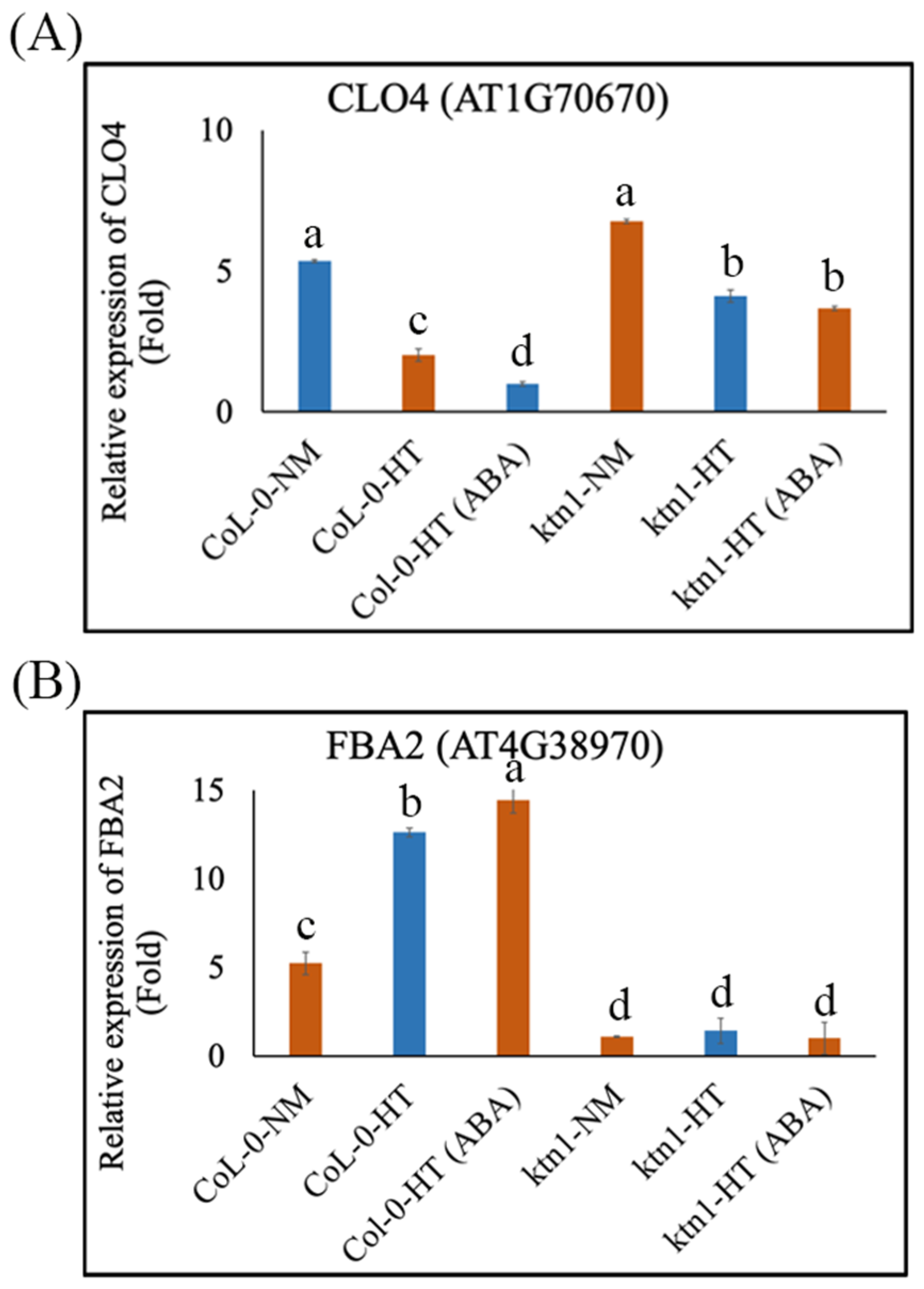
2.3. Katanin-Regulated Microtubule Ordering Might Work Downstream of ABA in Root Hydrotropism
3. Materials and Methods
3.1. Plant Material and Growth Conditions
3.2. RNA-Sequencing Assay
3.3. Quantitative (Q) RT-PCR
3.4. Drug Treatments
3.5. Microscopy
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kobayashi, A.; Takahashi, A.; Kakimoto, Y.; Miyazawa, Y.; Fujii, N.; Higashitani, A.; Takahashi, H. A gene essential for hydrotropism in roots. Proc. Natl. Acad. Sci. USA 2007, 104, 4724–4729. [Google Scholar] [CrossRef] [PubMed]
- Antoni, R.; Gonzalez-Guzman, M.; Rodriguez, L.; Peirats-Llobet, M.; Pizzio, G.A.; Fernandez, M.A.; De Winne, N.; De Jaeger, G.; Dietrich, D.; Bennett, M.J.; et al. PYRABACTIN RESISTANCE1-LIKE8 plays an important role for the regulation of abscisic acid signaling in root. Plant Physiol. 2013, 161, 931–941. [Google Scholar] [CrossRef]
- Dietrich, D.; Pang, L.; Kobayashi, A.; Fozard, J.A.; Boudolf, V.; Bhosale, R.; Antoni, R.; Nquyen, T.; Hiratsuka, S.; Fujii, N.; et al. Root hydrotropism is controlled via a cortex-specific growth mechanism. Nat. Plants 2017, 3, 17057. [Google Scholar] [CrossRef] [PubMed]
- Miao, R.; Russinova, E.; Rodriguez, P.L. Tripartite hormonal regulation of plasma membrane H+-ATPase activity. Trends Plant Sci. 2022, 13, S1360–S1385. [Google Scholar] [CrossRef]
- Miao, R.; Wang, M.; Yuan, W.; Ren, Y.; Li, Y.; Zhang, N.; Zhang, J.; Kronzucker, H.J.; Xu, W. Comparative analysis of Arabidopsis ecotypes reveals a role for brassinosteroids in root hydrotropism. Plant Physiol. 2018, 176, 2720–2736. [Google Scholar] [CrossRef] [PubMed]
- Miao, R.; Yuan, W.; Wang, Y.; Garcia-Maquilon, I.; Dang, X.; Li, Y.; Zhang, J.; Zhu, Y.; Rodriguez, P.L.; Xu, W. Low ABA concentration promotes root growth and hydrotropism through relief of ABA INSENSITIVE 1-mediated inhibition of plasma membrane H+-ATPase 2. Sci. Adv. 2021, 7, eabd4113. [Google Scholar] [CrossRef] [PubMed]
- Yuan, W.; Li, Y.; Li, L.; Siao, W.; Zhang, Q.; Zhang, Y.; Liu, J.; Xu, W.; Miao, R. BR-INSENSITIVE1 regulates hydrotropic response by interacting with plasma membrane H+-ATPases in Arabidopsis. Plant Signal. Behav. 2018, 13, e1486147. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.; Li, X.; Fu, W.; Wang, J.; Yong, Y.; Shi, H.; Ding, Z.; Kui, H.; Gou, X.; He, K.; et al. Asymmetric distribution of cytokinins determines root hydrotropism in Arabidopsis thaliana. Cell Res. 2019, 29, 984–993. [Google Scholar] [CrossRef]
- Huang, S.; Miao, R. An irew1 mutant intensifies lateral root branching for adaption upon water stress. Biotechniques 2020, 69, 242–244. [Google Scholar] [CrossRef]
- Yuan, W.; Zhang, Q.; Li, Y.; Wang, Q.; Xu, F.; Dang, X.; Xu, W.; Zhang, J.; Miao, R. Abscisic acid is required for root elongation associated with Ca2+ influx in response to water stress. Plant Physiol. Biochem. 2021, 169, 127–137. [Google Scholar] [CrossRef]
- Nakayama, M.; Kaneko, Y.; Miyazawa, Y.; Fujii, N.; Higashitani, N.; Wanda, S.; Ishida, H.; Yoshimoto, K.; Shirasu, K.; Yamada, K.; et al. A possible involvement of autophagy in amyloplast degradation in columella cells during hydrotropic response of Arabidopsis roots. Planta 2012, 236, 999–1012. [Google Scholar] [CrossRef] [PubMed]
- Krieger, G.; Shkolnik, D.; Miller, G.; Fromm, H. Reactive oxygen species tune root tropic responses. Plant Physiol. 2016, 172, 1209–1220. [Google Scholar] [CrossRef] [PubMed]
- Shkolnik, D.; Nuriel, R.; Bonza, M.C.; Costa, A.; Fromm, H. MIZ1 regulates ECA1 to generate a slow, long-distance phloem-transmitted Ca2+ signal essential for root water tracking in Arabidopsis. Proc. Natl. Acad. Sci. USA 2018, 115, 8031–8036. [Google Scholar] [CrossRef] [PubMed]
- Mitchison, T.; Kirschner, M. Dynamic instability of microtubule growth. Nature 1984, 312, 237–242. [Google Scholar] [CrossRef]
- Chaaban, S.; Jariwala, S.; Hsu, C.T.; Redemann, S.; Kollman, J.M.; Muller-Reicher, T.; Sept, D.; Huy Bui, K.; Brouhard, G.J. The Structure and Dynamics of C. elegans Tubulin Reveals the Mechanistic Basis of Microtubule Growth. Dev. Cell 2018, 47, 191–204. [Google Scholar]
- Paredez, A.R.; Somerville, C.R.; Ehrhardt, D.W. Visualization of cellulose synthase demonstrates functional association with microtubules. Science 2006, 312, 1491–1495. [Google Scholar] [CrossRef]
- Gutierrez, R.; Lindeboom, J.J.; Paredez, A.R.; Emons, A.M.C.; Ehrhardt, D.W. Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nat. Cell Biol. 2009, 11, 797–806. [Google Scholar] [CrossRef]
- Crowell, E.F.; Timpano, H.; Desprez, T.; Franssen-Verheijen, T.; Emons, A.-M.; Höfte, H.; Vernhettes, S. Differential regulation of cellulose orientation at the inner and outer face of epidermal cells in the Arabidopsis hypocotyl. Plant Cell 2011, 23, 2592–2605. [Google Scholar] [CrossRef]
- Lindeboom, J.J.; Nakamura, M.; Hibbel, A.; Shundyak, K.; Gutierrez, R.; Ketelaar, T.; Emons, A.M.C.; Mulder, B.M.; Kirik, V.; Ehrhardt, D.W. A mechanism for reorientation of cortical microtubule arrays driven by microtubule severing. Science 2013, 342, 1245533. [Google Scholar] [CrossRef]
- Sampathkumar, A.; Gutierrez, R.; Mcfarlane, H.; Bringmann, M.; Lindeboom, J.; Emons, A.M.; Samuels, L.; Ketelaar, T.; Ehrhardt, D.W.; Persson, S. Patterning and life-time of plasma membrane localized cellulose synthase is dependent on actin organization in Arabidopsis interphase cells. Plant Physiol. 2013, 162, 675–688. [Google Scholar] [CrossRef]
- Oda, Y. Cortical microtubule rearrangements and cell wall patterning. Front. Plant Sci. 2015, 6, 236. [Google Scholar] [CrossRef] [PubMed]
- Blancaflor, E.B. The cytoskeleton and gravitropism in higher plants. J. Plant Growth Regul. 2002, 21, 120–136. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Wu, S.; Liu, Z.; Friml, J. Environmental and endogenous control of cortical microtubule orientation. Trends Cell Biol. 2016, 26, 409–419. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Mao, T. Understanding the functions and mechanisms of plant cytoskeleton in response to environmental signals. Curr. Opin. Plant Biol. 2019, 52, 86–96. [Google Scholar] [CrossRef]
- Morita, M.T. Directional gravity sensing in gravitropism. Annu. Rev. Plant Biol. 2010, 61, 705–720. [Google Scholar] [CrossRef]
- Chen, X.; Grandont, L.; Li, H.; Hauschind, R.; Paque, S.; Abuzeineh, A.; Rakusova, H.; Benkova, E.; Perrot-Rechenmann, C. Inhibition of cell expansion by rapid ABP1-mediated auxin effect on microtubules. Nature 2014, 516, 90–93. [Google Scholar] [CrossRef]
- Allen, N.S.; Chattaraj, P.; Collings, D.; Johannes, E. Gravisensing: Ionic responses, cytoskeleton and amyloplast behavior. Adv. Space Res. 2003, 32, 1631–1637. [Google Scholar] [CrossRef]
- Sedbrook, J.C.; Kaloriti, D. Microtubules, MAPs and plant directional cell expansion. Trends Plant Sci. 2008, 13, 303–310. [Google Scholar] [CrossRef]
- Roll-Mecak, A.; McNally, F.J. Microtubule-severing enzymes. Curr. Opin. Cell Biol. 2010, 22, 96–103. [Google Scholar] [CrossRef]
- Lin, D.; Cao, L.; Zhou, Z.; Zhu, L.; Ehrhardt, D.; Yang, Z.; Fu, Y. Rho GTPase signaling activates microtubule severing to promote microtubule ordering in Arabidopsis. Curr Biol. 2013, 23, 290–297. [Google Scholar] [CrossRef]
- Uyttewaal, M.; Burian, A.; Alim, K.; Landrein, B.; Borowska-Wykręt, D.; Dedieu, A.; Peaucelle, A.; Ludynia, M.; Traas, J.; Boudaoud, A.; et al. Mechanical stress acts via katanin to amplify differences in growth rate between adjacent cells in Arabidopsis. Cell 2012, 149, 439–451. [Google Scholar] [CrossRef] [PubMed]
- Baral, A.; Aryal, B.; Jonsson, K.; Morris, E.; Demes, E.; Takatani, S.; Verger, S.; Xu, T.; Bennett, M.; Hamant, O.; et al. External Mechanical Cues Reveal a Katanin-Independent Mechanism behind Auxin-Mediated Tissue Bending in Plants. Dev. Cell. 2021, 56, 67–80. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Ma, Q.; Mao, T. Ethylene Regulates the Arabidopsis Microtubule-Associated Protein WAVE-DAMPENED2-LIKE5 in Etiolated Hypocotyl Elongation. Plant Physiol. 2015, 169, 325–337. [Google Scholar] [CrossRef] [PubMed]
- Hasenstein, K.H.; Blancaflor, E.B.; Lee, J.S. The microtubule cytoskeleton does not integrate auxin transport and gravitropism in maize roots. Physiol. Plant 1999, 105, 729–738. [Google Scholar] [CrossRef]
- Hou, G.; Mohamalawari, D.R.; Blancaflor, E.B. Enhanced gravitropism of roots with a disrupted cap actin cytoskeleton. Plant Physiol. 2003, 131, 1360–1373. [Google Scholar] [CrossRef][Green Version]
- Nakamura, M.; Ehrhardt, D.W.; Hashimoto, T. Microtubule and katanin-dependent dynamics of microtubule nucleation complexes in the acentrosomal Arabidopsis cortical array. Nat. Cell Biol. 2010, 12, 1064–1070. [Google Scholar] [CrossRef]
- Ueda, K.; Sakaguchi, S.; Kumagai, F.; Hasezawa, S.; Quader, H.; Kristen, U. Development and disintegration of phragmoplasts in living cultured cells of a GFP:TUA6 transgenic Arabidopsis thaliana plant. Protoplasma 1999, 220, 111–118. [Google Scholar] [CrossRef]
- Kim, Y.Y.; Jung, K.W.; Yoo, K.S.; Jeung, J.U.; Shin, J.S. A stress-responsive caleosin-like protein, AtCLO4, acts as a negative regulator of ABA responses in Arabidopsis. Plant Cell Physiol. 2011, 52, 874–884. [Google Scholar] [CrossRef]
- Lu, W.; Tang, X.; Huo, Y.; Xu, R.; Qi, S.; Huang, J.; Zheng, C.; Wu, C.A. Identification and characterization of fructose 1,6-bisphosphate aldolase genes in Arabidopsis reveal a gene family with diverse responses to abiotic stresses. Gene 2012, 503, 65–74. [Google Scholar] [CrossRef]
- Ghelis, T.; Bolbach, G.; Clodic, G.; Habricot, Y.; Miginiac, E.; Sotta, B.; Jeannette, E. Protein tyrosine kinases and protein tyrosine phosphatases are involved in abscisic acid-dependent processes in Arabidopsis seeds and suspension cells. Plant Physiol. 2008, 148, 1668–1680. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miao, R.; Siao, W.; Zhang, N.; Lei, Z.; Lin, D.; Bhalerao, R.P.; Lu, C.; Xu, W. Katanin-Dependent Microtubule Ordering in Association with ABA Is Important for Root Hydrotropism. Int. J. Mol. Sci. 2022, 23, 3846. https://doi.org/10.3390/ijms23073846
Miao R, Siao W, Zhang N, Lei Z, Lin D, Bhalerao RP, Lu C, Xu W. Katanin-Dependent Microtubule Ordering in Association with ABA Is Important for Root Hydrotropism. International Journal of Molecular Sciences. 2022; 23(7):3846. https://doi.org/10.3390/ijms23073846
Chicago/Turabian StyleMiao, Rui, Wei Siao, Na Zhang, Zuliang Lei, Deshu Lin, Rishikesh P. Bhalerao, Congming Lu, and Weifeng Xu. 2022. "Katanin-Dependent Microtubule Ordering in Association with ABA Is Important for Root Hydrotropism" International Journal of Molecular Sciences 23, no. 7: 3846. https://doi.org/10.3390/ijms23073846
APA StyleMiao, R., Siao, W., Zhang, N., Lei, Z., Lin, D., Bhalerao, R. P., Lu, C., & Xu, W. (2022). Katanin-Dependent Microtubule Ordering in Association with ABA Is Important for Root Hydrotropism. International Journal of Molecular Sciences, 23(7), 3846. https://doi.org/10.3390/ijms23073846




