1. Introduction
The SID histo-blood group system was acknowledged by the International Society of Transfusion Medicine (ISBT) when an association to the
B4GALNT2 gene was shown [
1,
2]. There is only one antigen in the system, namely the Sd
a antigen, which had already been discovered in 1967 [
3,
4]. Approximately 91% of the European population carry the antigen on their erythrocytes, although 96–98% express it in tissues such as colon and kidney, or soluble in urine and saliva [
5,
6]. Among pregnant women, the incidence of Sd(a+) erythrocytes is lower than in normal blood donors [
3,
6,
7], which is similar to the Lewis system, where glycolipid antigens are passively adsorbed from plasma by the erythrocytes [
8]. No Sd
a is detected on the erythrocytes of newborns, while high levels are detected in their saliva [
6]. The 2–4% of adults who are truly Sd
a negative may produce naturally-occurring antibodies against the antigen. Although rare, hemolytic transfusion reactions have occurred [
9,
10], and rising IgG titers have been seen in patients after incompatible transfusions [
7]. Especially, transfusion of erythrocytes from donors with particularly high Sd
a expression should therefore be avoided in patients with the Sd(a−) phenotype and antibodies to Sd
a, since this could lead to hemolytic transfusion reactions. The rare Sd(a++) phenotype is defined by erythrocytes being highly reactive with anti-Sd
a. The most striking example of such a high-expressing phenotype is Cad, first detected in 1968 when erythrocytes of blood group B and O were strongly agglutinated by the
Dolichos biflorus agglutinin (DBA). This is a plant lectin otherwise used to type for the blood group A antigen, as it binds to terminal GalNAc residues [
11]. The structure of the anti-Sd
a binding epitope was defined in erythrocytes from Cad individuals as GalNAcβ1-4(NeuAcα2-3)Gal-R, which is found on O-linked core 1 structures carried on glycophorin A (GLPA) and on glycolipids in the shape of elongated sialyl-paragloboside [
12,
13]. In the urine of Sd(a+) individuals, Sd
a is carried on N-linked glycans attached to the Tamm–Horsfall glycoprotein (uromodulin) and on core 3
O-GalNAc glycans on mucins in colon [
14,
15]. However, Sd
a epitopes on N-glycans have not, to our knowledge, been described on other major glycoproteins on erythrocytes, such as, for instance, the band 3 anion transport protein (B3AT) [
16]. The antigen has been suggested to be used as a colon tumor marker due to its downregulation in cancerous tissue in favor of the sialyl-Lewis X (sLe
x) epitope, the synthesis of which utilizes the same sialylated precursors as those of the Sd
a epitope [
17]. Additionally, the structure seems to inhibit various pathogens from adhering or invading human cells [
18,
19,
20].
The
B4GALNT2 gene was cloned in 2003 and found to encode a
β-1,4-
N-acetylgalactosaminyltransferase (β4GalNAc-T2) able to synthesize the Sd
a-specific glycan epitope [
21,
22]. Two transcripts were detected, differing only in the use of a longer or a shorter alternative exon 1 and thereby generating enzymes with different lengths of the cytoplasmic tail [
22]. The gene seems not to be expressed in the erythroid lineage [
23] and, despite the revealed identity of the implicated glycosyltransferase, genetic differences between the three main phenotypes, Sd(a−), Sd(a+), and Sd(a++)/Cad, have remained unresolved for decades. In 2019, the nonsynonymous single nucleotide polymorphism (SNP) rs7224888:T>C (GenBank accession no. MK765047) in
B4GALNT2 was found to dominate two different cohorts of nine Sd(a−) individuals each, making up 13 of 18 and 17 of 18 alleles, respectively [
1,
2]. Even if these data were mainly correlative and descriptive, they led to the decision by the International Society of Blood Transfusion (ISBT) Working Party on Red Cell Immunogenetics and Blood Group Terminology to promote the high-frequency antigen Sd
a from the 901 series of blood group antigens to form an independent, new blood group system of its own, SID (ISBT no. 038, established on June 22, 2019). These studies also found one allele each with an intron 8 splice-site polymorphism, rs72835417:G>A (MK797056). In addition, our study found one Sd(a−) individual compound heterozygous for two rare missense mutations, rs148441237:A>G (MK765048) and rs61743617:C>T (MK765049), and one individual in whom no variation was found in the
B4GALNT2 regions investigated [
1].
In the present study, we aimed to provide conclusive proof that these missense variants cause Sda deficiency in humans. In addition, we added glycopeptide LC-MS/MS analyses to expand our knowledge on Sda-carrying glycoproteins and, more specifically, describe the epitope in its different glycoforms. Furthermore, we hypothesized and searched for a possible correlation between B4GALNT2 sequences, the Sda antigen, and its erythrocyte glycoprotein carriers in individuals with the Cad phenotype.
3. Discussion
The Sd
a histo-blood group antigen and its null phenotype has long withstood attempts to understand its underlying genetic basis. In addition, structural and functional aspects of this glycan are poorly understood. After the cloning of a candidate Sd
a synthase more than 15 years ago, we have now proven that polymorphism in this gene affects Sd
a expression. Our results demonstrate that rs7224888:T>C is not only commonly found in Sd(a−) individuals but truly is the causative alteration that abolishes the formation of the Sd
a epitope. This missense mutation is situated in exon 10 and leads to the amino acid change where a cysteine is replaced by arginine at p.406 (in AJ517771) or p.466 in the enzyme utilizing the long exon 1 (AJ517770) in the globular catalytic domain of the enzyme. While the transferase is detected in cells transfected with this allele, but not its expected carbohydrate product, it is likely that the amino acid shift causes a functional alteration, severely damaging the enzymatic activity of the glycosyltransferase. The β4GalNAc-T2 homologue, human β4GalNAc-T1 (also called GM2 synthase) encoded by
B4GALNT1, has been evaluated for its structural conformation dependence on its cysteine residues [
24]. The homologue occurs as a homodimer and can be found in a soluble form, [
25], in analogy with β4GalNAc-T2 where soluble forms are found in urine and plasma from humans [
27,
28]. All the cysteines of the soluble β4GalNAc-T1 are involved in disulfide bonds, four in intersubunit (dimer) formation and two in intrasubunit bonds. The spacing between all these cysteine residues is conserved, comparing the two enzymes (
Supplementary Figure S5). The p.406Cys affected by rs7224888:T>C is highly conserved among several species, as we showed by sequence alignments from multiple species in our previous study [
1], and this cysteine corresponds to one of the intrasubunit disulfide bonds in soluble β4GalNAc-T1 (
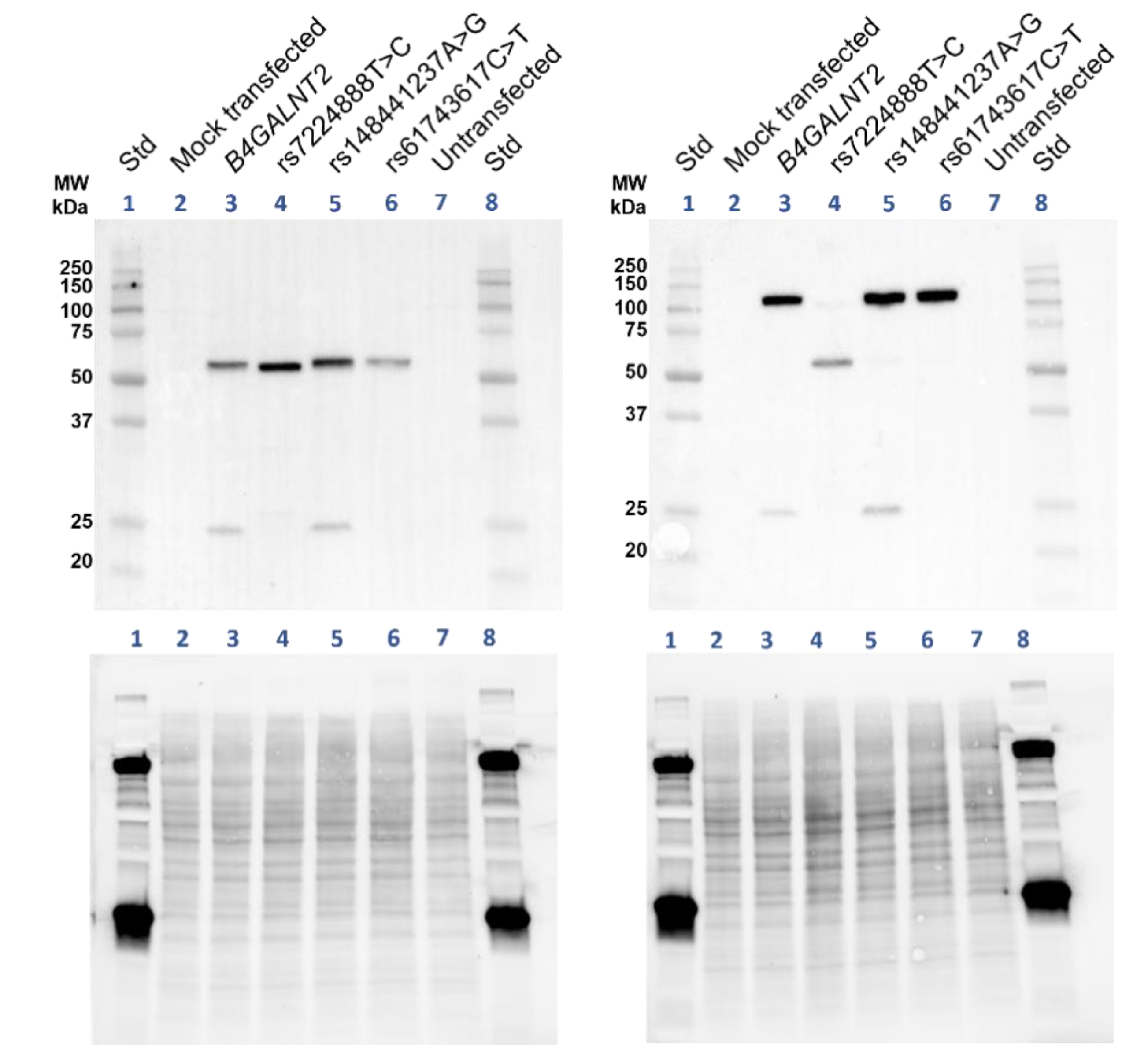
Supplementary Figure S5). The results of the nonreduced immunoblot on lysates from transfected cells support that β4GalNAc-T2 is also involved in homodimer formation, which is almost entirely lost when p.Cys406Arg is introduced (
Figure 3). It is therefore reasonable to hypothesize that the role of this cysteine is quite plausibly the same in Sd
a synthase as that described for GM2 synthase, and replacement with the large and positively charged arginine must have major consequences for structure and function of the
B4GALNT2-encoded enzyme.
The structure of β4GalNAc-T2 has not yet been determined by x-ray crystallography or other types of in vitro experiments. To visualize the location of p.Cys406 and other residues affected in the mutants studied here, we interrogated the AlphaFold protein structure database and found a structure model for the long form of β4GalNAc-T2. The catalytic domain is predicted with a high degree of confidence (>90) as judged by the per residue confidence metric (predicted local distance difference test, pLDDT), given on a scale from 0 to 100. In
Supplementary Figure S6, an overview of the molecule (
Figure S6A–D), its pLDDT values (
Figure S6C,D), DXD motif (
Figure S6C) and the implicated amino acid positions studied here (
Figure S6A–I) are shown, as well as close-ups of each residue which is altered in the mutants. It is particularly interesting to note that AlphaFold predicts a disulfide bridge formed between p.466Cys and p.513Cys, in full analogy with GM synthase (
Supplementary Figures S5 and S6B,E,F).
The two rare SNPs, rs148441237:A>G and rs61743617:C>T, that were found in a compound heterozygous Sd(a−) individual, cause amino acid changes not too far from the predominant SNP just discussed (p.406Cys>Arg), namely p.376Glu>Arg and p.463Arg>Trp (
Supplementary Figure S6G–I, respectively). Surprisingly, our study shows that overexpression of these two variants produces Sd
a epitopes in HEK293 cells with profiles similar to the wt β4GalNAc-T2. This is in sharp contrast to the predominant Sd(a−) allele with rs7224888:T>C, for which all Sd
a expression was abolished. Accordingly, the situation for the two rare alleles remains to be explored. If these two SNPs truly do not affect the synthesis of Sd
a antigen, the explanation may be found elsewhere. It is possible that a causative regulatory element variant would occur in linkage disequilibrium with one or both of these missense mutations, as recently reported for another carbohydrate blood group antigen [
29]. In our previous study, we identified one Sd(a−) individual without any crucial mutation in the coding region of
B4GALNT2, which may also hint at a regulatory, noncoding defect beyond the scope of this study [
1]. One could also speculate as to whether the β4GalNAc-T2 catalytic ability is dependent on homodimerization, as seems to be the case for β4GalNAc-T1 [
24]. If so, one could even speculate that the combination of the two different rare alleles could be so unique that the two different β4GalNAc-T2 variants together are incapable of forming a functional unit. However, we would have to assume that if such dimers are required, homodimer formation would not be affected. The co-transfection of the two rare alleles did not indicate that their products would interfere with the functionality of Sd
a synthesis. The co-transfection experiment was only performed once and did not include mass spectrometric evaluation or immunoblotting, but the flow cytometric result was virtually identical to the wt construct and the results obtained when each of the rare mutations was assessed separately.
Our LC-MS/MS data show that the Sd
a epitope is found on a broad range of membrane glycoproteins from the
B4GALNT2-transfected HEK293 cells and carried on both
N- and O-linked glycans (
Table 1). This confirms what is known about the human transferase in different tissues, i.e., that it is capable to build upon Neu5Acα2,3Gal both on N-glycans, as in the case of Tamm–Horsfall protein in urine [
15], and upon
O-GalNAc initiated glycans, as seen in the colon as well as on Cad erythrocytes [
12,
14]. A hypothesis about qualitative differences between the wt enzyme and the two rare variants discussed above could be that they have slightly different acceptor preferences. However, LC-MS/MS did not pinpoint any major differences between the glycan structures of glycoproteins identified in these cell preparations. If anything, the wt showed slightly fewer targets than the two Sd
a-synthesizing but mutated constructs (
Figure 5).
The Cad samples displayed no common crucial genetic alteration in the coding gene sequence or 2000 bp upstream of the gene. We cannot yet rule out that the genetic background of Cad lies in unknown regulatory regions of the
B4GALNT2 gene. Such regions can stretch much further than what has been investigated here and, in the case of Cad, it would be of interest to search systematically for enhancer elements. Methylation in areas of the gene and the transcription factors ETS1 and SP1 have been associated with regulation of
B4GALNT2 in malignant colon tissue [
30,
31,
32] However, it is not known how this may affect Sd
a expression on erythrocytes. Among the Cad samples (Cad-a and Cad-b), it was interesting to see that one individual was heterozygous for rs72835417:G>A, the splice-site mutation in intron 8, found earlier heterozygously in two Sd(a−) individuals (1,2), and thereby suggested as a reason to lack the antigen. This is of course still possible, as the genotype that causes the Cad phenotype could be a dominant trait carried on the other allele. Besides, further studies of the rs72835417:G>A allele are needed to determine its functional consequences on Sd
a synthesis.
It is also possible that Cad depends on a
B4GALNT2-independent genetic trait, a hypothesis that would require a genome-wide approach, which is beyond the scope of this study. One can also speculate on whether the access to precursor substrate for β4GalNAc-T2 can differ in some people. It is interesting to note that Cad is more common in Asia and that there is an association between Sd
a expression and malaria susceptibility.
B4GALNT2 was also recently identified as the key inhibitory factor for avian influenza A [
20], so the variable frequency of Cad in different parts of the world may have multiple evolutionary backgrounds.
The Sd
a antigen was also detected in the glycoproteome of membranes from erythrocytes with the Cad phenotype, but not in erythrocytes with the common Sd(a+) phenotype. This confirms earlier studies, as does our finding of the Sd
a epitope on GLPA from Cad erythrocytes [
12,
33]. Additionally, we identified B3AT and S29A1 as carriers of the Sd
a epitope on N- and O-glycans, respectively. These erythroid membrane proteins are the carriers of antigens in the Diego and Augustine blood group systems, respectively [
34,
35]. The peptide epitope has thereby been found on three proteins that are abundant in the erythrocyte membrane and considered to be endogenously synthesized [
34,
35]. Finding the Sd
a epitope on these proteins suggests an erythroid origin, which appears to contradict data that indicate
B4GALNT2 not to be expressed in erythroid tissue [
23]. Based on this, we hypothesized that erythroid
B4GALNT2 expression could be restricted to individuals with the Cad phenotype. We were, however, not able to detect transcripts of the gene in whole blood from Cad individuals, but this could be a matter of sensitivity of the method and the available material.
The Sd
a-decorated proportion of glycopeptides differed substantially between the two samples investigated. For the Cad-a and Cad-b samples, the glycoproteomic analysis showed a relative peak intensity of Sd
a glycans for GLPA glycopeptides of 10–12% vs. <0.5%, respectively. This mirrors the Sd
a histogram patterns of the stained erythrocytes in
Figure 7C,D, where the latter express the epitope on a much smaller number of cells. In line with this observation, the Cad phenotype has historically been divided into subtypes 1–3, where Cad-1 defines erythrocytes that are most reactive and polyagglutinable, Cad-2 has been found in individuals of Asian origin and are only weakly (if at all) polyagglutinable. Finally, Cad-3 was found in two European families in whom polyagglutination does not appear to occur [
36,
37], although this has been disputed [
38]. A previous study of the Sd
a epitope on GLPA in three different Cad donors also detected individual variations between the donors in the amount of glycans that carried the epitope. The strongest immunoreactive cells had indeed GLPA decorated with a larger number of Sd
a [
12]. The two Cad samples in our study are both from Australian individuals of Chinese origin. Although the origin suggests both samples to be of subtype Cad-2, their subtype was not further characterized. It is nevertheless interesting that the relative amounts of Sd
a epitopes in erythrocytes of our Cad-a and Cad-b samples vary considerably and that the glycoproteomic methodology could be used so efficiently to distinguish them.
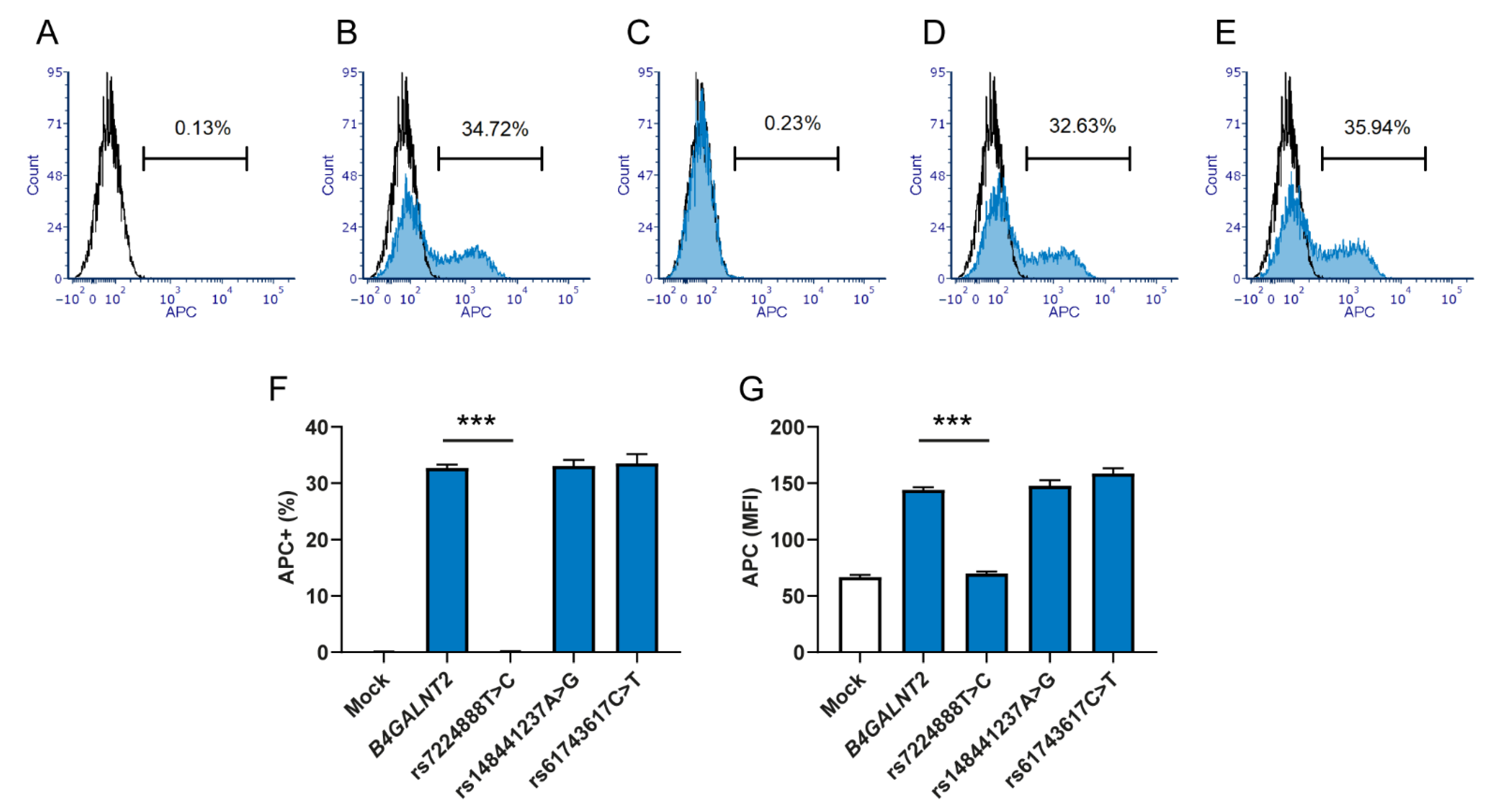
In summary, we evaluated the consequences of three B4GALNT2 alleles previously found in Sd(a−) individuals, and we were able to present data that conclusively point out a relatively common polymorphism in B4GALNT2 (rs7224888:T>C) as the main reason underlying complete deficiency of Sda-containing glycans in humans. On the other hand, two rare variants (rs148441237:A>G and rs61743617:C>T) previously associated with the null phenotype did not appear to affect the synthesis of Sda, as detected here. A more detailed investigation may be required to understand their role. We also provide genetic and biochemical insight into the high-expressing Sd(a++)/Cad variant, indicating that future studies of the erythroid regulation of B4GALNT2 expression may offer an explanation to the underlying basis of this enigmatic phenotype.
4. Experimental Procedures
4.1. B4GALNT2 Overexpression in HEK293 Cells
Constructs with the B4GALNT2 wt gene (GenBank accession no. AJ517771) or B4GALNT2 mutated constructs based on the same sequence, but with SNPs, B4GALNT2_rs7224888:T>C, B4GALNT2_rs148441237:A>G or B4GALNT2_rs61743617:C>T in the bicistronic GFP expressing vector CS-2719-M61, were synthesized by GeneCopeia (Rockville, MD, USA) and distributed by Labomics (Nivelles, Belgium). The plasmid constructs were transformed into One Shot Top10 chemically competent E. coli (Thermo Fisher Scientific, Waltham, MA, USA) by adding 50 ng DNA to 25 µL competent cells, incubated on ice for 30 min, followed by a heat shock (42 °C for 30 s), then back on ice for 2 min. The cells were incubated in 250 µL SOB medium (10 mM NaCl, 2,5 mM KCl, 10 mM MgCl2, 10 mM MgSO4, 20 g/L Tryptone and 5 g/L Yeast extract) for 60 min at 37 °C while shaking. Cells were then spread on SOB–agar plates with ampicillin and cultured overnight (16 h) at 37 °C. Single colonies were picked for further growth in a starter cultures of 2 mL SOB with ampicillin for ca 6 h at 37 °C. The cultures were diluted 1:500 in 25 mL SOB and cultured overnight. The plasmid constructs were extracted using the plasmid midi kit from Qiagen (Hilden, Germany). The sequences were controlled applying Sanger sequencing by Eurofins Genomics (Ebersberg, Germany), with primers M61-F (5´-GCGGTAGGCGTGTACGGT) and M61-R (5´-AGCAGTCCCCAAGTCAGT).
The constructs were transfected into human embryonic kidney cell line HEK293, as this cell line shows no or low levels of endogenous
B4GALNT2 expression and Sd
a [
17]. Transfections were performed on cell cultures at 80% confluency, plated 24 h previously. JetPRIME transfection reagent from Polyplus transfection (Illkirch, France) was used according to manufacturer’s protocol for cultures in 24- or 6-well plates. The cells were cultured in DMEM high glucose medium (Thermo Fisher Scientific) with 10% FBS. Transfected cells were harvested 24 h post transfection by detaching the adherent cells in TrypLE Express (Thermo Fisher Scientific) for 5 min at 37 °C after rinsing in PBS. For LC-MS/MS and sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE)/Western blot GFP+ cells were sorted on BD FACSAria III (Becton, Dickinson and Company, Franklin Lakes, NJ, USA) 48 h post transfection and, once readily growing, cultured with selection pressure at 500 µL/mL Geneticin (Thermo Fisher Scientific).
4.2. Blood Samples
Anonymized whole blood samples were obtained from the Australian Red Cross Lifeblood (Brisbane, Queensland, Sydney, New South Wales and Perth, Western Australia, Australia) and from in-house collection reagent test erythrocytes and anonymized donor blood from the blood group reference laboratory in Lund, Sweden. A Sd(a−) test erythrocyte preparation from a commercially available rare cell panel (special panel from the German Red Cross Blood Donor Service Baden-Wuerttemberg—Hessen, Baden-Baden, Germany) was used as a negative control in flow cytometry experiments. Four Australian Cad samples (from individuals of Asian origin) were initially identified in the routine automated PK Olympus blood grouping reaction (DIAGAST, Loos, France) and further investigated at the Red Cell Reference Laboratory, Clinical Services and Research, Australian Red Cross Lifeblood, Kelvin Grove, Australia and in the blood group reference laboratory at the Department of Clinical Immunology and Transfusion Medicine in Lund, Sweden. Briefly, an apparent discrepancy in the routine ABO blood grouping reactions between forward (erythrocytes) and reverse (plasma) typing led to further evaluation of the underlying phenotype. In Australia, the serology was first investigated with a polyagglutination kit (Inverclyde Biologicals, Bellshill, Scotland, UK), DBA, papain treatment, Hydatid cyst fluid (HCF) and guinea pig urine (GPU) from Serum, Cells and Rare Fluids (SCARF) distribution scheme, Helix pomatia agglutinin (HPA) from Immucor Medizinische Diagnostik GmbH (Dreieich, Germany) and 10 different polyclonal Anti-A (in-house from donor plasma). Subsequent investigation of the erythrocytes and Cad phenotyping in Sweden included additional reagents: DBA (Immucor, Norcross, GA, USA), monoclonal Seraclone anti-A1 (Bio-Rad), a panel of lectins from Gamma Biologicals/Immucor including Salvia horminum, Arachis hypogea, Glycine soy and Salvia sclarea. Additionally, monoclonal anti-Sda antibody (clone KM694, Kyowa, BioFrontier Laboratories, Tokyo, Japan) and pooled human urine was used. In flow cytometry, the FITC-conjugated lectin Helix pomatia-stained proportion of the erythrocyte population was used as a measure to differentiate common Sd(a+) from Cad status. The serological investigation ruled out other causes of polyagglutination, as well as ABO and FORS blood group systems’ variants, and concluded the irregular, original reaction to be due to the Cad phenotype. Erythrocytes from Sd(a+) donors were analyzed for comparison in glycoproteomic analysis, flow cytometry experiments and detection of B4GALNT2 transcripts. These were from random Swedish donors or draw-date-matched donors sent together with the Cad-b sample from Australia.
In-house reagents consisting of human plasma samples from Sd(a−) individuals with anti-Sda, used for flow cytometry, were obtained as a kind gift from the Hoxworth Blood Center´s Immunohematology Reference Laboratory (ORL, Cincinnati, OH, USA).
4.3. Preparation of Erythrocyte (White Ghost) Membranes
The erythrocytes were lysed and repeatedly washed in cold hypotonic buffer (310 buffer, 100 mM NaH2PO4, 155 mM Na2HPO4 and diluted 1:20.5 in H2O), until the supernatant was neutral in color and a white fluffy pellet of membranes remained. The membranes were stored at −80 °C.
4.4. Flow Cytometry
Transfected HEK293 cells (approximately 4 × 105) were stained with biotinylated DBA (Vector laboratories Inc., Burlingame, CA, USA), diluted 1:400,000 for 30 min at 4 °C in 50 µL FACS buffer (PBS with 1% BSA). The cells were washed and incubated with APC-conjugated streptavidin (Thermo Fisher Scientific), diluted 1:1000 for 30 min at 4 °C. The experiment was repeated three times with three biological replicates in each experiment. For Sda staining, HEK293 cells or glutaraldehyde (0.1%) fixed erythrocytes were incubated in 50 µL plasma (diluted 1:1 in PBS) for 30 min at room temperature while agitating. After washing in PBS, secondary antibody was added, polyclonal Alexa fluor 647 or phycoerythrin (PE)- conjugated goat anti-human IgM from Jackson ImmunoResearch (West Grove, PA, USA). Incubation conditions were 10 min at room temperature. The staining protocol with human anti-Sda included a separate incubation with ABO-matched control plasma without anti-Sda. In prior analysis, the HEK293 cells were washed and filtered through 50 µm cup filters (Becton, Dickinson and Company). The GFP and APC/Alexa fluor 647 signals were recorded with BD FACSCanto II (Becton, Dickinson and Company) and the data analyzed in FCS express 6 (De Novo Software, Glendale, CA, USA). For statistical analysis, nonparametric Mann–Whitney U test was performed using the GraphPad prism software v. 8.4.2. (San Diego, CA, USA).
4.5. SDS-PAGE and Western Blotting of β4GalNAc-T2
Transfected cells were washed in PBS and one million were pelleted and lysed directly in 50 µL sample buffer, Laemmli (Bio-Rad, Hercules, CA, USA), with or without 5% β-mercaptoethanol (Sigma Aldrich, St. Louis, MA, USA). The lysates were sonicated and incubated for 5 min at 99 °C and SDS-PAGE performed as described elsewhere [
39]. In brief, the proteins, samples and standard (precision plus protein dual xtra prestained standard) were separated at 300 V in SDS-PAGE on Mini-PROTEAN any-kD Tris-Glycine extended stain-free precast gel (Bio-Rad), activated by ultraviolet light (UV) in a ChemiDoc Touch camera and transferred to low-fluorescence polyvinylidene difluoride (PVDF) membranes. Total protein image was obtained by UV-light exposure of the PVDF-membrane, which then was subjected to blocking overnight in Casein ×1 solution (Vector laboratories) diluted in H
2O. Blotting was achieved by incubating the membrane in blocking buffer and the polyclonal rabbit IgG anti-β4GalNAc-T2 (Sigma-Aldrich) diluted to 1:1000 at room temperature for two hours. The membrane was washed in TBS-T and incubated one hour at room temperature with Horseradish peroxidase (HRP)-conjugated polyclonal goat anti-Rabbit IgG (Bio-Rad) diluted to 1:3000. Enhanced chemiluminescent reagents (Bio-Rad) were used to develop the blot, which then was visualized in the ChemiDoc Touch camera as well as a colorimetric image of the standards. The images were annotated in Image Lab Software v 6.1. The SDS-PAGE equipment, camera and software were from Bio-Rad.
4.6. Sample Preparation of Cells for LC- MS/MS Analysis
Transfected cells were plated at 5 million cells per 10-cm-diameter dish and cultured until cells reached circa 80% confluency. The adhering cells were rinsed three times in PBS, lysed with 50 mM triethylammonium bicarbonate (TEAB) buffer with 2% SDS and the cell debris detached with a scraper, transferred to tubes and stored at −80 °C. The frozen cell lysates were sent to BioMS, the national infrastructure node for glycomics and glycoproteomics (University of Gothenburg, Sweden), for further sample preparation.
The lysates from either transfected cells or erythrocyte membranes were subjected to filter-aided sample preparation (FASP) with trypsin digestion. Briefly, samples were reduced with 100 mM dithiothreitol at 60 °C for 30 min, and then applied to 30 kDa MWCO filters (Pall Nanosep, Sigma-Aldrich). After centrifugation, the retentate was washed with 8 M urea and then with 0.5% sodium deoxycholate (SDC) in TEAB buffer. Cys residues were methylthio derivatized with 10 mM methyl methanethiosulfonate in 0.5% SDC in TEAB buffer at room temperature for 20 min. The trypsin digestion was performed on the MWCO filter with 4 µg trypsin (Pierce MS grade, Thermo Fisher Scientific) in 0.5% SDC in TEAB buffer at 37 °C overnight, with a new trypsin addition over 2 h the following day. Cleavages were also performed with chymotrypsin and proteinase K (sequencing grade, Promega) on selected samples. After centrifugation of the MWCO spin column, the filtrate was saved and SDC was precipitated by acidification using 10% trifluoroacetic acid (TFA), and the supernatant was saved after centrifugation.
The cleaved samples were desalted with C18 spin columns and dissolved in 80% acetonitrile/1% TFA in water, and glycopeptides enriched with hydrophilic interaction liquid chromatography (HILIC), with slight modifications [
40]. Briefly, the samples were applied to in-house packed cartridges containing 20 mg Zic-HILIC particles (10 μm, 200 Å; Sequant/Merck). The flow-through was collected and re-applied three times. The column was then washed with 1.2 mL of 80% acetonitrile and 1% TFA in water. Enriched glycopeptides were eluted with 4× of 50 µL 0.1% TFA, 50 μL of 25 mM ammonium bicarbonate in water and, finally, 50 μL of 50% (
v/
v) acetonitrile in water and dried by vacuum centrifugation.
4.7. LC-MS/MS Analysis
The samples were dissolved in 20 µL 0.2% formic acid, 3% acetonitrile in water and analyzed on an Orbitrap Lumos Tribrid mass spectrometer interfaced with Easy-nLC1200 liquid chromatography system (Thermo Fisher Scientific). An Acclaim PepMap 100 C18 trap column (100 μm × 2 cm, particle size 5 μm, Thermo Fischer Scientific) was used and peptides were separated at 300 nL/min on an in-house packed analytical column (75 μm × 30 cm, particle size 3 μm, Reprosil-Pur C18, Dr. Maisch) using a linear gradient of 7–35% of solvent B in solvent A over 75 min, then an increase to 100% of solvent B for 5 min and, finally, 100% of solvent B for 10 min. Solvent A was 0.2% formic acid in water and solvent B was 80% acetonitrile and 0.2% formic acid in water. The precursor ion scans were performed at a resolution of 120,000 and the m/z range was 600–2000. The most abundant precursor ions with a positive charge (z) of 2–7 were selected for MS/MS (MS2) over 3 s with a 5 m/z unit isolation window and subjected sequentially to higher-energy collision induced dissociation (HCD) at normalized collision energies (NCEs) of 20%, 30% and 40%. The MS2 spectra were detected in the Orbitrap in centroid mode at 30,000 resolution.
4.8. Glycoproteomic Analysis
The LC-MS/MS raw files were analyzed with the Byonic software (Protein Metrics) using a modified list of the modifications “182 human N-glycans” and “6 most common O-glycans”. For instance, N-glycan modifications with NeuAc2Hex5HexNAc6 and NeuAc2Hex5HexNAc6dHex compositions were added, and O-glycan modifications of NeuAcHexHexNAc2 and NeuAc2HexHexNAc2 were added. Additional search criteria included use of the UniProt Homo sapiens database (20,369 sequences), C-terminal cleavage after Lys and Arg, two missed cleavages were allowed, accuracy for the MS1 precursor ion was set to 10 ppm and for MS2 ions it was 20 ppm static modification was a methylthio group on Cys (+45.9877 u), and variable modification was oxidation of Met. For the chymotrypsin digested samples, C-terminal cleavage after Phe, Tyr, Trp, and Leu, with two allowed missed cleavages, were applied. For the pronase digested samples, all possible cleavages were considered, and the protein database was composed of selected glycoproteins: GLPA_HUMAN, GLPB_HUMAN, GLPC_HUMAN, GLPE_HUMAN, B3AT_HUMAN, TFR1_HUMAN, AQP1_HUMAN, RHD_HUMAN, and ACKR1_HUMAN.
For the trypsin and chymotrypsin Byonic analyses, the reversed sequences (decoy database) were included in the analysis, and a cut off score of 300 was used, which resulted in false discovery rates of 0.46–2.00% for the glycopeptide hits (
Supplementary Tables S1, S2, S4 and S5). In addition, lower scores were also accepted for alternative glycoforms, which shared the same peptide and had the expected glycosidic fragmentation profile, with glycopeptides having Byonic scores >300 from the same sample. In addition, all glycopeptide MS/MS spectra were verified to contain the correct peptide + HexNAc ion for N-glycopeptides and the correct peptide ion for O-glycopeptides. For NeuAc hits, the diagnostic NeuAc oxonium ions at
m/
z 274.09 and
m/
z 292.10 had to be present. Extracted ion chromatograms (XICs) were produced with the Xcalibur software (Thermo Fisher Scientific) for the diagnostic ion (NeuAc HexHexNAc
2)
+ at
m/
z 860.31 to identify additional glycopeptides.
4.9. Gene Sequencing
Whole blood derived DNA was prepared by a simple salting out method [
41]. Amplification was achieved using modified Expand high-fidelity PCR system (Roche Basel Switzerland) [
42]. Briefly, DNA (ca 150 ng) was mixed with 0.2 U Taq Polymerase and 7 pmol of each primer, previously published [
1], with the addition of SID-2301F (5′-TAGTTTCTGCCTGTAGCCC), in a total reaction volume of 20 µL. The PCR protocol was initiated for 3 min at 95 °C followed by 35 thermal cycles of 95 °C (20 s), 58 °C (30 s), and 72 °C (40 s or 3 min, depending on amplicon size). The PCR products were run on a 3% gel from which the correct sized amplicons were excised and purified with QIAquick gel extraction kit (Qiagen). Sanger sequencing was performed in house [
42] or by Eurofins Genomics (Ebersberg, Germany). Sequences were analyzed in CodonCode aligner software v. 4.2.7 (Barnstable, MA, USA).
4.10. Real-Time Quantitative Polymerase Chain Reaction (qPCR)
Frozen whole blood in Trizol® (Invitrogen) was thawed and RNA extracted according to the manufacturer’s instructions, followed by cDNA synthesis with SuperScript IV Vilo (Thermo Fisher Scientific). Real-time qPCR was performed using Taqman gene expressing assays Hs00963127_m1, Hs01060665_g1, and Hs02758991_g1 (Thermo Fisher Scientific), targeting B4GALNT2, ACTB (reference), and GAPDH (control), respectively. The gastric adenocarcinoma cell line MKN-45 cDNA was used as positive control.
4.11. β4GalNAc-T2 Structure Prediction
Three-dimensional molecular modelling of β4GalNAc-T2 (identifier AF-Q8NHY0-F1 including amino acids 1–566) was performed using the AlphaFold Protein Structure Database (developed by DeepMind and EMBL-EBI), and the resulting PDB file was downloaded and analyzed in PyMOL (version 2.3.4) [
43,
44,
45]. Numbering of amino acids in the model is based on the long form, i.e., p.466Cys>Arg corresponds to p.406Cys>Arg in the short form.
4.12. Ethical Consideration
Ethical review and approval were not required for this study since only leftover, anonymized blood samples taken as part of routine blood donation from healthy blood donors were used. This is in accordance with the Swedish law (2003:460) on research on humans and biological material from humans.