Abstract
Lipid-derived jasmonates (JAs) play a crucial role in a variety of plant development and defense mechanisms. In recent years, significant progress has been made toward understanding the JA signaling pathway. In this review, we discuss JA biosynthesis, as well as its core signaling pathway, termination mechanisms, and the evolutionary origin of JA signaling. JA regulates not only plant regeneration, reproductive growth, and vegetative growth but also the responses of plants to stresses, including pathogen as well as virus infection, herbivore attack, and abiotic stresses. We also focus on the JA signaling pathway, considering its crosstalk with the gibberellin (GA), auxin, and phytochrome signaling pathways for mediation of the trade-offs between growth and defense. In summary, JA signals regulate multiple outputs of plant defense and growth and act to balance growth and defense in order to adapt to complex environments.
1. Introduction
The plant lipid-derived hormone jasmonic acid (JA), a critical mediator of the plant defense response, is an important regulator of plant growth and development. It was in the 1980s that the first physiological processes caused by JA or methyl jasmonate (MeJA) were described, such as growth inhibition and senescence promotion [1,2]. In the early 1990s, researchers found that JA induced the accumulation of vegetative storage proteins (VSPs) after wounding in soybean leaves [3] and proteinase inhibitors (PIs) in tomato plants injured by herbivores. Turning to recent years, JA has become a research hotspot due to its diverse, complex, and specific functions.
During the last decades, much work has been performed to investigate the structure, function, and regulation of the enzymes involved in JAs biosynthesis, primarily contributing to explaining how JA is accumulated. An appropriate level of JA is essential for its biological functions [4,5,6,7], while excessive JA accumulation would trigger an overactivation of the defense machinery, which in turn comes at the expense of plant growth. Thus, a negative feedback regulation mechanism is required to delay the termination of JA signaling [8,9,10]. What has been proven beyond dispute is that plants have evolved strategies to balance both growth and reproduction with the need for defense in order to optimize their fitness in diverse environments [11]. In fact, JA modulates a major branch in the growth-defense balance [12,13,14], as a key hub of crosstalk between and with the salicylic acid (SA), ethylene (ET), gibberellin (GA), and phytochrome signaling pathways, among others [15,16,17]. Through these sophisticated JA-mediated mechanisms, plants have evolved to relocate nutrition in response to diverse and complex environmental conditions; as such, exploitation of these mechanisms may enrich both plant breeding and engineering strategies for maximizing plant fitness as well as yield.
In this paper, we review and summarize the biosynthetic pathway of JAs, a core signaling pathway, its evolutionary origin, and the multiple terminal mechanisms of JA signals, as well as the research progress considering JA in plant growth and development, stress tolerance, and recent mechanistic insights into the close relationship between growth and immunity, in order to explain and explore the possibility of maximizing the growth-defense balance.
2. Biosynthesis of Jasmonate
JAs are members of the family of oxylipins. There are at least three JA biosynthesis pathways in plants (Figure 1), either initiating with α-linolenic acid (shorthand notation, 18:3), starting from 7, 10, 13-hexadecatrienoic acid (shorthand notation, 16:3), or through an OPR3-independent pathway [8,18,19].
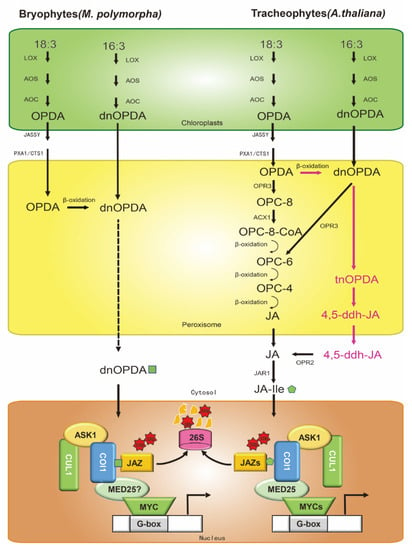
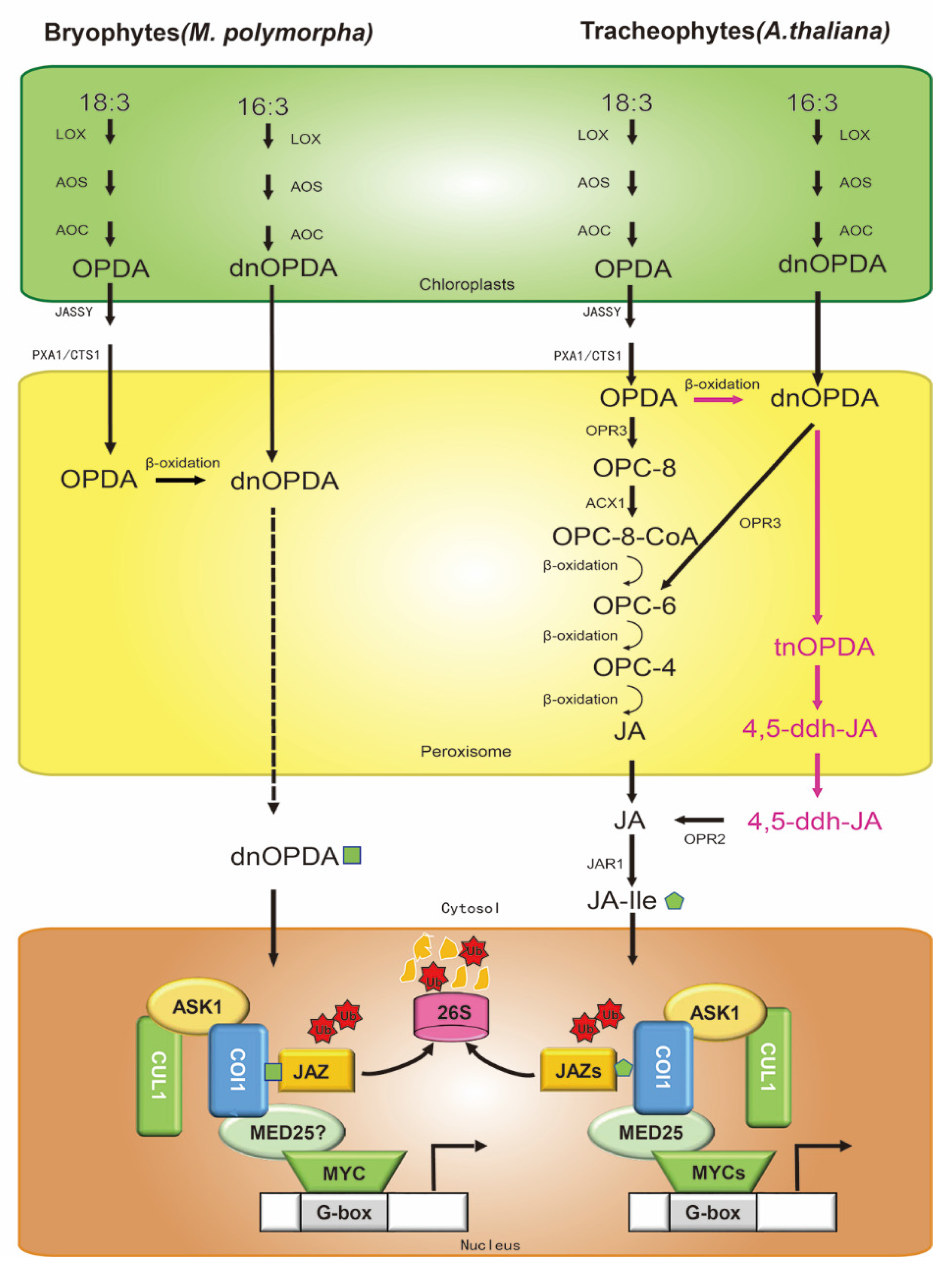
Figure 1.
Biosynthesis and core signaling pathways of jasmonic acid (JA) in Arabidopsis and Marchantia polymorpha. There are three synthetic pathways in Arabidopsis (right): (1) the 18:3 pathway; (2) the 16:3 pathway; and (3) the independent OPR3 pathway, in which the cytoplasm JAR1 catalyzes JA to form JA-Ile with biological activity. JA-Ile interacts with the COI1-JAZ complex to degrade JAZ(s) by ubiquitination through the 26S proteasome degradation pathway. The difference in jasmonic acid evolution between Marchantia polymorpha (left) and Arabidopsis is mainly reflected in (1) the number of JAZ and MYC genes in M. polymorpha and Arabidopsis; (2) the bioactive COI1-JAZ ligand is dnOPDA in M. polymorpha, but JA-Ile in Arabidopsis. The mechanisms of MYC and MED25 in the JA signaling pathway in M. polymorpha are not proven (question mark). Ub, ubiquitin. Arrows: activations; bar-headed arrows: repressions.
The first step of the 18:3 biosynthetic pathway occurs at the chloroplast membranes (Figure 1) [8,20]. Galactolipids release α-linolenic acid (18:3) by lipases [21]. Then, 13-lipoxygenase (LOX) catalyzes 18:3 to 13(S)-hydroperoxyoctadecatrienoic acid (13-HPOT). The coupled dehydration-cyclization of 13-HPOT, promoted by allene oxide synthase (AOS) and allene oxide cyclase (AOC), then forms 12-oxophytodienoic acid (OPDA). OPDA is transported from chloroplasts to the peroxisomes by the chloroplast envelope-localized transporter JASSY [22], peroxisomal ABC-transporter1 (PXA1), and COMATOSE1 (CTS1) [23], and is catalyzed by OPDA reductase 3 (OPR3) to 3-oxo-2-(20(Z)-pentenyl)-cyclopentane-1-octanoic acid (OPC8:0), which yields JA after undergoing three cycles of β-oxidation [24]. In a parallel pathway, dinor-OPDA (dnOPDA) is produced by the consecutive action of LOX, AOS, and AOC from 16:3 in chloroplast membranes (Figure 1) [25]. dnOPDA is transported into the peroxisome and reduced to hexanoic acid (OPC6:0) by OPR3, following which JA is generated by two cycles of β-oxidation. Recent studies have found an alternative pathway (OPR3-independent) in Arabidopsis (Figure 1) [19,26]. It is the same as the 18:3 biosynthetic pathway in chloroplast membranes, but in the peroxisome, OPDA yields dnOPDA through a single round of β-oxidation. Then, dnOPDA converts to 4, 5-didehydro-JA (4, 5-ddh-JA) in two further rounds of β-oxidation, which is subsequently reduced to JA by OPR2 in cytosol. After that, Jasmonoyl-isoleucine synthetase (JAR1) conjugates Ile to JA to form the receptor-active ligand (3R, 7S)-jasmonoyl-L-isoleucine (JA-Ile) [19].
3. Jasmonate Signaling Pathway
3.1. The Core JA-Ile Pathway
The core jasmonate signaling pathway consists of interconnected functional modules that regulate the transcriptional state of JA-responsive genes. The bioactive JA-Ile is perceived by cognate nuclear receptors CORONATINE INSENSITIVE 1 (COI1). As a typical F-box protein, COI1 first binds to SKP1-LIKE proteins (ASK1 or ASK2) and subsequently forms the Skp1/Cul1/F-box complex (SCFCOI1) with Arabidopsis Cullin1 to recruit JASMONATE-ZIM DOMAIN (JAZ) proteins for ubiquitination and degradation [24,27,28]. JAZ proteins are significant repressors that connect early responsive MYCs (MYC2/3/4/5) transcription factors (TFs) and, later, responsive downstream genes [8,29]. Actually, 1 COI, 13 JAZ, and 4 MYC proteins had been described in Arabidopsis [30,31,32]. In the absence of biotic effectors in plants, JA-Ile is typically at a lower level, and the JAZ family proteins accumulate, which physically bind to and inhibit MYCs [33,34] through two distinct mechanisms [29,33,35]. First, MYC-bound JAZ proteins recruit the co-repressors TOPLESS (TPL), either directly by ETHYLENE-RESPONSE FACTOR-associated amphiphilic repression (EAR) motifs located at the N terminus of a subset of JAZ proteins (JAZ5/6/7/8/13), or indirectly through NOVEL INTERACTOR OF JAZ (NINJA), an EAR motif-containing protein [36,37,38] (Figure 1). Constitutively, it prevents MYCs from binding to the G-box sequence of downstream gene promoters, thereby blocking the JA signaling pathway [9]. JA-Ile is rapidly activated in response to various stresses, followed by the generation of the JAZ-JA-IIe-SCFCOI1 E3 protein complex, which ubiquitinates and degrades JAZs proteins [33]. After the degradation of JAZ proteins, TPL is disintegrated to release MYCs, inducing the transcription of the responsive genes in the JA signaling pathway [39].
In contrast to vascular plants, the bioactive COI1-JAZ ligand in Marchantia polymorpha is not JA-Ile, but rather dnOPDA -the precursor of JA-Ile. Both the 18:3 and 16:3 pathways in chloroplasts are the same as in vascular plants, whereas OPDA produced by the 18:3 pathway in the peroxisome passes through one round of β-oxidation to dnOPDA. In the nucleus, the receptor COI1 directly recognizes dnOPDA and exerts its functional activity (Figure 1). The ligand specificity of M. polymorpha is due to a single residue substitution in COI1, which explains the evolutionary events between the bryophytes and vascular plants [40,41]. The Mpcoi1 mutation is functionally complemented by AtCOI1 and gains JA-Ile responsiveness [40]. Similarly, in contrast to all of the embryophyta studied so far, the M. polymorpha genome has single JAZ and MYC orthologs (Figure 1), which are functionally conserved in other land plants [40,41,42]. Mpjaz mutants show similar developmental phenotypes to Arabidopsis JAZ-depleted mutants (Table 1), such as growth inhibition, rhizoid emergence, gemma cup formation, cell size, or antheridiophore development [13,42,43], and the defects of Mpjaz-1 mutant are complemented by overexpression of AtJAZ3 [42]. As in Arabidopsis (Table 1), JAZ deficiency in Marchantia compromises fertility, perhaps as a consequence of constitutive activation of MYCs along with MYCs interacting with MED25 to activate JA-responsive genes, which requires further confirmation [12,13,44]. Similar to AtJAZs, for the proper regulation of JAZ function, MpJAZ transcripts exist splicing (elimination of the Jas domain, MpJAZ∆Jas), which is stabilized against hormone-induced degradation and responsible for terminating the JA response [42,45]. The trade-off between overactivation of jasmonate signaling and reproductive fitness is widely conserved, suggesting that the JA core signaling pathway has been maintained since plants first colonized terrestrial habitats.

Table 1.
Mutants and overexpression lines of JA biosynthesis pathways and core signaling components in Arabidopsis, Oryza sativa, and Solanum lycopersicum.
3.2. The Termination of JA Signaling
The JA signaling pathway is terminated to create time-delayed negative feedback regulation mechanisms, which serve to ensure sufficient initiation of the JA signal [45].
In varying stress responses and different growth stages of plants, JA is metabolized into active, partially active, or inactive components through different approaches to balance the homeostasis of JA, including conjugation, hydrogenation, carboxylation, decarboxylation, methylation, esterification, sulfonation, glycosylation, and the formation of 12-OH-JA lactone [8]. JA-signaled transcriptional responses are tightly integrated with the accumulation of JA and JA-Ile [9]. A key feature of JA-Ile, as the molecular trigger of signaling, is its rapid accumulation induced by herbivores, wounding, necrotrophic pathogens, or other types of stress [100]. Furthermore, JA-Ile promotes the expression of two kinds of COI-dependent JA-Ile deactivated enzymes: CYP94 oxidase and amidohydrolases, including IAR3, ILL5, and ILL6, and so on (Figure 2A) [8]. First, JAR1 transforms JA into JA-Ile. Then JA-Ile is oxidized twice by CYP94 P450 family enzymes to 12-OH-JA-Ile and 12-COOH-JA-Ile in turn. Next, JA-Ile and 12-OH-JA-Ile generate JA and 12-OH-JA through the amidohydrolases IAR3, ILL5, and ILL6 [101]. This process usually leads to a decrease in JA-Ile. In addition, JA can be catalyzed by jasmonic acid carboxyl methyltransferase (JMT) to produce MeJA [8,102]. In reality, plant growth, including seeding growth and hypocotyl elongation, under warm temperature conditions requires tight control of available JA-Ile levels, mainly through JASMONATE-INDUCED OXYGENASES (JOXs) and ST2A-mediated JA catabolism [103,104,105]. This co-operative action of metabolic and catabolic enzymes results in a highly dynamic balance of JA and JA-Ile associated with the corresponding biological responses [106].
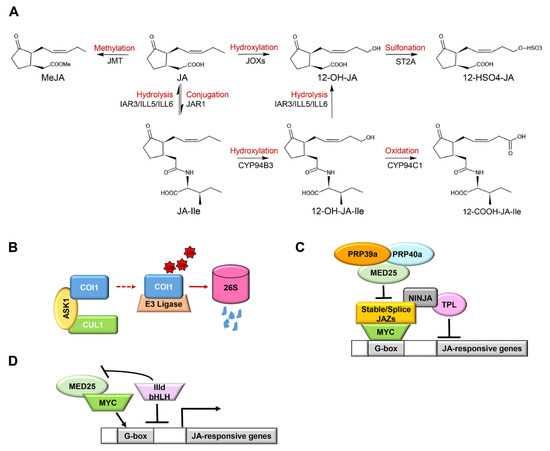
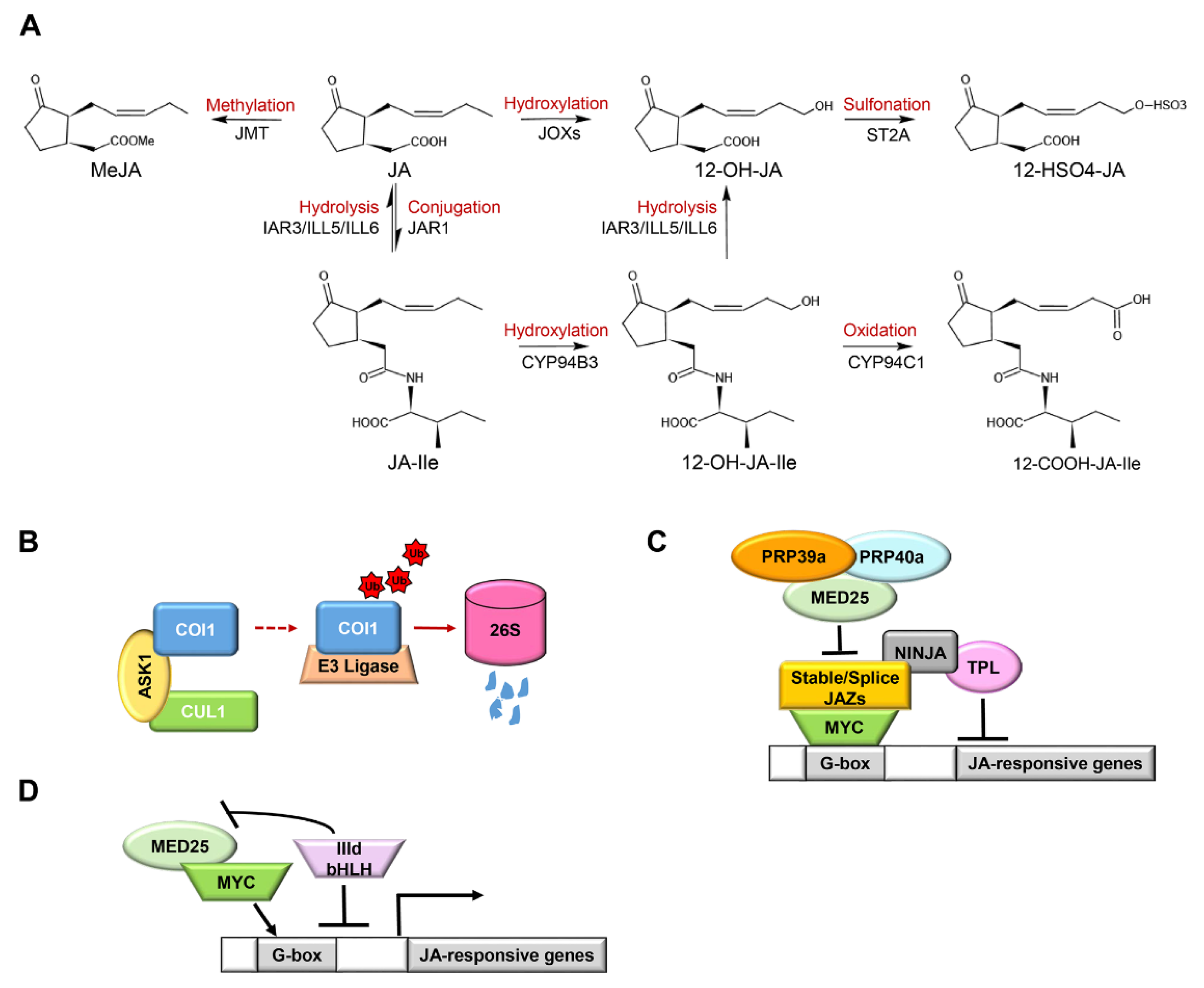
Figure 2.
The terminal regulation mechanism of jasmonic acid (JA) signaling: (A) the dynamic banlance of JAs. Metabolic and catabolic enzymes maintain a dynamic balance between JA and JA-Ile. Methyl jasmonate (MeJA) is formed by JA catalyzed by jasmonic acid carboxyl methyltransferase (JMT). JAR1 transforms JA into JA-Ile. JA-Ile is continuously oxidized twice by CYP94 P450 family enzymes to generate 12-OH-JA-Ile and 12-COOH-JA-Ile in turn. The amide bond of JA-Ile/12-OH-JA-Ile is broken by amide hydrolases (IAR3/ILL5/ILL6) to generate JA/12-OH-JA, respectively. JA can be directly hydroxylated to 12-OH-JA and further sulfated to form 12-HSO4-JA; (B) COI1 degradation pattern. Jasmonate receptor COI1 forms the SCFCOI1 complex with ASK1 and CUL1. Excessive COI1 protein is recruited and degraded by ubiquitination through the 26S proteasome pathway; (C) the inert and alternative splicing of JAZ-mediated JA attenuation pattern. JA signal suppressor JAZ protein can bind to NINJA protein and directly or indirectly recruit co-inhibitor TPL to inhibit the transcription of MYC2, thereby inhibiting the expression of JA-responsive genes. Inert JAZs (JAZ7, JAZ8, and JAZ13) have a conserved EAR domain that directly interacts with TPL. JAZ10.4, a variable splice of JAZ10, lacks the Jas motif and mediates desensitization to JA. MED25-recruited splicing factors PRP39a and PRP40a promote the correct splicing of JAZ genes and prevent the overproduction of splicing variants, thereby regulating the activation of JA signal; (D) bHLH-like protein-mediated JA attenuation pattern. bHLH IIId (JAM1/2/3, bHLH14) proteins compete with MYC-like TFs to bind the G-box element on the promoter of the JA response gene, inhibit the formation of the MYC2-MED25 complex, and negatively regulate the JA response. Arrows: activations; bar-headed arrows: repressions.
The COI1 protein is strictly regulated by a dynamic balance of SCFCOI1-mediated stabilization and 26S proteasome-mediated degradation (Figure 2B). The stable formation of SCFCOI1 and degradation of excessive COI1 maintain the stable content of COI1 in plants, which is of great significance for biological functions [107].
COI1-dependent removal of pre-existing JAZ proteins can provide for rapid activation of defenses and other JA-dependent processes, whereas rapid synthesis of new JAZ proteins ensures attenuation of the JA response soon after transmission of the signal [29]. Arabidopsis group IV JAZ proteins (AtJAZ7, AtJAZ8, and AtJAZ13), a particular sub-family of JAZs, harbors a divergent Jas motif that interacts weakly with COI1 [31]. Although the atypical ZIM domain of JAZ7, JAZ8, and JAZ13 fails to mediate the interaction with NINJA, they have a conserved EAR domain that directly interacts with the co-repressor TPL [31,107,108]. Following their interaction with MYC2 through the Jas-like motif, they behave as a constitutive repressor in the JA pathway (Figure 2C) [37,107,108]. For example, overexpression of JAZ13 confers JA-insensitivity and decreased resistance to insect herbivory [108]. JAZ10.4, a variable splice of JAZ10, lacks the Jas motif and mediates desensitization to JA through interacting with MYC2, MYC3, and MYC4 TFs through the CMID region, which can form extensive contact with the transcriptional activation domain (TAD) of MYCs (Figure 2C) [45]. Arabidopsis plants overexpressing JAZ10.4 were found to be insensitive to exogenous JA application and exhibited high tolerance to JA-induced degradation [79]. The inert JAZ proteins and alternative splicing of JAZ10 pre-RNA create part of the regulatory circuit to attenuate JA responses. It is intriguing that plants have developed a mechanism to prevent excessive desensitization to JA responses mediated by JAZ splice variants [63]. Following the generation of JAZ splice variants depending on the mediator subunit MED25, MED25 recruited the splicing factors RPR39a and PRP40a, which promoted the correct splicing of JAZ genes (full splicing of Jas intron) and prevented the overproduction of JAZ splice variants (Figure 2C).
Arabidopsis bHLH subclade IIId proteins, including bHLH17/JAM1, bHLH13/JAM2, bHLH3/JAM3, and bHLH14, compete with MYC2-like TFs to bind to the G-box elements of its target gene promoters and impair the formation of the MYC2-MED25 complex, thereby deactivating MYC2-like TFs-dependent gene transcription (Figure 2D) [9,109,110], which has also been proved in tomato MTB (MYC2-targeted bHLH) TFs, homologs of Arabidopsis JAM TFs. The MID region in the TAD domain of MYC2 is essential for the interaction between MYC2 and MED25. Tomato MTB proteins lack a canonical MED25-interacting domain due to an altered MID domain (AMID) and therefore fail to interact with MED25. Moreover, the AMID plays a significant role in the MTB1-JAZ interaction, which possibly helps to repress gene expression and to terminate JA signaling [9]. As a result, MYC2 and bHLH IIId proteins generate an auto-regulatory negative feedback circuit to terminate JA signaling in a highly organized manner [9,10].
Therefore, multiple highly organized strategies have been developed to control the accurate termination of JA signaling in plants in order to avoid over-stunted growth as a result of excess defense response. Obviously, both the activation and the deactivation/desensitization of JA responses must be under tight control.
4. The Functions of JA in Growth and Development
4.1. Promotion of Plant Regeneration
Recent studies have suggested new roles for JA in promoting plant regeneration [111,112,113]. JA reduces the quiescent center (QC) quiescence in the root stem cell niche (SCN) through the RBR-SCR network and stress response protein ETHYLENE RESPONSE FACTOR115 (ERF115) [113]. Furthermore, JAs serve as wound signals during de novo root regeneration (DNRR) by activating ERF109 (Figure 3). On the one hand, ERF109 co-operates with SET DOMAIN GROUP8 (SDG8)-mediated histone H3 lysine 36 trimethylation (H3K36me3) to upregulate ANTHRANILATE SYNTHASE α1 (ASA1), a tryptophan biosynthesis gene in the auxin production pathway and promotes cell fate transition to form the root primordium [5,112]. On the other hand, JA-induced ERF109 transcription stimulates CDK interactor CYCLIND6;1 (CYCD6;1) expression, functions upstream of ERF115 and promotes regeneration [111,113]. Therefore, the JA tissue damage response pathway induces stem cell activation and regeneration, as well as activating growth after environmental stress.
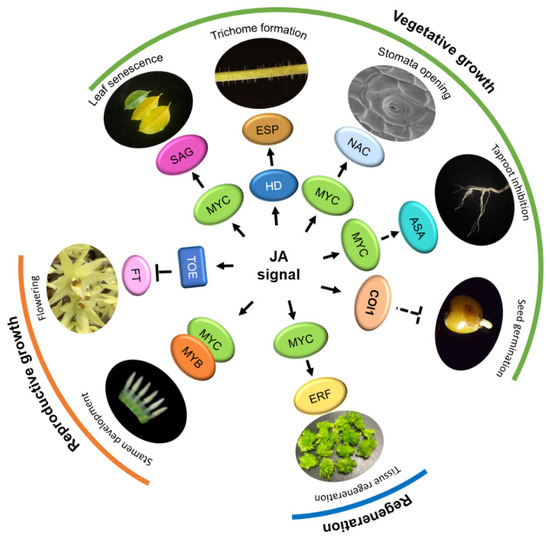
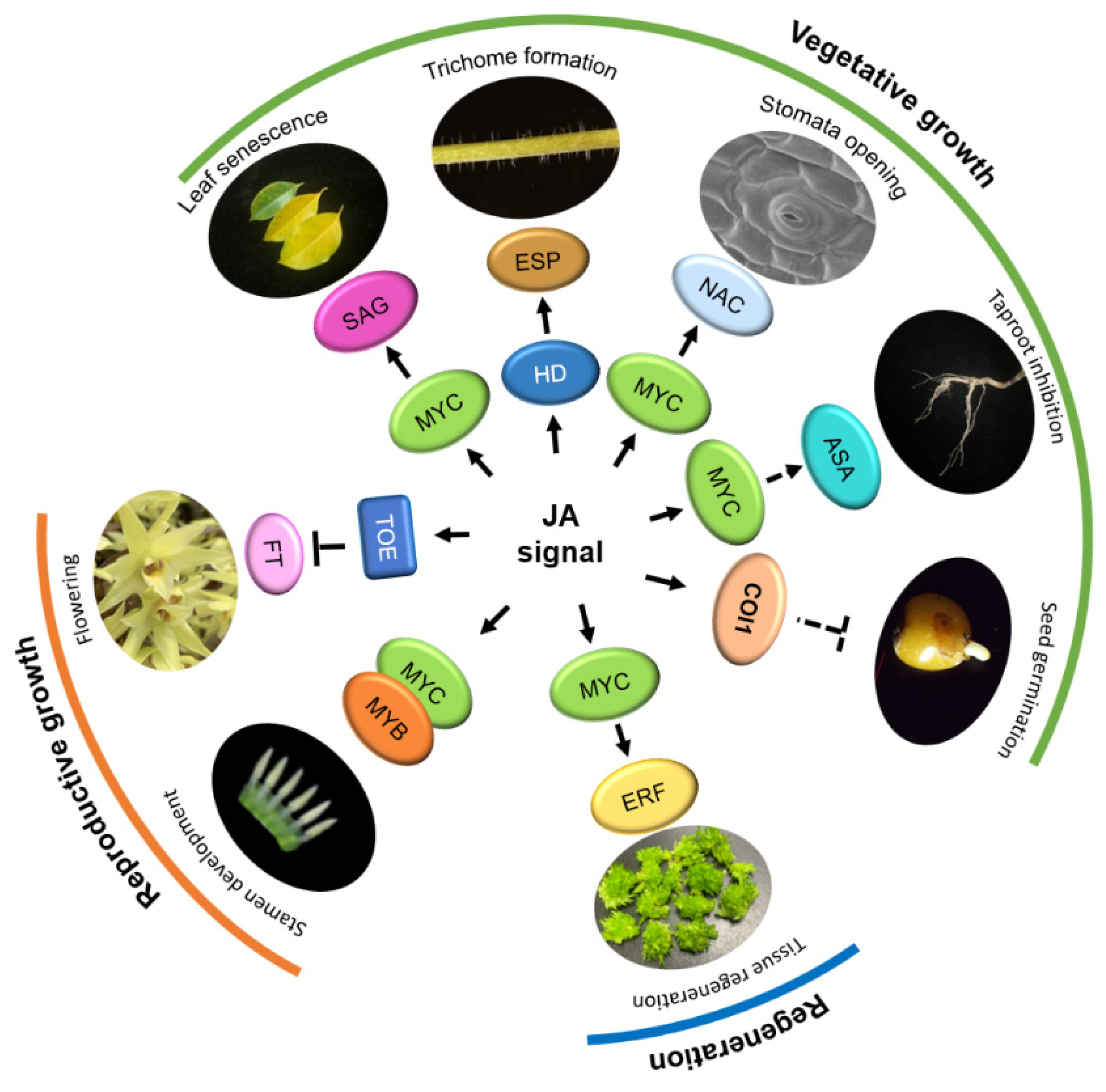
Figure 3.
The function of Jasmonic acid (JA) in plant growth and development. In the reproductive growth stage, bHLH IIIe transcription factors (TFs), MYCs, and MYBs promote stamen development. JA interacts with AP2 family TOEs to inhibit FT transcription and regulate plant flowering. In the vegetative growth stage, MYCs and ASA jointly inhibit the elongation of the taproot; MYCs interact with SAG to activate JA-induced leaf senescence; MYC and downstream NAC TFs promote stomatal opening; HD-ZIP family members (HDs) activate the expression of expansion proteins gene (ESP) and regulate trichome formation; JA also inhibits seed germination through COI1. In addition, MYC and ERF jointly promote tissue regeneration. Arrows: Activations; Bar-headed arrows: Repressions.
4.2. Regulation of Reproductive Growth
JA has been found to coordinate stamen filament elongation, anther dehiscence, and pollen viability [6]. Male sterility is recognized as one of the typical characteristics of JA disability. The Arabidopsis JA biosynthesis mutant opr3, as well as aos, showed defects in anther and pollen development resulting in male sterility (Table 1) [48,114,115]. A maize double mutant opr7opr8 with dramatically reduced JA has displayed reproductive deficiency and strong developmental defects, which were rescued by exogenous JA (Table 1) [116]. Mutations in the Arabidopsis JA receptor COI1 (coi1-1) caused abnormal anthers and pollen, leading to reproductive deficiency (Table 1) [30,61,63]. coi1-2 and coi1-8 displayed a partial fertility phenotype [27,63]. coi1-16 exhibited fertility in a temperature-sensitive manner [28]. Overexpression of rice COI1 genes (either OsCOI1a or OsCOI1b) could restore the fertility of an Arabidopsis coi1-1 mutant, while OsCOI2 failed to [101]. Overexpression of OsJAZ6, which interacts with OsJAZ1, altered JA signaling and led to abnormal spikelet development [85]. However, tomato JAI1, a homolog of Arabidopsis COI1, is required for the maternal control of seed maturation. A Sljai1 mutant displayed reduced viability of seeds as a result of a defect in female reproductive development, which was associated with the loss of accumulation of JA-regulated proteinase inhibitor proteins in reproductive tissues (Table 1) [100]. Furthermore, JA regulates petal and stamen growth by releasing the inhibitory effect of JAZ on the downstream R2R3-MYB TFs MYB21/MYB24 (Figure 3) [117]. Overexpression of MYB21 in coi1-1 plants restored stamen development [8,118]. MYB21 and MYB24 connect physically with MYC2, MYC3, MYC4, and MYC5 to control stamen development [8,110]. The myc2 myc3 myc4 myc5 quadruple mutant exhibited short filament, delayed anther dehiscence, and unviable pollen grains at the floral stage (Table 1) [8], while coi1-1 plants overexpressing the MYC5 and MYC3 exhibited restored stamen maturation and productivity [8,110].
JA represses the vegetative-reproductive maturation transition as well. In Arabidopsis, JA acts through COI1-JAZ/TOE-FT to inhibit flowering (Figure 3). A coi1-2 mutant, JAZ1Δ3A transgenic plants, and JAZ9 overexpression plants displayed early flowering (Table 1) [119]. JAZ proteins interact with APETALA2 (AP2) family TFs TARGET OF EAT1 (TOE1) and TOE2 and repress the transcription of FLOWERING LOCUS T (FT) [119]. In tomatoes, SlJAZ2 regulates plant morphology and accelerates flower initiation. Plants overexpressing SlJAZ2 exhibited quicker leaf initiation, shorter plant height and internode length, earlier lateral bud emergence, and more advanced flowering transition (Table 1) [99].
4.3. Actions of JA in Vegetative Growth
JA plays a dual role in seed germination in co-operation with abscisic acid (ABA). In cold-stimulated germination of wheat seeds, JA content rapidly increased after up-regulation of JA biosynthesis-related gene expression and further suppressed ABA biosynthesis by repressing two key ABA biosynthesis genes, TaNCED1 and TaNCED2 [120]. During the rice germination period, ABA acts upstream of JA and cooperatively inhibits rice seed germination through the SAPK10-bZIP72-AOC regulation pathway. It directly activates the transcription of AOC by phosphorylating bZIP72 through SAPK10 in order to promote the biosynthesis of JA and inhibit rice seed germination [121].
JA acts through COI1 to induce leaf senescence [110]. Arabidopsis coi1 mutants showed a stay-green phenotype under dark-induced senescence conditions (coi1-1) [62] and under MeJA treatment (coi1-2) [64], indicating that COI1 plays a role in leaf senescence (Table 1). A jaz7-1 (WiscDsLox7H11) mutant displayed severer dark-induced leaf yellowing, as well as quicker chlorophyll degradation (Table 1) [73]. In addition, MYC2, MYC3, and MYC4 function redundantly, binding to and activating the promoter of their target gene, SENESCENCE-ASSOCIATED GENE29 (SAG29), to activate JA-induced leaf senescence (Figure 3). However, MYC2/3/4-activated JA-induced leaf senescence is attenuated by the bHLH sub-group IIId factors (bHLH03/13/14/17), competitively binding to the promoter of SAG29 and repressing its expression [110]. A recent study has found that DNA binding-with-one-finger 2.1 (Dof2.1), a JA-inducible gene, acts as an enhancer of JA-induced leaf senescence through the MYC2-Dof2.1-MYC2 feed-forward transcriptional loop [122]. In reality, JA signaling-mediated leaf senescence is also regulated by the circadian clock. For instance, evening complex (EC), a core component of the circadian oscillator, negatively regulates leaf senescence by directly binding the promoter of MYC2 and repressing its expression, as well as gating JA signaling in order to regulate leaf senescence [123].
As has been widely accepted, stomata are cavities surrounded by guard cells on the leaf epidermis, which regulate water balance, gas exchange, and immune response to pathogens. JA has the ability to control the opening and closing of stomata in a COI1-independent manner. Pseudomonas syringae pv. tomato (Pto) DC3000 is a pathogenic factor of tomato bacterial spot disease, producing coronatine (COR) as a virulent factor to activate the JA pathway, promoting the formation of JAZ2-COI1 complexes and triggering JAZ degradation through the 26S proteasome to inhibit SA-dependent defense responses to P. syringae, thus inducing stomata opening and facilitating the entrance of the pathogen into the leaf apoplast [67]. Consistently, the dominant mutant SlJAZ2Δjas, lacking the C-terminal jas domain, presents inhibiting effects toward the reopening of stomata induced by COR (Table 1) [98]. Similarly, Arabidopsis jaz2-3 mutants are partially impaired in pathogen-induced stomatal closing and, thus, are more susceptible to Pseudomonas (Table 1). Remarkably, Arabidopsis dominant jaz2∆jas mutants are resistant to P. syringae but retain unaltered resistance against necrotrophs (Table 1) [67]. Furthermore, a recent study has demonstrated that AvrB, a type III effector protein of P. syringae, induces stomatal opening through a canonical JA signaling pathway involving COI1 and NAC TFs (Figure 3). It promotes the interaction between COI1 and JAZ by the RPM1-INTERACTING4 (RIN4)-Arabidopsis plasma membrane H+-ATPase (AHA1) pathway and induces the degradation of multiple JAZs to open stomata [124].
Trichomes are epidermal appendages with different forms, structures, and functions, such as protecting plants against herbivores. The JA/COI1 signaling pathway plays an important role in the promotion of glandular trichomes. In tomatoes, deficiencies in JA perception block glandular trichome formation [100]. The trichome-preferentially expressed SlJAZ4 is the critical component in JA-triggered tomato trichome elongation by interaction with HOMEODOMAIN PROTEIN8 (SlHD8). SlHD8 promotes elongation by activating the expression of expansin genes (EXPs; Figure 3) [125]. Similarly, Artemisia annua AaJAZ8 interacts with a positive regulatory factor, AaHD1, repressing its transcriptional activity and inhibiting the formation of glandular trichomes [126]. It is worth mentioning that JA and GA hormonal signals synergistically regulate plant development. The Arabidopsis GL3/EGL3/TT8 complex in the bHLH family binds to WD-Repeat and MYB proteins in order to form the WD-repeat/bHLH/MYB complex. Both JAZ and DELLA target the WD-repeat/bHLH/MYB complex to repress trichomes formation. The initiation of trichomes is regulated by the WD-repeat/bHLH/MYB complex in a JA-dependent manner, which was attenuated in the JA signaling-deficient mutant coi1-1, along with low expression of GL2 and MYB23 [127].
JA inhibits primary root growth in Arabidopsis through MYC2-mediated repression of PLETHORA1 (PLT1) and PLT2, which are known as the key TFs of the auxin-regulated root meristem activity and maintenance [128]. It is also reported that two Arabidopsis YUCCA genes, YUC8 and YUC9, which participate in auxin homeostasis and root development, are regulated by oxylipins dependent on the COI1 signal transduction pathway [129]. In fact, mutations in COI1 lead to insensitivity to JA-inhibitory primary root elongation [27]. The triple mutant myc2 myc3 myc4 (myc2/3/4) showed an obvious reduction in JA-dependent primary root growth inhibition (Table 1) but less severe than that in coi1-1 [32]. However, the role of JA in lateral root development is different. The exogenous application of MeJA up-regulates the expression of ERF109, which stimulates ASA1 gene expression and increases the content of auxin in Arabidopsis, thus promoting the occurrence of lateral roots and inhibiting taproot elongation (Figure 3) [130]. The defect of lateral root formation of asa1-1 mutants after MeJA treatment is closely related to the significant down-regulation of PIN2 protein levels by JA [131].
As described in Table 1, researchers have also uncovered other roles for JA in developmental and growth-related processes. For example, coi1-37 displayed leaf epinasty, dark green leaves, strong apical dominance, and enhanced meristem longevity (Table 1) [66]. Overexpression of JAZ9 in plants led to phenocopy of the coi1 mutant with longer hypocotyls and petioles under low-in-tensity light conditions and early flowering, GA-hypersensitivity phenotype (Table 1) [29]. Over-expressed JMT rice plants showed reduced height and yield (Table 1) [82].
5. Role of JA during Plant Defense Responses
5.1. JA Mediates Plant Defense against Pathogens
In natural environments, plants may be infested by different types of pathogens, including biotrophic, hemi-biotrophic, and necrotrophic pathogens. Accumulating evidence points to the JA signaling pathway as mainly corresponding to plant immunity against necrotrophic fungal pathogens, including Alternaria brassicicola, Botrytis cinerea, Plectosphaerella cucumerina, Pythium spp., and so on [95,132,133]. In Arabidopsis, the JA biosynthetic mutants fad3fad7fad8 and jar1 showed increased susceptibility to A. brassicicola [46,47] and P. irregular [134], respectively (Table 1). The coi1-1 mutants exhibited increased susceptibility to the necrotrophic fungi A. brassicicola, B. cinerea, and P. cucumerina (Table 1) [132,133]. JAZ6 has been proven to be a crucial component in time-of-day defense against B. cinerea. When responding to B. cinerea, jaz6-1 lost the time-of-day difference due to failure to induce JAZ6 expression and did not exhibit enhanced susceptibility at subjective night while retaining resistance at dawn (Table 1) [71]. The maize double mutant opr7opr8 showed extreme susceptibility to a root-rotting oomycete (Pythium spp.) [116]. A tomato mutant (jai1) suffered 100% mortality from root-rot disease [95] and exhibited increased susceptibility to B. cinerea and Fusarium species (Table 1) [96,97]. A recent study has reported that WRKY75 exploits the JA signaling pathway by directly binding to downstream target genes such as ORA59 in order to positively regulate the Arabidopsis defense response against B. cinerea and A. brassicicola (Figure 4A) [135]. IQ-MOTIF-CONTAINING PROTEIN 1(AtIQM1), a Ca2+-independent CaMBP, increases the activity of the JA biosynthetic enzymes ACX2 and ACX3 by interacting with CATALASE2 (CAT2), thereby positively regulating JA content and the B. cinerea resistance of Arabidopsis [136]. In Rosa chinensis, upon JA treatment, free forms of RcMYB84 and RcMYB123 were released due to JAZ1 degradation, further activating the plant’s defense against B. cinerea [137].
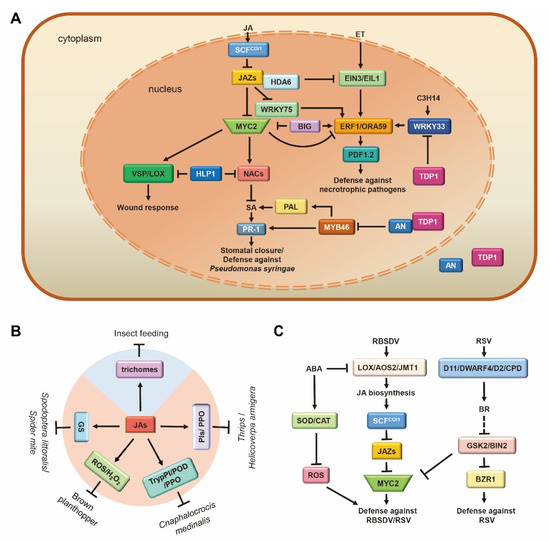
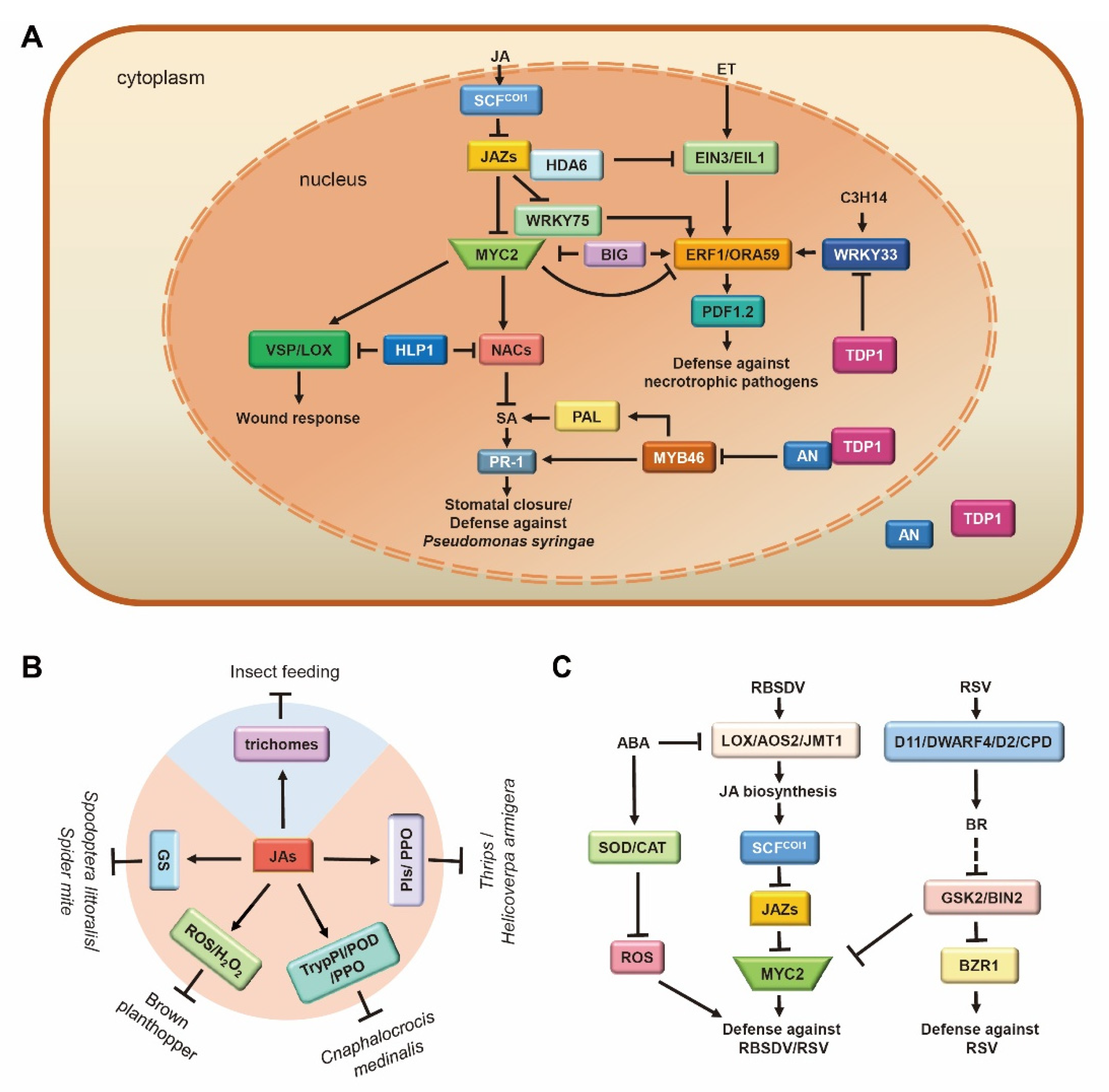
Figure 4.
Multi-functional role of jasmonic acid (JA) interaction module in stress. (A) JA-ET-SA network modulates the defense against pathogens in Arabidopsis. JA and ET synergistically regulate the defense against necrotrophic pathogens. BIG gene positively/negatively regulates MYC2/ERF1 and coordinates the defense of plants against pathogens and insects. NACs transcription factors in the JA pathway and SA pathway antagonize the regulation of stomatal dynamics and defense response against P. syringae. (B) JA defense against herbivorous insects through physical (blue) and chemical (orange) means. JA promotes the accumulation of GS (defense against Spodoptera littoralis/Spider mite), ROS/H2O2 (defense against brown planthopper), TrypPI/POD/PPO (defense against Cnaphalocrocis medinalis), and PIs/PPO (defense against Thrips/Helicoverpa armigera), as well as the initiation of trichomes to defend against a variety of herbivorous insects. (C) JA-BR-ABA crosstalk-regulated defense response to viruses. JA and BR synergistically defend against RSV through MYC2/BZR1. ABA antagonistically regulates the defense response to RBSDV by inhibiting the production of ROS and the synthesis of JA. Arrows: Activations; Bar-headed arrows: Repressions. JA and ET are two crucial plant hormones that co-operate to activate defenses against necrotrophic pathogens, whereas the SA pathway triggers defenses against biotrophic and hemi-biotrophic pathogens.
Although transcriptional regulatory elements involved in the JA signaling pathway (e.g., COI1, JAZs, and MYC2) are relatively conserved in plants, the manner in which MYC2 regulates downstream genes is species-specific. Arabidopsis defenses against B. cinerea are regulated by the expression of two groups of genes through AtMYC2 [138,139]. The first group of genes involved in JA-mediated systemic responses to wounding is activated by AtMYC2 through the direct regulation of NAC019. The second group includes genes involved in defense against pathogens, which are negatively regulated by AtMYC2 through suppression of ERF1 (Figure 4A). Arabidopsis myc2/jin1 mutants showed increased resistance to B. cinerea and Fusarium oxysporum but attenuated resistance to insects (Table 1). However, in tomatoes, SlMYC2 both positively regulates wounding-responsive genes by activating JA2L and pathogen-responsive genes by activating ERF.C3 [140]. A B. cinerea infection assay in tomatoes presented significantly larger necrotic lesions in MYC2-RNAi plants than in the wild type [140].
In Arabidopsis ein2 or coi1 mutant, exogenous JA and ET alone or in combination failed to induce the expression of downstream defense genes (such as ERF1 and PLANT DEFENSIN 1.2 (PDF1.2)) [139,141], illustrating that these two signaling pathways are concomitantly essential for the activation of plant defense responses (Figure 4A). JA enhances the transcriptional activity of EIN3/EIL1 through the removal of JAZ proteins, which recruit RPD3-type histone deacetylase (HDA6) as a co-repressor to repress EIN3/EIL1 transcriptional activity and further activates downstream ERF1/ORA59PDF1.2 cascades, thereby defending against necrotrophic pathogens [139,141,142]. Arabidopsis CCCH protein C3H14 regulates the activation of WRKY33-ORA59 cascades to correspondingly promote JA/ET signal transduction and camalexin biosynthesis in order to increase the plant’s tolerance to B. cinerea [143]. A recent study has reported that the Arabidopsis BIG gene orchestrates the antagonism between two parallel ERF1/ORA59 and MYC2 branches in the JA pathway that determine resistance to pathogens and wound response. BIG deficiency promotes JA-dependent gene induction and increases JA production but restricts the accumulation of both ET and SA. Eventually, JA-induced stomatal immunity is impaired after BIG disruption [16]. Moreover, JA acts in a complex signaling network combined with SA signaling pathways after P. syringae infection. COR produced by P. syringae hijacks a signaling module, COI1-JAZ2-MYC2/3/4-ANAC19/55/72, to control stomatal dynamics during the invasive process (Figure 4A). In detail, COR hijacks the JA pathway to suppress the SA pathway by directly activating the expression of SA biosynthesis enzyme inhibitor NACs TFs (ANAC19/55/72) through MYC2/3/4, which is targeted by JAZ2, thereby inhibiting the SA-mediated defense response against P. syringae [67]. In rice, following infection by Magnaporthe oryzae, the SA signaling regulator OsNPR1 sequesters OsbHLH6 in the cytosol (activating JA signaling when localized to the nucleus) and activates SA signaling but represses JA signaling to control rice resistance to M. oryzae [15]. The PRC1 protein LHP1 is involved in the repression of the MYC2 branch of the JA/ET pathway of immunity by inhibiting the expression of NACs and their target BSMT1 and promoting the accumulation of SA, thereby enhancing the defense against P. syringae (Figure 4A) [144]. The Arabidopsis C-terminal binding protein ANGUSTIFOLIA (AN) antagonistically regulates plant defense against the hemi-biotrophic pathogen P. syringae and the necrotrophic pathogen B. cinerea (Figure 4A). AN interacts with TYROSYL-DNA PHOSPHODIESTERASE1 (TDP1) and imposes transcriptional repression on MYB46, which encodes a transcriptional activator of the SA biosynthesis gene PHENYLALANINE AMMONIA-LYASE (PAL) while releasing TDP1-imposed transcriptional repression on WRKY33, a master regulator of the JA/ET signaling pathway. The antagonistic effect of MYB46 and WRKY33 through AN regulation suggests a transcriptional co-regulatory mechanism of SA and JA/ET pathways, indicating a transcriptional node regulating the trade-offs between (hemi)biotrophic and necrotrophic defenses [145].
In contrast to the biotroph/necrotroph dichotomy mentioned above, it is noteworthy that JA is also required for the induction of immunity against biotrophic and hemi-biotrophic pathogens, including the fungi F. oxysporum [146] and Verticillium dahlia [96], as well as the bacteria Pectobacterium atrospecticum and Xanthomonas oryzae pv. oryzae (Xoo) [88,147]. In rice, OsJAZ8∆C-overexpressing plants, which lack the Jas domain, exhibited a JA-insensitive phenotype and reduced JA-induced resistance to Xoo (Table 1) [88]. ABA-inducible SnRK2-type kinase SAPK10-mediated phosphorylation on Thr129 of WRKY72 weakens its DNA-binding ability to AOS1, promotes the endogenous JA level, and finally enhances Xoo resistance, which highlights the role of ABA-JA interplay in post-translational modification and an epigenetic regulation mechanism [147].
5.2. JA Acts as a Double Agent in Plant Defense against Herbivorous Insects
Plants usually have the intrinsic ability to resist attacks from herbivorous insects through a combination of constitutive and inducible defenses. JA is employed as a central defense signal for plant resistance against herbivory. Upon insect feeding, JA synthesis is rapidly triggered, inducing massive defense-related genes, the production of diverse secondary metabolites (terpenoids, phenolics, as well as nitrogenous and sulfur-containing compounds), specific defense proteins (protease inhibitors, polyphenol oxidases, leucine aminopeptidase, lectins, and chitinases), and the formation of a physical barrier (e.g., trichomes) to suppress or prevent the feeding (Figure 4B) [148,149]. Arabidopsis plants deficient in JA biosynthesis and signaling typically suffers more damage from molluscan herbivores [149]. MeJA treatment induced the expression of glucosinolate (GS) synthesis genes, as well as GS accumulation [150,151]. MYC2/3/4 directly binds to the promoters of GS biosynthetic genes and interacts with GS-related MYBs, thereby promoting the JA-mediated synthesis of secondary metabolites and defense against external assaults [76]. Consistent with this, the Arabidopsis myc2/3/4 triple mutant is completely devoid of GS and is extremely susceptible to the generalist herbivore Spodoptera littoralis [76] and spider mite herbivory (Table 1) [77]. In rice, the concentrations of JAs were dramatically increased after a brown planthopper (BPH) attack, along with an increase in H2O2 level [82]. BPH performed better on JA-deficient lines (AOC and MYC2 knockout) than on wild-type (WT) plants due to the attenuation of defensive secondary metabolites accumulation (Table 1) [80]. Additionally, rice COI1 RNAi lines increase susceptibility to chewing insect Cnaphalocrocis medinalis as a result of impairing inducible defense by induction of trypsin protease inhibitor (TrypPI), peroxidase (POD), and polyphenol oxidase (PPO) [10]. Tomato plants treated with JA showed reduced numbers of Frankliniella occidentalis (thrips), Helicoverpa armigera, flea beetles, and aphids due to an increase in the activities of PIs and polyphenol oxidase [152]. Furthermore, the development of glandular trichomes in tomato leaves is controlled, in part, by the JA pathway [153], providing an important anti-insect defense layer [154]. Tomato jai1 plants exhibited several defense-related features, including the inability to express JA-responsive genes, severely compromised resistance to two-spotted spider mites, and reduced monoterpene production due to the abnormal development of glandular trichomes (Table 1) [100]. Upon herbivore attack, JA signaling is activated, and subsequently, MYC2, MYC3, and MYC4 mediate indole-3-acetic acid (IAA) biosynthesis through the activation of YUCCA9 expression [155,156].
As a countermeasure, many specialized herbivores manipulate their defense by modulating the JA signaling pathway. For instance, Whitefly (Bemisia tabaci)-induced tomato plant volatiles prime SA-dependent defenses and suppress JA-dependent defenses, thus rendering neighboring tomato plants more susceptible to whiteflies, which has been confirmed by experiments with volatiles from caterpillar-damaged and pathogen-infected plants [157]. The effector HARP1, which is released from the cotton bollworm during feeding, also attenuates the plant defense by interacting with JAZ repressors to restrain COI1-mediated JAZ degradation, therefore blocking JA signaling [158]. Interactions between brassinosteroids (BRs) and SA/JA have been reported in the insect-resistance response in rice. BR biosynthesis is induced after BPH infestation by upregulating the expression of OsBRI1/OsBZR1, followed by the JA synthesis-related gene OsLOX1/OsAOS2. The JA signal down-regulates the expression of the SA biosynthesis gene ISOCHORISMATE SYNTHASE 1 (OsICS1) and OsPAL to inhibit the SA-mediated defense response to BPH. The suppressive effects of BRs on the SA pathway were eliminated in JA-deficient and JA-insensitive mutants. These results indicate that BRs obviously promote the susceptibility of rice host plants to BPH by modulating the SA/JA co-action defense responses [159].
5.3. JA Plays Vital Roles in the Arms Race between Plants and Viruses
BRs and JAs finely participated in building the plant defense system in a synergistic or antagonistic manner. The synergistic effect of BR and JA enhances the resistance to rice stripe virus (RSV) (Figure 4C). BR-induced RSV resistance is blocked in osmyc2 knockout plants (Table 1). RSV reduces JA-mediated defense by increasing the accumulation of the BR signaling negative regulator OsGSK2 [86] and by making it physically interact with the JA positive regulator OsMYC2, resulting in the degradation of OsMYC2 by phosphorylation, thus promoting its infection [87]. Collectively, these results demonstrate that BRs positively contributed to regulating JA-mediated resistance. However, an antagonistic relationship between BR and JA effects in viral defense has been reported. Rice black-streaked dwarf virus (RBSDV)-infected rice plants show that genes of the JA pathway (LOX1, AOS2, JMT1, and MYC2) are up-regulated, while genes in the BR pathway (D11, OsDWARF4, D2, CPDs, BRI1, and BZR1) are down-regulated. The line Go (a mutant overexpressing OsGSK2, which can block the BR signaling pathway) plants showed a marked decrease in susceptibility to RBSDV (Figure 4C). coi1-13 mutant infection experiments and application of exogenous hormones indicated that JA-mediated defense can suppress the BR-mediated susceptibility to RBSDV infection in a manner dependent on the JA co-receptor OsCOI1 [83]. Furthermore, ABA is also involved in the JA-mediated resistance to the virus. ABA negatively regulates rice defense against RBSDV by preventing JA-mediated accumulation of reactive oxygen species (ROS) [160]. In recent years, an increasing number of studies have shown that viruses perform host-plant manipulation on their specific host plants. Rice viruses in the genera Fijivirus, Tenuivirus, and Cytorhabdovirus all possess transcriptional repressors that directly disassociate the OsMED25-OsMYC3 complex, inhibit the transcriptional activation of OsMYC3, and then combine with OsJAZ proteins to cooperatively overcome the JA pathway in a manner that benefits viral infection and the feeding activity of their vectors [161]. RBSDV-encoded P5-1 regulates the ubiquitination activity of SCF E3 ligases and inhibits JA signaling to benefit its infection [162]. RSV, transmitted by the small brown planthopper (SBPH), is the type member of the genus Tenuivirus. MeJA treatment attracted SBPHs to feed on rice plants, where a JA-deficient mutant was less attractive than wild-type rice. This is because the JA pathway, induced by the coat protein, activates the plant defense against RSV while attracting SBPHs to feed, thus benefiting viral transmission [163].
5.4. JA Regulates Plant Tolerance against Abiotic Stresses
JAs have been implicated in the control of plant responses to abiotic stimuli, such as mechanical stress [35], salt [164], drought [84,88,165], UV irradiation [166], and ozone exposure [167]. JA synthesis genes, including LOX2, LOX3, AOS, AOC, and OPR3 in Arabidopsis, and TomLoxD and AOS in tomato, are significantly up-regulated by exogenous JA and mechanical damage [168,169]. Tomato res mutants, which accumulate JA and show remarkable growth inhibition and important morphological alterations, can restore cell structure alterations under salty stress (Table 1) [164]. In rice, OsJAZ1-overexpressing plants were more sensitive to drought stress treatment, while jaz1 mutant plants indicated increased drought tolerance (Table 1) [84]. In wheat, when under drought stress, JA acts on the upstream of ABA in response to initiating the drought tolerance of plants. Plants first promote the biosynthesis of ABA and JA and subsequently induce related signal pathway genes, such as SnRK2 and MYC2, which further activate the transcriptional activities of downstream genes, including calmodulin, disease resistance protein RPM1, CAT, SOD, and HSP70 [170]. Recent evidence in apple (Malus domestica) has revealed that E3 ligase MdMIEL1 (MIEL1, MYB30-interacting E3 Ligase1) and MdJAZ proteins directly modulated the BBX37-ICE1-CBF module to achieve the dual regulation of JA-mediated cold stress [171].
6. JA Mediates the Trade-Offs between Growth and Defense
JA controls a multitude of transcriptional programs affecting plant regeneration, reproductive process, phenotype formation, and stress defense and exerts strong control over the growth-defense balance. In most cases, it contains multiple biological processes rather than a single hormone pathway to shape plant growth and their response to defenses. However, how do interactions among JA core pathway components and crosstalk between JA and other biological processes control myriad aspects of growth, development, reproduction, and immunity?
6.1. Interactions among JA Core Components
In Arabidopsis, the functions of MYC2/3/4, including restricting leaf and root growth, activating leaf defense, and enhancing susceptibility to the pathogen P. syringae and promoting defense against insect herbivory, have been validated through phenotypic comparison of the quintuple mutant jazQ (JAZ1/3/4/9/10) (Table 1) and a jazQ myc2 myc3 myc4 octuple mutant. Therefore, MYC TFs exert epistatic control over JAZ-repressible transcriptional processes that govern JA-mediated growth-defense trade-offs [12]. The researchers further combined 13 JAZ gene mutants to examine the effects of long-term JAZ deletion on defense, growth, and reproductive output. The results considering an uncovered jaz decuple mutant (jazD) of 10 JAZ genes (JAZ1-7, -9, -10, and -13) showed that it possessed resistance to insect herbivores and fungal pathogens but had slow vegetative growth and poor fertility (Table 1). The absence of the remaining JAZ repressors in jazD mutant plants further aggravated growth arrest, led to almost no seed production, and even facilitated the spread of necrotic lesions and tissue death under extreme conditions [43]. Therefore, the dual role of the inhibitory effect of JA on growth and the enhancement of defense provides evidence of the antagonistic relationship between growth and immunity.
6.2. Crosstalk between JA and Other Phytohormones
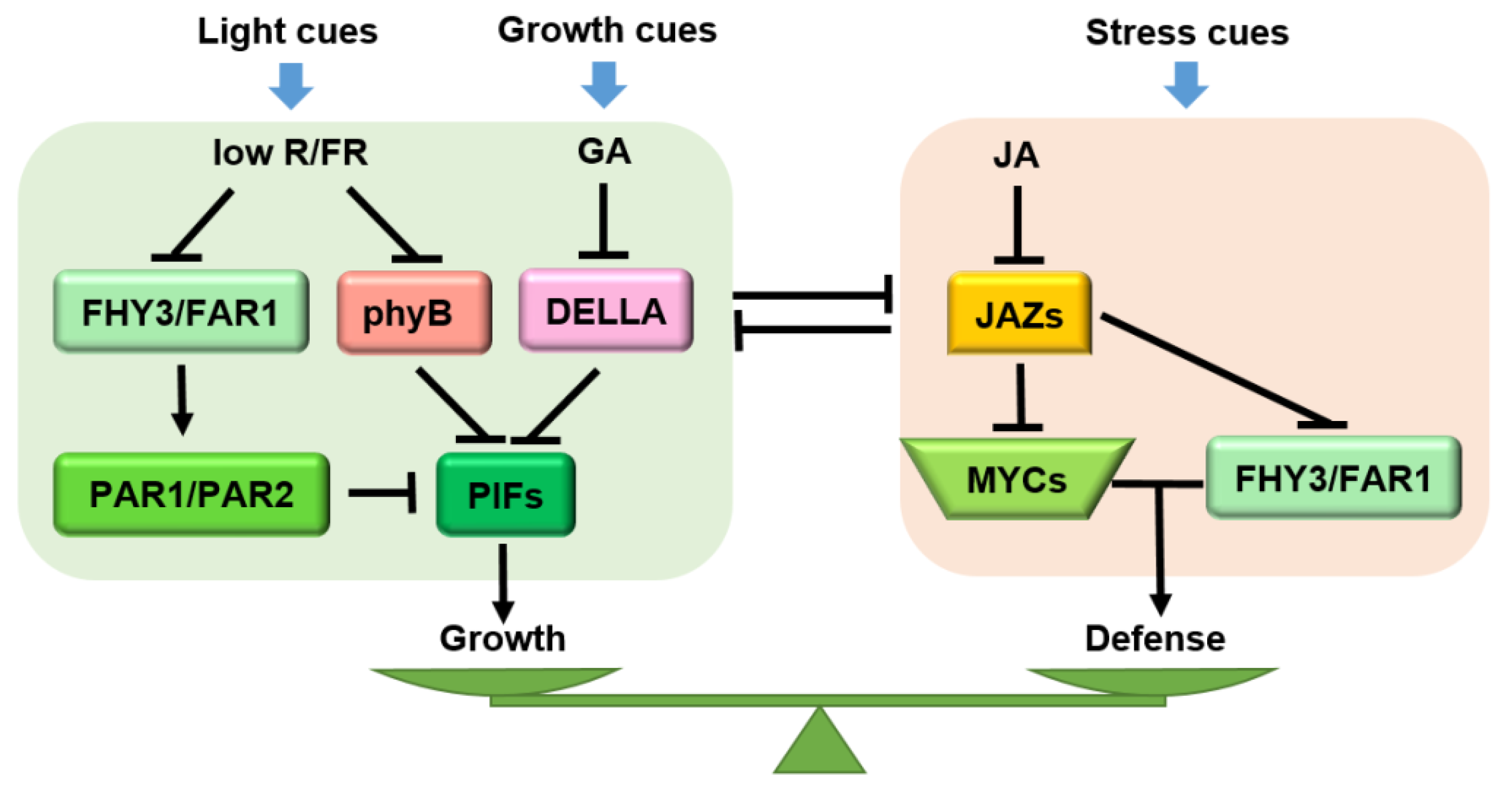
JA is an important hormone related to plant defense, while gibberellin (GA) is an important hormone mediating plant growth. Antagonistic signal crosstalk between the binding of bioactive JAs or GAs to cognate receptors leads to proteasome-dependent degradation of JAZ/DELLA proteins that, at the resting state, represses cognate TFs involved in defense (e.g., MYCs) or growth (e.g., phytochrome-interacting factors, PIFs) (Figure 5). DELLAs, serving as central regulators linking the crosstalk between JA and GA, inhibit MYC2-JAZ interactions, thus liberating MYC2 to promote the JA response. Meanwhile, in the presence of GA, DELLAs are eliminated to compromise the JA response [172]. In rice, the JA-induced defense against rice root knot nematode (Meloidogyne graminicola) requires SLENDER RICE1 (SLR1, a DELLA protein in rice) accumulation to repress the GA pathway [17]. In Arabidopsis, the interaction between RGA (a DELLA protein) and PIF3 is inhibited by JAZ9, thereby promoting hypocotyl elongation. Both a della quintuple mutant and a pif quadruple mutant displayed insensitivity to JA-induced hypocotyl inhibition, indicating that the DELLA-PIF interaction is required for JA-mediated growth inhibition during the response. Additionally, overexpression of PIF3 could partially overcome JA-induced growth inhibition. Therefore, a molecular cascade involving the COI1-JAZ-DELLA-PIF signaling module elucidates that the antagonistic effects of JA and GA reconcile the growth-defense dilemma [68].
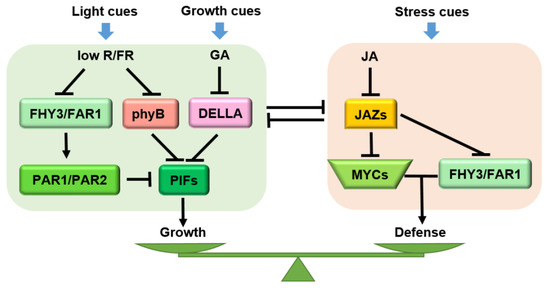
Figure 5.
Molecular mechanism of JA-GA-phyB crosstalk that governs growth and defense. When defense cues are generated, JA dominates the defense response and regulates the growth-defense balance together with GA/phyB. GA and phyB jointly inhibit plant growth under shade conditions, while JA and GA antagonize and regulate the plant growth response under growth status. Arrows: activations; bar-headed arrows: repressions; lines: interactions.
More recent works have described the interaction between JA and auxin as of particular relevance for the control of plant growth-defense trade-offs [173,174]. Wound-inducible amidohydrolases (IAH) contribute to JA and auxin levels to coordinate stress responses and development by controlling JA-Ile and IAA contents [175]. Both JA and auxin perception depend on SCF-type ubiquitin protein ligase (E3) complexes [173,176], and both the arx1 and arx6 mutants result not only in reduced auxin response but also a reduction in JA sensitivity [177], which might reduce JA responsiveness through the recruitment of such shared components by auxin, thus leading to a limitation of JA-mediated defense responses and amplification of auxin-mediated growth responses, and vice versa [173,174]. Therefore, the interaction between JA and auxin contributes to the fine-tuning of plant stress responses and development.
6.3. Crosstalk between JA and Phytochrome Signaling Pathway
Light is one of the most significant signals for plants to respond to the external environment. JA, in conjunction with phytochrome photoreceptors, is able to affect a variety of plant growth, development, and defense processes.
In Arabidopsis, the activities of FARRED ELONGATED HYPOCOTYLS3 (FHY3) and FARRED IMPAIRED RESPONSE1 (FAR1) are induced by shading (low R/FR) (Figure 5). On the one hand, FHY3/FAR1 activates the expression of atypical bHLH transcriptional co-factors PHYTOCHROME RAPIDLY REGULATED (PAR1) and PAR2, which inhibit the expression of downstream growth-related genes by forming heterodimers with PIF. On the other hand, FHY3/FAR1 and MYC2 jointly promote the expression of defense-related genes. In these two processes, JAZ proteins inhibit the activities of FHY3 and MYC2 from maintaining the balance between growth and defense [178]. Recent evidence has revealed that antagonistic crosstalk between JA and the red-light receptor phytochrome B (phyB) participates in the plant growth-defense balance. phyB mutation completely rescued the growth and reproductive defects in a jazQ mutant without affecting the defense level (Figure 5) [13]. Uncoupling of growth-defense antagonism in jazQ phyB plants has been attributed, in part, to simultaneous activation of the MYC and PIF modules [13] which, in wild-type plants, antagonizes one another partly through the interaction between JAZs and DELLAs [40]. However, unlike the jazQ phyB mutant, which both grows and defends, jazD phyB plants maintained the strong defense of the jazD but showed weak growth status, which reveals an independent pathway of phyB for the defense-related growth restriction. Moreover, the slow growth of jazD and jazD phyB plants is tightly correlated with up-regulation of the Trp biosynthetic pathway, together with enhanced expression of genes encoding enzymes for the conversion of Trp to defensive GSs [14,76]. In this case, it is possible that the growth-defense trade-offs do not rely only on transcriptional networks but also depend on strong metabolic constraints due to the reallocation of metabolites for defense [179].
7. Conclusions and Perspectives
JA has various physiological effects. As a signal molecule, on the one hand, it plays a key role in plant growth and development, including plant regeneration, anther and pollen development, flowering time, seed germination, leaf senescence, stomata closure, and trichome development [73,98,113,118,119,121,125]. On the other hand, it also provides plants with a strong defensive capability to ward off the majority of their natural enemies, including necrotrophic pathogens [139], herbivorous insects [80], and viruses [160]. Intriguingly, some cunning enemies-particularly herbivorous insects [159] and viruses [83], have evolved ingenious mechanisms to hijack the JA signal network in order to suppress or evade host defense responses.
Although a host of studies in Arabidopsis and tomato have described the synthesis and regulation mechanisms of JA, there are still many problems to be solved, especially with regard to the complexity of transcriptional regulation of JA as a key role in co-regulation with other pathways. Unlike in higher plants, there are single COI1, JAZ, and MYCs orthologs in the liverwort M. polymorpha (Figure 1), which allows it to serve as a window to overcome some bottlenecks caused by genetic redundancy in vascular plants, as well as to unveil the evolutionary history of JAs in growth and defense. Recent evidence has supported the dn-OPDA (Figure 1), instead of JA-Ile, as the bioactive COI1-JAZ ligand in M. polymorpha [40]] and the function MpJAZ is conserved when compared with Arabidopsis [42]. However, JAZ-interacting TFs have not been studied in M. polymorpha. Exploration of the epistatic interactions within the COI1-JAZ-TFs module in M. polymorpha, instead of formidable members of the AtJAZ and AtMYC families in Arabidopsis, is expected to open new frontiers in the field of JA biology.
When plants are in defensive status, their growth will be inhibited, which is generally believed to be due to the transfer of resources for growth to the synthesis of defensive metabolites [11]. Previous evidence has demonstrated that the constitutive activation of jasmonate-mediated defenses can be achieved with minimal effects on growth [180,181,182]. Increased resistance to both necrotrophic fungi and herbivorous insects but unaffected growth status has been detected in Arabidopsis when after down-regulation of the JASMONATE-ASSOCIATED VQ MOTIF 1 (JAV1) repressor [180]. In addition to genetic strategies, it may be possible to use chemical tools or endophytes to break the antagonistic relationship between growth and defense. For instance, NOPh (a phenyloxime derivative of COR stereoisomer)-treated Arabidopsis showed a moderate defense response to necrotrophic pathogens without growth inhibition through selective activation of the ERF-ORA branch by binding with co-receptor COI1-JAZ9 [181]. Epichloë fungal endophytes in plants alleviate the trade-off between growth and defense by regulating GA, auxins, SA, or JA pathways [182]; however, the associated molecular basis remains to be determined. The uncoupling of growth-defense trade-offs has been observed in jazQ phyB plants but not jazD phyB plants [13,14], suggesting that the balance of growth and defense not only involves the interaction between JA and phytochrome signaling pathways but also that with other pathways. Additional research is required in order to determine how the complex networks between the JA signaling pathway and other pathways act to synergistically regulate in order to uncouple, or, at least, minimize, such trade-offs to produce plants with robust growth and defense simultaneously.
Previous efforts have provided several meaningful clues that JAs can induce the biosynthesis of terpenes, flavonoids, and other medicinally active ingredients, thus crucially contributing to plant secondary metabolism. The JAs-mediated biosynthesis of secondary metabolites is mainly modulated by the SCFCOI1 complex, JAZ protein, MYC2, and other TFs, such as WRKYs and MYBs [145,147], which activate or inhibit the expression of multiple important enzyme genes in plant secondary metabolic biosynthetic pathways by interacting with cis-acting elements in their promoters. For example, exogenous MeJA treatment of the hairy roots of Salvia miltiorrhiza induced the biosynthesis of tanshinone, phenolic acids, and other active ingredients by regulating the expression of the secondary metabolite synthesis gene SmWRKYs [183]. Therefore, there exists a bridge between TFs, biosynthetic genes, and secondary metabolites. However, in most medicinal plants that are rich in metabolites, the JA-mediated biosynthetic pathways of secondary metabolites have not yet been clearly elucidated. It is imperative that further study clarifying the molecular mechanism of JAs regulating the synthesis of secondary metabolites in medicinal herbs be carried out, which will broaden our knowledge of the functions and signaling events associated with JAs, and which may even benefit human health.
Author Contributions
C.L. and D.C. conceived the topic. C.L., M.X., X.C. and J.S. reviewed and finalized the manuscript. C.L., M.X. and Z.H. integrated the information of tables, analyzed the data, and created the pictures. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (31901235), the Major Science and Technology Projects in Yunnan Province (202102AE090042), and the Scientific R & D Foundation for Talent Start-up Project of Zhejiang A & F University (2019FR010).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We appreciate Lin Xu (Institute of Plant Physiology and Ecology, Chinese Academy of Sciences) for revising and Dun Si (Zhejiang A & F University) for language modification.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ueda, J.; Kato, J. Isolation and Identification of a Senescence-promoting Substance from Wormwood (Artemisia absinthium L.). Plant Physiol. 1980, 66, 246–249. [Google Scholar] [CrossRef] [PubMed]
- Dathe, W.; Preiss, A.; Schade, W.; Sembdner, G.; Schreiber, K. Endogenous plant hormones of the broad bean, Vicia faba L. (-)-jasmonic acid, a plant growth inhibitor in pericarp. Planta 1981, 153, 530–535. [Google Scholar] [CrossRef] [PubMed]
- Staswick, P.E. Novel Regulation of Vegetative Storage Protein Genes. Plant Cell. 1990, 2, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Kashihara, K.; Onohata, T.; Yariuchi, R.; Tanaka, S.; Akimitsu, K.; Gomi, K. The overexpression of OsSRO1a, which encodes an OsNINJA1- and OsMYC2-interacting protein, negatively affects OsMYC2-mediated jasmonate signaling in rice. Plant Cell Rep. 2020, 39, 489–500. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.-T.; Xu, P.; Zhao, P.-X.; Liu, R.; Yu, L.-H.; Xiang, C.-B. Arabidopsis ERF109 mediates cross-talk between jasmonic acid and auxin biosynthesis during lateral root formation. Nat. Commun. 2014, 5, 5833. [Google Scholar] [CrossRef]
- Thines, B.; Mandaokar, A.; Browse, J. Characterizing Jasmonate Regulation of Male Fertility in Arabidopsis. In Jasmonate Signaling; Springer Nature: Cham, Switzerland, 2013; Volume 1011, pp. 13–23. [Google Scholar] [CrossRef]
- He, Y.; Fukushige, H.; Hildebrand, D.F.; Gan, S. Evidence Supporting a Role of Jasmonic Acid in Arabidopsis Leaf Senescence. Plant Physiol. 2002, 128, 876–884. [Google Scholar] [CrossRef] [PubMed]
- Wasternack, C.; Song, S. Jasmonates: Biosynthesis, metabolism, and signaling by proteins activating and repressing transcription. J. Exp. Bot. 2017, 68, 1303–1321. [Google Scholar] [CrossRef]
- Liu, Y.; Du, M.; Deng, L.; Shen, J.; Fang, M.; Chen, Q.; Lu, Y.; Wang, Q.; Li, C.; Zhai, Q. MYC2 Regulates the Termination of Jasmonate Signaling via an Autoregulatory Negative Feedback Loop. Plant Cell 2019, 31, 106–127. [Google Scholar] [CrossRef]
- Fonseca, S.; Calvo, P.F.; Fernández, G.M.; Diez-Diaz, M.; Gimenez-Ibanez, S.; López-Vidriero, I.; Godoy, M.; Fernández-Barbero, G.; Van Leene, J.; De Jaeger, G.; et al. bHLH003, bHLH013 and bHLH017 Are New Targets of JAZ Repressors Negatively Regulating JA Responses. PLoS ONE 2014, 9, e86182. [Google Scholar] [CrossRef]
- Ballaré, C.L.; Austin, A.T. Recalculating growth and defense strategies under competition: Key roles of photoreceptors and jasmonates. J. Exp. Bot. 2019, 70, 3425–3434. [Google Scholar] [CrossRef]
- Major, I.T.; Yoshida, Y.; Campos, M.L.; Kapali, G.; Xin, X.F.; Sugimoto, K.; de Oliveira Ferreira, D.; He, S.Y.; Howe, G.A. Regulation of growth-defense balance by the Jasmonate Zim-Domain (Jaz)-Myc transcriptional module. New Phytol. 2017, 215, 1533–1547. [Google Scholar] [CrossRef] [PubMed]
- Campos, M.L.; Yoshida, Y.; Major, I.T.; de Ferreira, D.O.; Weraduwage, S.M.; Froehlich, J.E.; Johnson, B.F.; Kramer, D.M.; Jander, G.; Sharkey, T.D.; et al. Rewiring of jasmonate and phytochrome B signalling uncouples plant growth-defense tradeoffs. Nat. Commun. 2016, 7, 12570. [Google Scholar] [CrossRef] [PubMed]
- Major, I.T.; Guo, Q.; Zhai, J.; Kapali, G.; Kramer, D.M.; Howe, G.A. A Phytochrome B-Independent Pathway Restricts Growth at High Levels of Jasmonate Defense. Plant Physiol. 2020, 183, 733–749. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.; Yang, C.; Cao, J.; Chen, H.; Pang, J.; Zhao, Q.; Wang, Z.; Fu, Z.Q.; Liu, J. A bHLH transcription activator regulates defense signaling by nucleo-cytosolic trafficking in rice. J. Integr. Plant Biol. 2020, 62, 1552–1573. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.-X.; Ge, S.; He, J.; Li, S.; Hao, Y.; Du, H.; Liu, Z.; Cheng, R.; Feng, Y.-Q.; Xiong, L.; et al. BIG regulates stomatal immunity and jasmonate production in Arabidopsis. New Phytol. 2018, 222, 335–348. [Google Scholar] [CrossRef] [PubMed]
- Yimer, H.Z.; Nahar, K.; Kyndt, T.; Haeck, A.; Van Meulebroek, L.; Vanhaecke, L.; Demeestere, K.; Hofte, M.; Gheysen, G. Gibberellin antagonizes jasmonate-induced defense against Meloidogyne graminicola in rice. New Phytol. 2018, 218, 646–660. [Google Scholar] [CrossRef]
- Bonaventure, G.; Schuck, S.; Baldwin, I.T. Revealing complexity and specificity in the activation of lipase-mediated oxylipin biosynthesis: A specific role of the Nicotiana attenuata GLA1 lipase in the activation of jasmonic acid biosynthesis in leaves and roots. Plant Cell Environ. 2011, 34, 1507–1520. [Google Scholar] [CrossRef]
- Chini, A.; Monte, I.; Zamarreño, A.M.; Hamberg, M.; Lassueur, S.; Reymond, P.; Weiss, S.; Stintzi, A.; Schaller, A.; Porzel, A.; et al. An OPR3-independent pathway uses 4,5-didehydrojasmonate for jasmonate synthesis. Nat. Chem. Biol. 2018, 14, 171–178. [Google Scholar] [CrossRef]
- Ahmad, P.; Rasool, S.; Gul, A.; Sheikh, S.A.; Akram, N.A.; Ashraf, M.; Kazi, A.M.; Gucel, S. Jasmonates: Multifunctional Roles in Stress Tolerance. Front. Plant Sci. 2016, 7, 813. [Google Scholar] [CrossRef]
- Ellinger, D.; Stingl, N.; Kubigsteltig, I.I.; Bals, T.; Juenger, M.; Pollmann, S.; Berger, S.; Schuenemann, D.; Mueller, M.J. DONGLE and DEFECTIVE IN ANTHER DEHISCENCE1 lipases are not essential for wound- and pathogen-induced jasmonate biosynthesis: Redundant lipases contribute to jasmonate formation. Plant Physiol. 2010, 153, 114–127. [Google Scholar] [CrossRef]
- Guan, L.; Denkert, N.; Eisa, A.; Lehmann, M.; Sjuts, I.; Weiberg, A.; Soll, J.; Meinecke, M.; Schwenkert, S. JASSY, a chloroplast outer membrane protein required for jasmonate biosynthesis. Proc. Natl. Acad. Sci. USA 2019, 116, 10568–10575. [Google Scholar] [CrossRef] [PubMed]
- Theodoulou, F.L.; Job, K.; Slocombe, S.P.; Footitt, S.; Holdsworth, M.; Baker, A.; Larson, T.R.; Graham, I.A. Jasmonic Acid Levels Are Reduced in COMATOSE ATP-Binding Cassette Transporter Mutants. Implications for Transport of Jasmonate Precursors into Peroxisomes. Plant Physiol. 2005, 137, 835–840. [Google Scholar] [CrossRef] [PubMed]
- Sheard, L.B.; Tan, X.; Mao, H.; Withers, J.; Ben-Nissan, G.; Hinds, T.R.; Kobayashi, Y.; Hsu, F.-F.; Sharon, M.; Browse, J.; et al. Jasmonate perception by inositol-phosphate-potentiated COI1–JAZ co-receptor. Nature 2010, 468, 400–405. [Google Scholar] [CrossRef] [PubMed]
- Schaller, A.; Stintzi, A. Enzymes in jasmonate biosynthesis-structure, function, regulation. Phytochemistry 2009, 70, 1532–1538. [Google Scholar] [CrossRef]
- Howe, G.A. Plant hormones: Metabolic end run to jasmonate. Nat. Chem. Biol. 2018, 14, 109–110. [Google Scholar] [CrossRef]
- Xu, L.; Liu, F.; Lechner, E.; Genschik, P.; Crosby, W.L.; Ma, H.; Peng, W.; Huang, D.; Xie, D. The SCF(COI1) ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell. 2002, 14, 1919–1935. [Google Scholar] [CrossRef]
- Devoto, A.; Nieto-Rostro, M.; Xie, D.; Ellis, C.; Harmston, R.; Patrick, E.; Davis, J.; Sherratt, L.; Coleman, M.; Turner, J.G. COI1 links jasmonate signalling and fertility to the SCF ubiquitin-ligase complex in Arabidopsis. Plant J. 2002, 32, 457–466. [Google Scholar] [CrossRef]
- Thines, B.; Katsir, L.; Melotto, M.; Niu, Y.; Mandaokar, A.; Liu, G.; Nomura, K.; He, S.Y.; Howe, G.A.; Browse, J. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 2007, 448, 661–665. [Google Scholar] [CrossRef]
- Xie, D.-X.; Feys, B.F.; James, S.; Nieto-Rostro, M.; Turner, J.G. COI1: An Arabidopsis Gene Required for Jasmonate-Regulated Defense and Fertility. Science 1998, 280, 1091–1094. [Google Scholar] [CrossRef]
- Howe, G.A.; Major, I.T.; Koo, A.J. Modularity in Jasmonate Signaling for Multistress Resilience. Annu. Rev. Plant Biol. 2018, 69, 387–415. [Google Scholar] [CrossRef]
- Fernandez-Calvo, P.; Chini, A.; Fernandez-Barbero, G.; Chico, J.M.; Gimenez-Ibanez, S.; Geerinck, J.; Eeckhout, D.; Schweizer, F.; Godoy, M.; Franco-Zorrilla, J.M.; et al. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell. 2011, 23, 701–715. [Google Scholar] [CrossRef] [PubMed]
- Chini, A.; Fonseca, S.; Fernández, G.; Adie, B.; Chico, J.M.; Lorenzo, O.; García-Casado, G.; López-Vidriero, I.; Lozano, F.M.; Ponce, M.R.; et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 2007, 448, 666–671. [Google Scholar] [CrossRef] [PubMed]
- Melotto, M.; Mecey, C.; Niu, Y.; Chung, H.S.; Katsir, L.; Yao, J.; Zeng, W.; Thines, B.; Staswick, P.; Browse, J.; et al. A critical role of two positively charged amino acids in the Jas motif of Arabidopsis JAZ proteins in mediating coronatine- and jasmonoyl isoleucine-dependent interactions with the COI1 F-box protein. Plant J. 2008, 55, 979–988. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Stolz, S.; Chételat, A.; Reymond, P.; Pagni, M.; Dubugnon, L.; Farmer, E.E. A Downstream Mediator in the Growth Repression Limb of the Jasmonate Pathway. Plant Cell 2007, 19, 2470–2483. [Google Scholar] [CrossRef]
- Shyu, C.; Figueroa, P.; DePew, C.L.; Cooke, T.F.; Sheard, L.B.; Moreno, J.; Katsir, L.; Zheng, N.; Browse, J.; Howe, G.A. JAZ8 Lacks a Canonical Degron and Has an EAR Motif That Mediates Transcriptional Repression of Jasmonate Responses in Arabidopsis. Plant Cell 2012, 24, 536–550. [Google Scholar] [CrossRef]
- Pauwels, L.; Barbero, G.F.; Geerinck, J.; Tilleman, S.; Grunewald, W.; Pérez, A.C.; Chico, J.M.; Bossche, R.V.; Sewell, J.; Gil, E.; et al. NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 2010, 464, 788–791. [Google Scholar] [CrossRef]
- Ke, J.; Ma, H.; Gu, X.; Thelen, A.; Brunzelle, J.S.; Li, J.; Xu, H.E.; Melcher, K. Structural basis for recognition of diverse transcriptional repressors by the TOPLESS family of corepressors. Sci. Adv. 2015, 1, e1500107. [Google Scholar] [CrossRef]
- Kazan, K.; Manners, J.M. MYC2: The Master in Action. Mol. Plant 2013, 6, 686–703. [Google Scholar] [CrossRef]
- Monte, I.; Ishida, S.; Zamarreño, A.M.; Hamberg, M.; Franco-Zorrilla, J.M.; García-Casado, G.; Gouhier-Darimont, C.; Reymond, P.; Takahashi, K.; García-Mina, J.M.; et al. Ligand-receptor co-evolution shaped the jasmonate pathway in land plants. Nat. Chem. Biol. 2018, 14, 480–488. [Google Scholar] [CrossRef]
- Howe, G.A.; Yoshida, Y. Evolutionary Origin of JAZ Proteins and Jasmonate Signaling. Mol. Plant 2019, 12, 153–155. [Google Scholar] [CrossRef]
- Monte, I.; Franco-Zorrilla, J.M.; García-Casado, G.; Zamarreño, A.M.; García-Mina, J.M.; Nishihama, R.; Kohchi, T.; Solano, R. A Single JAZ Repressor Controls the Jasmonate Pathway in Marchantia polymorpha. Mol. Plant 2018, 12, 185–198. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Yoshida, Y.; Major, I.T.; Wang, K.; Sugimoto, K.; Kapali, G.; Havko, N.E.; Benning, C.; Howe, G.A. JAZ repressors of metabolic defense promote growth and reproductive fitness in Arabidopsis. Proc. Natl. Acad. Sci. USA 2018, 115, E10768–E10777. [Google Scholar] [CrossRef] [PubMed]
- Schluttenhofer, C. Origin and evolution of jasmonate signaling. Plant Sci. 2020, 298, 110542. [Google Scholar] [CrossRef] [PubMed]
- Moreno, J.; Shyu, C.; Campos, M.; Patel, L.C.; Chung, H.S.; Yao, J.; He, S.Y.; Howe, G.A. Negative Feedback Control of Jasmonate Signaling by an Alternative Splice Variant of JAZ10. Plant Physiol. 2013, 162, 1006–1017. [Google Scholar] [CrossRef]
- McConn, M.; Browse, J. The critical requirement for linolenic acid is pollen development, not photosynthesis, in an Arabidopsis mutant. Plant Cell. 1996, 8, 403–416. [Google Scholar] [CrossRef]
- Stintzi, A.; Weber, H.; Reymond, P.; Browse, J.; Farmer, E.E. Plant defense in the absence of jasmonic acid: The role of cyclopentenones. Proc. Natl. Acad. Sci. USA 2001, 98, 12837–12842. [Google Scholar] [CrossRef]
- Stintzi, A.; Browse, J. The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc. Natl. Acad. Sci. USA 2000, 97, 10625–10630. [Google Scholar] [CrossRef]
- Staswick, P.E.; Yuen, G.Y.; Lehman, C.C. Jasmonate signaling mutants of Arabidopsis are susceptible to the soil fungus Pythium irregulare. Plant J. 1998, 15, 747–754. [Google Scholar] [CrossRef]
- Staswick, P.E.; Su, W.; Howell, S.H. Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proc. Natl. Acad. Sci. USA 1992, 89, 6837–6840. [Google Scholar] [CrossRef]
- Ishiguro, S.; Kawai-Oda, A.; Ueda, J.; Nishida, I.; Okada, K. The DEFECTIVE IN ANTHER DEHISCIENCE gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell. 2001, 13, 2191–2209. [Google Scholar] [CrossRef]
- Sanders, P.M.; Lee, P.Y.; Biesgen, C.; Boone, J.D.; Beals, T.P.; Weiler, E.W.; Goldberg, R.B. The arabidopsis DELAYED DEHISCENCE1 gene encodes an enzyme in the jasmonic acid synthesis pathway. Plant Cell. 2000, 12, 1041–1061. [Google Scholar] [CrossRef] [PubMed]
- Park, J.-H.; Halitschke, R.; Kim, H.B.; Baldwin, I.; Feldmann, K.A.; Feyereisen, R. A knock-out mutation in allene oxide synthase results in male sterility and defective wound signal transduction in Arabidopsis due to a block in jasmonic acid biosynthesis. Plant J. 2002, 31, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Schilmiller, A.; Koo, A.; Howe, G.A. Functional Diversification of Acyl-Coenzyme A Oxidases in Jasmonic Acid Biosynthesis and Action. Plant Physiol. 2006, 143, 812–824. [Google Scholar] [CrossRef] [PubMed]
- Caldelari, D.; Wang, G.; Farmer, E.E.; Dong, X. Arabidopsis lox3 lox4 double mutants are male sterile and defective in global proliferative arrest. Plant Mol. Biol. 2010, 75, 25–33. [Google Scholar] [CrossRef]
- Delker, C.; Zolman, B.K.; Miersch, O.; Wasternack, C. Jasmonate biosynthesis in Arabidopsis thaliana requires peroxisomal beta-oxidation enzymes--additional proof by properties of pex6 and aim1. Phytochemistry. 2007, 68, 1642–1650. [Google Scholar] [CrossRef]
- Richmond, T.A.; Bleecker, A.B. A defect in beta-oxidation causes abnormal inflorescence development in Arabidopsis. Plant Cell. 1999, 11, 1911–1924. [Google Scholar] [PubMed]
- Wiszniewski, A.A.; Bussell, J.D.; Long, R.L.; Smith, S.M. Knockout of the two evolutionarily conserved peroxisomal 3-ketoacyl-CoA thiolases in Arabidopsis recapitulates the abnormal inflorescence meristem 1 phenotype. J. Exp. Bot. 2014, 65, 6723–6733. [Google Scholar] [CrossRef]
- Heitz, T.; Widemann, E.; Lugan, R.; Miesch, L.; Ullmann, P.; Désaubry, L.; Holder, E.; Grausem, B.; Kandel, S.; Miesch, M.; et al. Cytochromes P450 CYP94C1 and CYP94B3 Catalyze Two Successive Oxidation Steps of Plant Hormone Jasmonoyl-isoleucine for Catabolic Turnover. J. Biol. Chem. 2012, 287, 6296–6306. [Google Scholar] [CrossRef]
- Kloek, A.P.; Verbsky, M.L.; Sharma, S.B.; Schoelz, J.E.; Vogel, J.; Klessig, D.F.; Kunkel, B.N. Resistance to Pseudomonas syringae conferred by an Arabidopsis thaliana coronatine-insensitive (coi1) mutation occurs through two distinct mechanisms. Plant J. 2001, 26, 509–522. [Google Scholar] [CrossRef]
- Feys, B.; Benedetti, C.E.; Penfold, C.N.; Turner, J.G. Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 1994, 6, 751–759. [Google Scholar] [CrossRef]
- Castillo, M.C.; Leon, J. Expression of the beta-oxidation gene 3-ketoacyl-CoA thiolase 2(KAT2) is required for the timely onset of natural and dark-induced leaf senescence in Arabidopsis. J. Exp. Bot. 2008, 59, 2171–2179. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Deng, L.; Zhai, Q.; Zhao, J.; Chen, Q.; Li, C. Mediator Subunit MED25 Couples Alternative Splicing of JAZ Genes with Fine-Tuning of Jasmonate Signaling. Plant Cell 2019, 32, 429–448. [Google Scholar] [CrossRef] [PubMed]
- Shan, X.; Wang, J.; Chua, L.; Jiang, D.; Peng, W.; Xie, D. The Role of Arabidopsis Rubisco Activase in Jasmonate-Induced Leaf Senescence. Plant Physiol. 2010, 155, 751–764. [Google Scholar] [CrossRef] [PubMed]
- Alonso, J.M.; Stepanova, A.N.; Leisse, T.J.; Kim, C.J.; Chen, H.; Shinn, P.; Stevenson, D.K.; Zimmerman, J.; Barajas, P.; Cheuk, R.; et al. Genome-Wide Insertional Mutagenesis of Arabidopsis thaliana. Science 2003, 301, 653–657. [Google Scholar] [CrossRef]
- Kim, J.; Dotson, B.; Rey, C.; Lindsey, J.; Bleecker, A.B.; Binder, B.M.; Patterson, S.E. New Clothes for the Jasmonic Acid Receptor COI1: Delayed Abscission, Meristem Arrest and Apical Dominance. PLoS ONE 2013, 8, e60505. [Google Scholar] [CrossRef]
- Gimenez-Ibanez, S.; Boter, M.; Ortigosa, A.; García-Casado, G.; Chini, A.; Lewsey, M.G.; Ecker, J.; Ntoukakis, V.; Solano, R. JAZ 2 controls stomata dynamics during bacterial invasion. New Phytol. 2016, 213, 1378–1392. [Google Scholar] [CrossRef]
- Yang, D.-L.; Yao, J.; Mei, C.-S.; Tong, X.-H.; Zeng, L.-J.; Li, Q.; Xiao, L.-T.; Sun, T.-P.; Li, J.; Deng, X.-W.; et al. Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc. Natl. Acad. Sci. USA 2012, 109, E1192–E1200. [Google Scholar] [CrossRef]
- Demianski, A.J.; Chung, K.M.; Kunkel, B.N. Analysis of Arabidopsis JAZ gene expression during Pseudomonas syringae pathogenesis. Mol. Plant Pathol. 2011, 13, 46–57. [Google Scholar] [CrossRef]
- Yan, H.; Yoo, M.-J.; Koh, J.; Liu, L.; Chen, Y.; Acikgoz, D.; Wang, Q.; Chen, S. Molecular Reprogramming of Arabidopsis in Response to Perturbation of Jasmonate Signaling. J. Proteome Res. 2014, 13, 5751–5766. [Google Scholar] [CrossRef]
- Ingle, R.; Stoker, C.; Stone, W.; Adams, N.; Smith, R.; Grant, M.; Carre, I.; Roden, L.C.; Denby, K. Jasmonate signalling drives time-of-day differences in susceptibility of Arabidopsis to the fungal pathogen Botrytis cinerea. Plant J. 2015, 84, 937–948. [Google Scholar] [CrossRef]
- Sehr, E.M.; Agusti, J.; Lehner, R.; Farmer, E.E.; Schwarz, M.; Greb, T. Analysis of secondary growth in the Arabidopsis shoot reveals a positive role of jasmonate signalling in cambium formation. Plant J. 2010, 63, 811–822. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Zhang, Y.; Di, C.; Zhang, Q.; Zhang, K.; Wang, C.; You, Q.; Yan, H.; Dai, S.Y.; Yuan, J.S.; et al. JAZ7 negatively regulates dark-induced leaf senescence in Arabidopsis. J. Exp. Bot. 2016, 67, 751–762. [Google Scholar] [CrossRef] [PubMed]
- de Torres Zabala, M.; Zhai, B.; Jayaraman, S.; Eleftheriadou, G.; Winsbury, R.; Yang, R.; Truman, W.; Tang, S.; Smirnoff, N.; Grant, M. Novel JAZ co-operativity and unexpected JA dynamics underpin Arabidopsis defence responses to Pseudomonas syringae infection. New Phytol. 2016, 209, 1120–1134. [Google Scholar] [CrossRef]
- Qi, T.; Huang, H.; Song, S.; Xie, D. Regulation of Jasmonate-Mediated Stamen Development and Seed Production by a bHLH-MYB Complex in Arabidopsis. Plant Cell 2015, 27, 1620–1633. [Google Scholar] [PubMed]
- Schweizer, F.; Fernandez-Calvo, P.; Zander, M.; Diez-Diaz, M.; Fonseca, S.; Glauser, G.; Lewsey, M.G.; Ecker, J.R.; Solano, R.; Reymond, P. Arabidopsis basic helix-loop-helix transcription factors MYC2, MYC3, and MYC4 regulate glucosinolate biosynthesis, insect performance, and feeding behavior. Plant Cell 2013, 25, 3117–3132. [Google Scholar] [CrossRef] [PubMed]
- Zhurov, V.; Navarro, M.; Bruinsma, K.A.; Arbona, V.; Santamaria, M.E.; Cazaux, M.; Wybouw, N.; Osborne, E.J.; Ens, C.; Rioja, C.; et al. Reciprocal Responses in the Interaction between Arabidopsis and the Cell-Content-Feeding Chelicerate Herbivore Spider Mite. Plant Physiol. 2013, 164, 384–399. [Google Scholar] [CrossRef]
- Meng, L.; Zhang, T.; Geng, S.; Scott, P.B.; Li, H.; Chen, S. Comparative proteomics and metabolomics of JAZ7-mediated drought tolerance in Arabidopsis. J. Proteom. 2019, 196, 81–91. [Google Scholar] [CrossRef]
- Chung, H.S.; Howe, G.A. A Critical Role for the TIFY Motif in Repression of Jasmonate Signaling by a Stabilized Splice Variant of the JASMONATE ZIM-Domain Protein JAZ10 inArabidopsis. Plant Cell 2009, 21, 131–145. [Google Scholar] [CrossRef]
- Xu, J.; Wang, X.; Zu, H.; Zeng, X.; Baldwin, I.T.; Lou, Y.; Li, R. Molecular dissection of rice phytohormone signaling involved in resistance to a piercing-sucking herbivore. New Phytol. 2021, 230, 1639–1652. [Google Scholar] [CrossRef]
- Dhakarey, R.; Raorane, M.L.; Treumann, A.; Peethambaran, P.K.; Schendel, R.R.; Sahi, V.P.; Hause, B.; Bunzel, M.; Henry, A.; Kohli, A.; et al. Physiological and proteomic analysis of the rice mutant cpm2 suggests a negative regulatory role of jasmonic acid in drought tolerance. Front Plant Sci. 2017, 8, 1903. [Google Scholar] [CrossRef]
- Qi, J.; Li, J.; Han, X.; Li, R.; Wu, J.; Yu, H.; Hu, L.; Xiao, Y.; Lu, J.; Lou, Y. Jasmonic acid carboxyl methyltransferase regulates development and herbivory-induced defense response in rice. J. Integr. Plant Biol. 2015, 58, 564–576. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Zhang, H.; Sun, Z.; Li, J.; Hong, G.; Zhu, Q.; Zhou, X.; MacFarlane, S.; Yan, F.; Chen, J. Jasmonic acid-mediated defense suppresses brassinosteroid-mediated susceptibility to Rice black streaked dwarf virus infection in rice. New Phytol. 2016, 214, 388–399. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Wu, H.; Ma, S.; Xiang, D.; Liu, R.; Xiong, L. OsJAZ1 Attenuates Drought Resistance by Regulating JA and ABA Signaling in Rice. Front. Plant Sci. 2017, 8, 2108. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Tian, J.; Liu, Y.; Chen, X.; Li, S.; Persson, S.; Lu, D.; Chen, M.; Luo, Z.; Zhang, D.; et al. Ectopic expression of OsJAZ6, which interacts with OsJAZ1, alters JA signaling and spikelet development in rice. Plant J. 2021, 108, 1083–1096. [Google Scholar] [CrossRef]
- Xiao, Y.; Zhang, G.; Liu, D.; Niu, M.; Tong, H.; Chu, C. GSK2 stabilizes OFP3 to suppress brassinosteroid responses in rice. Plant J. 2020, 102, 1187–1201. [Google Scholar] [CrossRef]
- Hu, J.; Huang, J.; Xu, H.; Wang, Y.; Li, C.; Wen, P.; You, X.; Zhang, X.; Pan, G.; Li, Q.; et al. Rice stripe virus suppresses jasmonic acid-mediated resistance by hijacking brassinosteroid signaling pathway in rice. PLoS Pathog. 2020, 16, e1008801. [Google Scholar] [CrossRef]
- Yamada, S.; Kano, A.; Tamaoki, D.; Miyamoto, A.; Shishido, H.; Miyoshi, S.; Taniguchi, S.; Akimitsu, K.; Gomi, K. Involvement of OsJAZ8 in Jasmonate-Induced Resistance to Bacterial Blight in Rice. Plant Cell Physiol. 2012, 53, 2060–2072. [Google Scholar] [CrossRef]
- Feng, X.; Zhang, L.; Wei, X.; Zhou, Y.; Dai, Y.; Zhu, Z. OsJAZ13 Negatively Regulates Jasmonate Signaling and Activates Hypersensitive Cell Death Response in Rice. Int. J. Mol. Sci. 2020, 21, 4379. [Google Scholar] [CrossRef]
- Howe, G.A.; Lightner, J.; Browse, J.; Ryan, C.A. An octadecanoid pathway mutant (JL5) of tomato is compromised in signaling for defense against insect attack. Plant Cell 1996, 8, 2067–2077. [Google Scholar]
- Lightner, J.; Pearce, G.; Ryan, C.A.; Browse, J. Isolation of signaling mutants of tomato (Lycopersicon esculentum). Mol. Gen. Genet. MGG 1993, 241, 595–601. [Google Scholar] [CrossRef]
- Lee, G.I.; Howe, G.A. The tomato mutant spr1 is defective in systemin perception and the production of a systemic wound signal for defense gene expression. Plant J. 2003, 33, 567–576. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Liu, G.; Xu, C.; Lee, G.I.; Bauer, P.; Ling, H.-Q.; Ganal, M.W.; Howe, G.A. The Tomato Suppressor of prosystemin-mediated responses2 Gene Encodes a Fatty Acid Desaturase Required for the Biosynthesis of Jasmonic Acid and the Production of a Systemic Wound Signal for Defense Gene Expression. Plant Cell 2003, 15, 1646–1661. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Li, C.; Lee, G.I.; Howe, G.A. Distinct roles for jasmonate synthesis and action in the systemic wound response of tomato. Proc. Natl. Acad. Sci. USA 2002, 99, 6416–6421. [Google Scholar] [CrossRef] [PubMed]
- Campos, M.; Kang, J.-H.; Howe, G.A. Jasmonate-Triggered Plant Immunity. J. Chem. Ecol. 2014, 40, 657–675. [Google Scholar] [CrossRef]
- Thaler, J.S.; Owen, B.; Higgins, V.J. The Role of the Jasmonate Response in Plant Susceptibility to Diverse Pathogens with a Range of Lifestyles. Plant Physiol. 2004, 135, 530–538. [Google Scholar] [CrossRef]
- AbuQamar, S.; Chai, M.-F.; Luo, H.; Song, F.; Mengiste, T. Tomato Protein Kinase 1b Mediates Signaling of Plant Responses to Necrotrophic Fungi and Insect Herbivory. Plant Cell 2008, 20, 1964–1983. [Google Scholar] [CrossRef]
- Ortigosa, A.; Gimenez-Ibanez, S.; Leonhardt, N.; Solano, R. Design of a bacterial speck resistant tomato by CRISPR/Cas9-mediated editing of SlJAZ2. Plant Biotechnol. J. 2018, 17, 665–673. [Google Scholar] [CrossRef]
- Yu, X.; Chen, G.; Tang, B.; Zhang, J.; Zhou, S.; Hu, Z. The Jasmonate ZIM-domain protein gene SlJAZ2 regulates plant morphology and accelerates flower initiation in Solanum lycopersicum plants. Plant Sci. 2017, 267, 65–73. [Google Scholar] [CrossRef]
- Li, L.; Zhao, Y.; McCaig, B.C.; Wingerd, B.A.; Wang, J.; Whalon, M.E.; Pichersky, E.; Howe, G.A. The Tomato Homolog of CORONATINE-INSENSITIVE1 Is Required for the Maternal Control of Seed Maturation, Jasmonate-Signaled Defense Responses, and Glandular Trichome Development. Plant Cell 2004, 16, 126–143. [Google Scholar] [CrossRef]
- Widemann, E.; Miesch, L.; Lugan, R.; Holder, E.; Heinrich, C.; Aubert, Y.; Miesch, M.; Pinot, F.; Heitz, T. The Amidohydrolases IAR3 and ILL6 Contribute to Jasmonoyl-Isoleucine Hormone Turnover and Generate 12-Hydroxyjasmonic Acid Upon Wounding in Arabidopsis Leaves. J. Biol. Chem. 2013, 288, 31701–31714. [Google Scholar] [CrossRef]
- Wasternack, C.; Strnad, M. Jasmonate signaling in plant stress responses and development—Active and inactive compounds. New Biotechnol. 2016, 33, 604–613. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.; Herrfurth, C.; Xin, M.; Savchenko, T.; Feussner, I.; Goossens, A.; De Smet, I. Warm temperature triggers JOX and ST2A-mediated jasmonate catabolism to promote plant growth. Nat. Commun. 2021, 12, 1–8. [Google Scholar] [CrossRef]
- Fernández-Milmanda, G.L.; Crocco, C.D.; Reichelt, M.; Mazza, C.A.; Köllner, T.G.; Zhang, T.; Cargnel, M.D.; Lichy, M.Z.; Fiorucci, A.-S.; Fankhauser, C.; et al. A light-dependent molecular link between competition cues and defence responses in plants. Nat. Plants 2020, 6, 223–230. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Yu, G.; Cao, C.; Liu, P. Metabolism, signaling, and transport of jasmonates. Plant Commun. 2021, 2, 100231. [Google Scholar] [CrossRef] [PubMed]
- Glauser, G.; Dubugnon, L.; Mousavi, S.A.; Rudaz, S.; Wolfender, J.-L.; Farmer, E.E. Velocity Estimates for Signal Propagation Leading to Systemic Jasmonic Acid Accumulation in Wounded Arabidopsis. J. Biol. Chem. 2009, 284, 34506–34513. [Google Scholar] [CrossRef]
- Yan, J.; Li, H.; Li, S.; Yao, R.; Deng, H.; Xie, Q.; Xie, D. The Arabidopsis F-Box Protein Coronatine Insensitive1 Is Stabilized by SCFCOI1 and Degraded via the 26S Proteasome Pathway. Plant Cell 2013, 25, 486–498. [Google Scholar] [CrossRef]
- Thireault, C.; Shyu, C.; Yoshida, Y.; St Aubin, B.; Campos, M.L.; Howe, G.A. Repression of jasmonate signaling by a non-TIFY JAZ protein in Arabidopsis. Plant J. 2015, 82, 669–679. [Google Scholar] [CrossRef]
- Nakata, M.; Mitsuda, N.; Herde, M.; Koo, A.J.; Moreno, J.E.; Suzuki, K.; Howe, G.A.; Ohme-Takagi, M. A bHLH-type transcription factor, aba-inducible bhlh-type transcription factor/ja-associated myc2-like1, acts as a repressor to negatively regulate jasmonate signaling in arabidopsis. Plant Cell. 2013, 25, 1641–1656. [Google Scholar] [CrossRef]
- Qi, T.; Wang, J.; Huang, H.; Liu, B.; Gao, H.; Liu, Y.; Song, S.; Xie, D. Regulation of Jasmonate-Induced Leaf Senescence by Antagonism between bHLH Subgroup IIIe and IIId Factors in Arabidopsis. Plant Cell 2015, 27, 1634–1649. [Google Scholar] [CrossRef]
- Zhang, G.; Liu, W.; Gu, Z.; Wu, S.; Zhou, W.; Lin, J.; Xu, L. Roles of the wound hormone jasmonate in plant regeneration. J. Exp. Bot. 2021. [Google Scholar] [CrossRef]
- Zhang, G.; Zhao, F.; Chen, L.; Pan, Y.; Sun, L.; Bao, N.; Zhang, T.; Cui, C.-X.; Qiu, Z.; Zhang, Y.; et al. Jasmonate-mediated wound signalling promotes plant regeneration. Nat. Plants 2019, 5, 491–497. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Lozano-Torres, J.L.; Blilou, I.; Zhang, X.; Zhai, Q.; Smant, G.; Li, C.; Scheres, B. A Jasmonate Signaling Network Activates Root Stem Cells and Promotes Regeneration. Cell 2019, 177, 942–956.e14. [Google Scholar] [CrossRef] [PubMed]
- Weber, H. Fatty acid-derived signals in plants. Trends Plant Sci. 2002, 7, 217–224. [Google Scholar] [CrossRef]
- Raza, A.; Charagh, S.; Zahid, Z.; Mubarik, M.S.; Javed, R.; Siddiqui, M.H.; Hasanuzzaman, M. Jasmonic acid: A key frontier in conferring abiotic stress tolerance in plants. Plant Cell Rep. 2021, 40, 1513–1541. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Christensen, S.; Isakeit, T.; Engelberth, J.; Meeley, R.; Hayward, A.; Emery, R.N.; Kolomiets, M.V. Disruption of OPR7 and OPR8 Reveals the Versatile Functions of Jasmonic Acid in Maize Development and Defense. Plant Cell 2012, 24, 1420–1436. [Google Scholar] [CrossRef]
- Reeves, P.H.; Ellis, C.M.; Ploense, S.E.; Wu, M.-F.; Yadav, V.; Tholl, D.; Chételat, A.; Haupt, I.; Kennerley, B.J.; Hodgens, C.; et al. A Regulatory Network for Coordinated Flower Maturation. PLoS Genet. 2012, 8, e1002506. [Google Scholar] [CrossRef]
- Song, S.; Qi, T.; Huang, H.; Ren, Q.; Wu, D.; Chang, C.; Peng, W.; Liu, Y.; Peng, J.; Xie, D. The Jasmonate-ZIM Domain Proteins Interact with the R2R3-MYB Transcription Factors MYB21 and MYB24 to Affect Jasmonate-Regulated Stamen Development in Arabidopsis. Plant Cell 2011, 23, 1000–1013. [Google Scholar] [CrossRef]
- Zhai, Q.; Zhang, X.; Wu, F.; Feng, H.; Deng, L.; Xu, L.; Zhang, M.; Wang, Q.; Li, C. Transcriptional Mechanism of Jasmonate Receptor COI1-Mediated Delay of Flowering Time in Arabidopsis. Plant Cell 2015, 27, 2814–2828. [Google Scholar] [CrossRef]
- Xu, Q.; Truong, T.T.; Barrero, J.M.; Jacobsen, J.V.; Hocart, C.H.; Gubler, F. A role for jasmonates in the release of dormancy by cold stratification in wheat. J. Exp. Bot. 2016, 67, 3497–3508. [Google Scholar] [CrossRef]
- Wang, Y.; Hou, Y.; Qiu, J.; Wang, H.; Wang, S.; Tang, L.; Tong, X.; Zhang, J. Abscisic acid promotes jasmonic acid biosynthesis via a ‘SAPK10-bZIP72-AOC’ pathway to synergistically inhibit seed germination in rice (Oryza sativa). New Phytol. 2020, 228, 1336–1353. [Google Scholar] [CrossRef]
- Zhuo, M.; Sakuraba, Y.; Yanagisawa, S. A Jasmonate-activated MYC2-Dof2.1-MYC2 transcriptional loop promotes leaf senescence in Arabidopsis. Plant Cell 2020, 32, 242–262. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, Y.; Wei, H.; Li, N.; Tian, W.; Chong, K.; Wang, L. Circadian Evening Complex Represses Jasmonate-Induced Leaf Senescence in Arabidopsis. Mol. Plant 2018, 11, 326–337. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Wu, Y.; Yang, Y.; Du, M.; Zhang, X.; Guo, Y.; Li, C.; Zhou, J.-M. An Arabidopsis Plasma Membrane Proton ATPase Modulates JA Signaling and Is Exploited by the Pseudomonas syringae Effector Protein AvrB for Stomatal Invasion. Plant Cell 2015, 27, 2032–2041. [Google Scholar] [CrossRef]
- Hua, B.; Chang, J.; Xu, Z.; Han, X.; Xu, M.; Yang, M.; Yang, C.; Ye, Z.; Wu, S. HOMEODOMAIN PROTEIN8 mediates jasmonate-triggered trichome elongation in tomato. New Phytol. 2021, 230, 1063–1077. [Google Scholar] [CrossRef] [PubMed]
- Yan, T.; Chen, M.; Shen, Q.; Li, L.; Fu, X.; Pan, Q.; Tang, Y.; Shi, P.; Lv, Z.; Jiang, W.; et al. HOMEODOMAIN PROTEIN 1 is required for jasmonate-mediated glandular trichome initiation in Artemisia annua. New Phytol. 2016, 213, 1145–1155. [Google Scholar] [CrossRef] [PubMed]
- Qi, T.; Huang, H.; Wu, D.; Yan, J.; Qi, Y.; Song, S.; Xie, D. Arabidopsis DELLA and JAZ proteins bind the WD-repeat/bHLH/MYB complex to modulate gibberellin and jasmonate signaling synergy. Plant Cell 2014, 26, 1118–1133. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Sun, J.; Zhai, Q.; Zhou, W.; Qi, L.; Xu, L.; Wang, B.; Chen, R.; Jiang, H.; Qi, J.; et al. The basic helix-loop-helix transcription factor MYC2 directly represses PLETHORA expression during jasmonate-mediated modulation of the root stem cell niche in Arabidopsis. Plant Cell 2011, 23, 3335–3352. [Google Scholar] [CrossRef]
- Hentrich, M.; Böttcher, C.; Düchting, P.; Cheng, Y.; Zhao, Y.; Berkowitz, O.; Masle, J.; Medina, J.; Pollmann, S. The jasmonic acid signaling pathway is linked to auxin homeostasis through the modulation ofYUCCA8andYUCCA9gene expression. Plant J. 2013, 74, 626–637. [Google Scholar] [CrossRef]
- Taki, N.; Sasaki-Sekimoto, Y.; Obayashi, T.; Kikuta, A.; Kobayashi, K.; Ainai, T.; Yagi, K.; Sakurai, N.; Suzuki, H.; Masuda, T.; et al. 12-Oxo-Phytodienoic Acid Triggers Expression of a Distinct Set of Genes and Plays a Role in Wound-Induced Gene Expression in Arabidopsis. Plant Physiol. 2005, 139, 1268–1283. [Google Scholar] [CrossRef]
- Qi, L.; Yan, J.; Li, Y.; Jiang, H.; Sun, J.; Chen, Q.; Li, H.; Chu, J.; Yan, C.; Sun, X.; et al. Arabidopsis thaliana plants differentially modulate auxin biosynthesis and transport during defense responses to the necrotrophic pathogen Alternaria brassicicola. New Phytol. 2012, 195, 872–882. [Google Scholar] [CrossRef]
- Yan, C.; Xie, D. Jasmonate in plant defence: Sentinel or double agent? Plant Biotechnol. J. 2015, 13, 1233–1240. [Google Scholar] [CrossRef] [PubMed]
- Thomma, B.P.H.J.; Eggermont, K.; Penninckx, I.A.M.A.; Mauch-Mani, B.; Vogelsang, R.; Cammue, B.P.A.; Broekaert, W.F. Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc. Natl. Acad. Sci. USA 1998, 95, 15107–15111. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Jiang, Y.; Han, X.; Wang, H.; Pan, J.; Yu, D. Jasmonate regulates leaf senescence and tolerance to cold stress: Crosstalk with other phytohormones. J. Exp. Bot. 2017, 68, 1361–1369. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhang, L.; Xiang, S.; Chen, Y.; Zhang, H.; Yu, D. The transcription factor WRKY75 positively regulates jasmonate-mediated plant defense to necrotrophic fungal pathogens. J. Exp. Bot. 2020, 72, 1473–1489. [Google Scholar] [CrossRef] [PubMed]
- Lv, T.; Li, X.; Fan, T.; Luo, H.; Xie, C.; Zhou, Y.; Tian, C.-E. The Calmodulin-Binding Protein IQM1 Interacts with CATALASE2 to Affect Pathogen Defense. Plant Physiol. 2019, 181, 1314–1327. [Google Scholar] [CrossRef] [PubMed]
- Ren, H.; Bai, M.; Sun, J.; Liu, J.; Ren, M.; Dong, Y.; Wang, N.; Ning, G.; Wang, C. RcMYB84 and RcMYB123 mediate jasmonate-induced defense responses against Botrytis cinerea in rose (Rosa chinensis). Plant J. 2020, 103, 1839–1849. [Google Scholar] [CrossRef] [PubMed]
- Dombrecht, B.; Xue, G.P.; Sprague, S.J.; Kirkegaard, J.A.; Ross, J.J.; Reid, J.B.; Fitt, G.P.; Sewelam, N.; Schenk, P.M.; Manners, J.M.; et al. MYC2 Differentially Modulates Diverse Jasmonate-Dependent Functions in Arabidopsis. Plant Cell 2007, 19, 2225–2245. [Google Scholar] [CrossRef]
- Lorenzo, O.; Chico, J.M.; Saénchez-Serrano, J.J.; Solano, R. JASMONATE-INSENSITIVE1 Encodes a MYC Transcription Factor Essential to Discriminate between Different Jasmonate-Regulated Defense Responses in Arabidopsis. Plant Cell 2004, 16, 1938–1950. [Google Scholar] [CrossRef]
- Du, M.; Zhao, J.; Tzeng, D.T.; Liu, Y.; Deng, L.; Yang, T.; Zhai, Q.; Wu, F.; Huang, Z.; Zhou, M.; et al. MYC2 Orchestrates a Hierarchical Transcriptional Cascade That Regulates Jasmonate-Mediated Plant Immunity in Tomato. Plant Cell 2017, 29, 1883–1906. [Google Scholar] [CrossRef]
- Zhu, Z.; An, F.; Feng, Y.; Li, P.; Xue, L.; Mu, A.; Jiang, Z.; Kim, J.M.; To, T.K.; Li, W.; et al. Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 12539–12544. [Google Scholar] [CrossRef]
- Pre, M.; Atallah, M.; Champion, A.; De Vos, M.; Pieterse, C.M.; Memelink, J. The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol. 2008, 147, 1347–1357. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Xu, H.; Huang, J.; Kong, Y.; AbuQamar, S.; Yu, D.; Liu, S.; Zhou, G.; Chai, G. The Arabidopsis CCCH protein C3H14 contributes to basal defense against Botrytis cinerea mainly through the WRKY33-dependent pathway. Plant Cell Environ. 2020, 43, 1792–1806. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Prado, J.S.; Latrasse, D.; Rodriguez-Granados, N.Y.; Huang, Y.; Manza-Mianza, D.; Brik-Chaouche, R.; Jaouannet, M.; Citerne, S.; Bendahmane, A.; Hirt, H.; et al. The Polycomb protein LHP 1 regulates Arabidopsis thaliana stress responses through the repression of the MYC 2-dependent branch of immunity. Plant J. 2019, 100, 1118–1131. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.; Zhang, J.; Yao, T.; Bryan, A.C.; Pu, Y.; Labbé, J.; Pelletier, D.A.; Engle, N.; Morrell-Falvey, J.L.; Schmutz, J.; et al. Arabidopsis C-terminal binding protein ANGUSTIFOLIA modulates transcriptional co-regulation of MYB46 and WRKY33. New Phytol. 2020, 228, 1627–1639. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, Q.; Zhai, H.; Gao, S.; Yang, L.; Wang, Z.; Xu, Y.; Huo, J.; Ren, Z.; Zhao, N.; et al. IbBBX24 Promotes the Jasmonic Acid Pathway and Enhances Fusarium Wilt Resistance in Sweet Potato. Plant Cell 2020, 32, 1102–1123. [Google Scholar] [CrossRef]
- Hou, Y.; Wang, Y.; Tang, L.; Tong, X.; Wang, L.; Liu, L.; Huang, S.; Zhang, J. SAPK10-Mediated Phosphorylation on WRKY72 Releases Its Suppression on Jasmonic Acid Biosynthesis and Bacterial Blight Resistance. iScience 2019, 16, 499–510. [Google Scholar] [CrossRef]
- Boter, M.; Ruíz-Rivero, O.; Abdeen, A.; Prat, S. Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Genes Dev. 2004, 18, 1577–1591. [Google Scholar] [CrossRef]
- Falk, K.L.; Kastner, J.; Bodenhausen, N.; Schramm, K.; Paetz, C.; Vassao, D.G.; Reichelt, M.; von Knorre, D.; Bergelson, J.; Erb, M.; et al. The role of glucosinolates and the jasmonic acid pathway in resistance of Arabidopsis thaliana against molluscan herbivores. Mol. Ecol. 2014, 23, 1188–1203. [Google Scholar] [CrossRef]
- Mikkelsen, M.D.; Petersen, B.L.; Glawischnig, E.; Jensen, A.B.; Andreasson, E.; Halkier, B. Modulation of CYP79 Genes and Glucosinolate Profiles in Arabidopsis by Defense Signaling Pathways. Plant Physiol. 2003, 131, 298–308. [Google Scholar] [CrossRef]
- Reymond, P.; Bodenhausen, N.; Van Poecke, R.M.P.; Krishnamurthy, V.; Dicke, M.; Farmer, E.E. A conserved transcript pattern in response to a specialist and a generalist herbivore. Plant Cell. 2004, 16, 3132–3147. [Google Scholar] [CrossRef]
- Thaler, J.S.; Stout, M.J.; Karban, R.; Duffey, S.S. Jasmonate-mediated induced plant resistance affects a community of herbivores. Ecol. Èntomol. 2001, 26, 312–324. [Google Scholar] [CrossRef]
- Peiffer, M.; Tooker, J.; Luthe, D.S.; Felton, G.W. Plants on early alert: Glandular trichomes as sensors for insect herbivores. New Phytol. 2009, 184, 644–656. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.-H.; McRoberts, J.; Shi, F.; Moreno, J.; Jones, A.D.; Howe, G.A. The Flavonoid Biosynthetic Enzyme Chalcone Isomerase Modulates Terpenoid Production in Glandular Trichomes of Tomato. Plant Physiol. 2014, 164, 1161–1174. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Alonso, M.-M.; Sánchez-Parra, B.; Ortiz-García, P.; Santamaría, M.E.; Díaz, I.; Pollmann, S. Jasmonic Acid-Dependent MYC Transcription Factors Bind to a Tandem G-Box Motif in the YUCCA8 and YUCCA9 Promoters to Regulate Biotic Stress Responses. Int. J. Mol. Sci. 2021, 22, 9768. [Google Scholar] [CrossRef] [PubMed]
- Machado, R.A.R.; Robert, C.; Arce, C.C.M.; Ferrieri, A.P.; Xu, S.; Jimenez-Aleman, G.; Baldwin, I.T.; Erb, M. Auxin Is Rapidly Induced by Herbivore Attack and Regulates a Subset of Systemic, Jasmonate-Dependent Defenses. Plant Physiol. 2016, 172, 521–532. [Google Scholar] [CrossRef]
- Zhang, P.-J.; Wei, J.-N.; Zhao, C.; Zhang, Y.-F.; Li, C.-Y.; Liu, S.-S.; Dicke, M.; Yu, X.-P.; Turlings, T.C.J. Airborne host–plant manipulation by whiteflies via an inducible blend of plant volatiles. Proc. Natl. Acad. Sci. USA 2019, 116, 7387–7396. [Google Scholar] [CrossRef]
- Chen, C.-Y.; Liu, Y.-Q.; Song, W.-M.; Chen, D.-Y.; Chen, F.-Y.; Chen, X.-Y.; Chen, Z.-W.; Ge, S.-X.; Wang, C.-Z.; Zhan, S.; et al. An effector from cotton bollworm oral secretion impairs host plant defense signaling. Proc. Natl. Acad. Sci. USA 2019, 116, 14331–14338. [Google Scholar] [CrossRef]
- Pan, G.; Liu, Y.; Ji, L.; Zhang, X.; He, J.; Huang, J.; Qiu, Z.; Liu, D.; Sun, Z.; Xu, T.; et al. Brassinosteroids mediate susceptibility to brown planthopper by integrating with the salicylic acid and jasmonic acid pathways in rice. J. Exp. Bot. 2018, 69, 4433–4442. [Google Scholar] [CrossRef]
- Xie, K.; Li, L.; Zhang, H.; Wang, R.; Tan, X.; He, Y.; Hong, G.; Li, J.; Ming, F.; Yao, X.; et al. Abscisic acid negatively modulates plant defence against rice black-streaked dwarf virus infection by suppressing the jasmonate pathway and regulating reactive oxygen species levels in rice. Plant Cell Environ. 2018, 41, 2504–2514. [Google Scholar] [CrossRef]
- Li, L.; Zhang, H.; Chen, C.; Huang, H.; Tan, X.; Wei, Z.; Li, J.; Yan, F.; Zhang, C.; Chen, J.; et al. A class of independently evolved transcriptional repressors in plant RNA viruses facilitates viral infection and vector feeding. Proc. Natl. Acad. Sci. USA 2021, 118. [Google Scholar] [CrossRef]
- He, L.; Chen, X.; Yang, J.; Zhang, T.; Li, J.; Zhang, S.; Zhong, K.; Zhang, H.; Chen, J. Rice black-streaked dwarf virus -encoded P5-1 regulates the ubiquitination activity of SCF E3 ligases and inhibits jasmonate signaling to benefit its infection in rice. New Phytol. 2019, 225, 896–912. [Google Scholar] [CrossRef] [PubMed]
- Han, K.; Huang, H.; Zheng, H.; Ji, M.; Yuan, Q.; Cui, W.; Zhang, H.; Peng, J.; Lu, Y.; Rao, S.; et al. Rice stripe virus coat protein induces the accumulation of jasmonic acid, activating plant defence against the virus while also attracting its vector to feed. Mol. Plant Pathol. 2020, 21, 1647–1653. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Abellan, J.O.; Fernandez-Garcia, N.; Lopez-Berenguer, C.; Egea, I.; Flores, F.B.; Angosto, T.; Capel, J.; Lozano, R.; Pineda, B.; Moreno, V.; et al. The tomato res mutant which accumulates JA in roots in non-stressed conditions restores cell structure alterations under salinity. Physiol Plantarum. 2015, 155, 296–314. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.-S.; Joo, J.; Kim, M.-J.; Kim, Y.-K.; Nahm, B.H.; Song, S.I.; Cheong, J.-J.; Lee, J.S.; Kim, J.-K.; Choi, Y.D. OsbHLH148, a basic helix-loop-helix protein, interacts with OsJAZ proteins in a jasmonate signaling pathway leading to drought tolerance in rice. Plant J. 2011, 65, 907–921. [Google Scholar] [CrossRef]
- Demkura, P.V.; Abdala, G.; Baldwin, I.; Ballare, C.L. Jasmonate-Dependent and -Independent Pathways Mediate Specific Effects of Solar Ultraviolet B Radiation on Leaf Phenolics and Antiherbivore Defense. Plant Physiol. 2009, 152, 1084–1095. [Google Scholar] [CrossRef]
- Rao, M.V.; Lee, H.; Creelman, R.A.; Mullet, J.E.; Davis, K.R. Jasmonic acid signaling modulates ozone-induced hypersensitive cell death. Plant Cell 2000, 12, 1633–1646. [Google Scholar] [CrossRef]
- Sivasankar, S.; Sheldrick, B.; Rothstein, S.J. Expression of Allene Oxide Synthase Determines Defense Gene Activation in Tomato. Plant Physiol. 2000, 122, 1335–1342. [Google Scholar] [CrossRef]
- Howe, G.A.; Schilmiller, A. Oxylipin metabolism in response to stress. Curr. Opin. Plant Biol. 2002, 5, 230–236. [Google Scholar] [CrossRef]
- Wang, X.; Li, Q.; Xie, J.; Huang, M.; Cai, J.; Zhou, Q.; Dai, T.; Jiang, D. Abscisic acid and jasmonic acid are involved in drought priming-induced tolerance to drought in wheat. Crop J. 2020, 9, 120–132. [Google Scholar] [CrossRef]
- An, J.P.; Wang, X.F.; Zhang, X.W.; You, C.X.; Hao, Y.J. Apple B-box protein BBX37 regulates jasmonic acid mediated cold tolerance through the JAZ-BBX37-ICE1-CBF pathway and undergoes MIEL1-mediated ubiquitination and degradation. New Phytol. 2021, 229, 2707–2729. [Google Scholar] [CrossRef]
- Hou, X.; Lee, L.Y.C.; Xia, K.; Yan, Y.; Yu, H. DELLAs Modulate Jasmonate Signaling via Competitive Binding to JAZs. Dev. Cell 2010, 19, 884–894. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Hentrich, M.; Pollmann, S. Auxin-Oxylipin Crosstalk: Relationship of AntagonistsF. J. Integr. Plant Biol. 2011, 53, 429–445. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Alonso, M.-M. How Auxin May Contribute to the Regulation of Plant Defense Responses against Herbivory. Austin J. Plant Biol. 2018, 4, 1019. [Google Scholar]
- Zhang, T.; Poudel, A.N.; Jewell, J.B.; Kitaoka, N.; Staswick, P.; Matsuura, H.; Koo, A.J. Hormone crosstalk in wound stress response: Wound-inducible amidohydrolases can simultaneously regulate jasmonate and auxin homeostasis in Arabidopsis thaliana. J. Exp. Bot. 2015, 67, 2107–2120. [Google Scholar] [CrossRef] [PubMed]
- Risseeuw, E.P.; Daskalchuk, T.E.; Banks, T.W.; Liu, E.; Cotelesage, J.; Hellmann, H.; Estelle, M.; Somers, D.E.; Crosby, W.L. Protein interaction analysis of SCF ubiquitin E3 ligase subunits from Arabidopsis. Plant J. 2003, 34, 753–767. [Google Scholar] [CrossRef] [PubMed]
- Ren, C.; Pan, J.; Peng, W.; Genschik, P.; Hobbie, L.; Hellmann, H.; Estelle, M.; Gao, B.; Peng, J.; Sun, C.; et al. Point mutations in Arabidopsis Cullin1 reveal its essential role in jasmonate response. Plant J. 2005, 42, 514–524. [Google Scholar] [CrossRef]
- Liu, Y.; Wei, H.; Ma, M.; Li, Q.; Kong, D.; Sun, J.; Ma, X.; Wang, B.; Chen, C.; Xie, Y.; et al. Arabidopsis FHY3 and FAR1 Regulate the Balance between Growth and Defense Responses under Shade Conditions. Plant Cell 2019, 31, 2089–2106. [Google Scholar] [CrossRef]
- Savchenko, T.V.; Rolletschek, H.; Dehesh, K. Jasmonates-Mediated Rewiring of Central Metabolism Regulates Adaptive Responses. Plant Cell Physiol. 2019, 60, 2613–2620. [Google Scholar] [CrossRef]
- Zhu, X.; Zhu, J.-K. Double Repression in Jasmonate-Mediated Plant Defense. Mol. Cell 2013, 50, 459–460. [Google Scholar] [CrossRef][Green Version]
- Ueda, M.; Kaji, T.; Kozaki, W. Recent Advances in Plant Chemical Biology of Jasmonates. Int. J. Mol. Sci. 2020, 21, 1124. [Google Scholar] [CrossRef]
- Bastías, D.A.; Gianoli, E.; Gundel, P.E. Fungal endophytes can eliminate the plant growth-defence trade-off. New Phytol. 2021, 230, 2105–2113. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Guo, W.; Yang, D.; Hou, Z.; Liang, Z. Transcriptional Profiles of SmWRKY Family Genes and Their Putative Roles in the Biosynthesis of Tanshinone and Phenolic Acids in Salvia miltiorrhiza. Int. J. Mol. Sci. 2018, 19, 1593. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).