Jasmonate Signaling Pathway Modulates Plant Defense, Growth, and Their Trade-Offs
Abstract
:1. Introduction
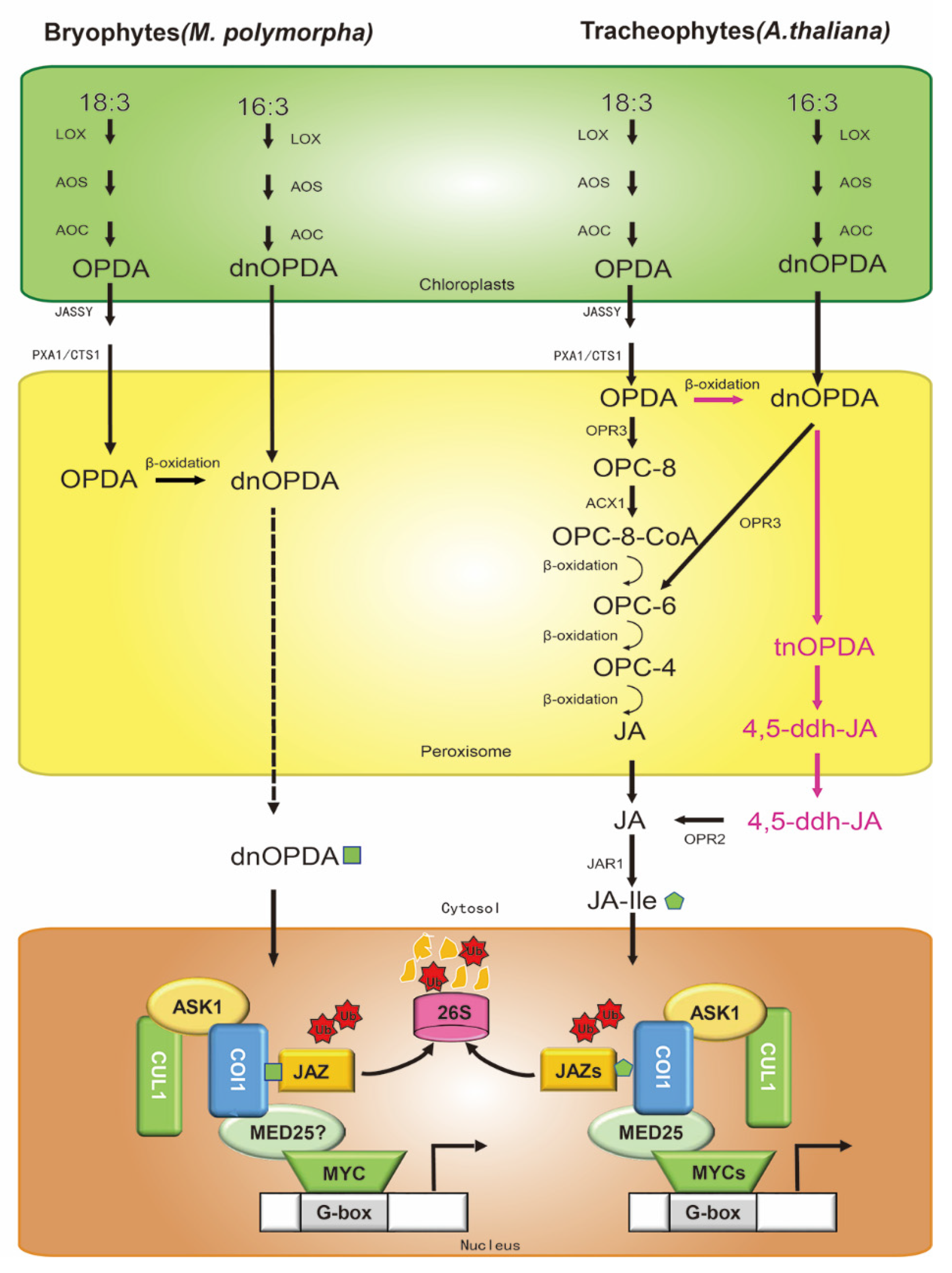
2. Biosynthesis of Jasmonate
3. Jasmonate Signaling Pathway
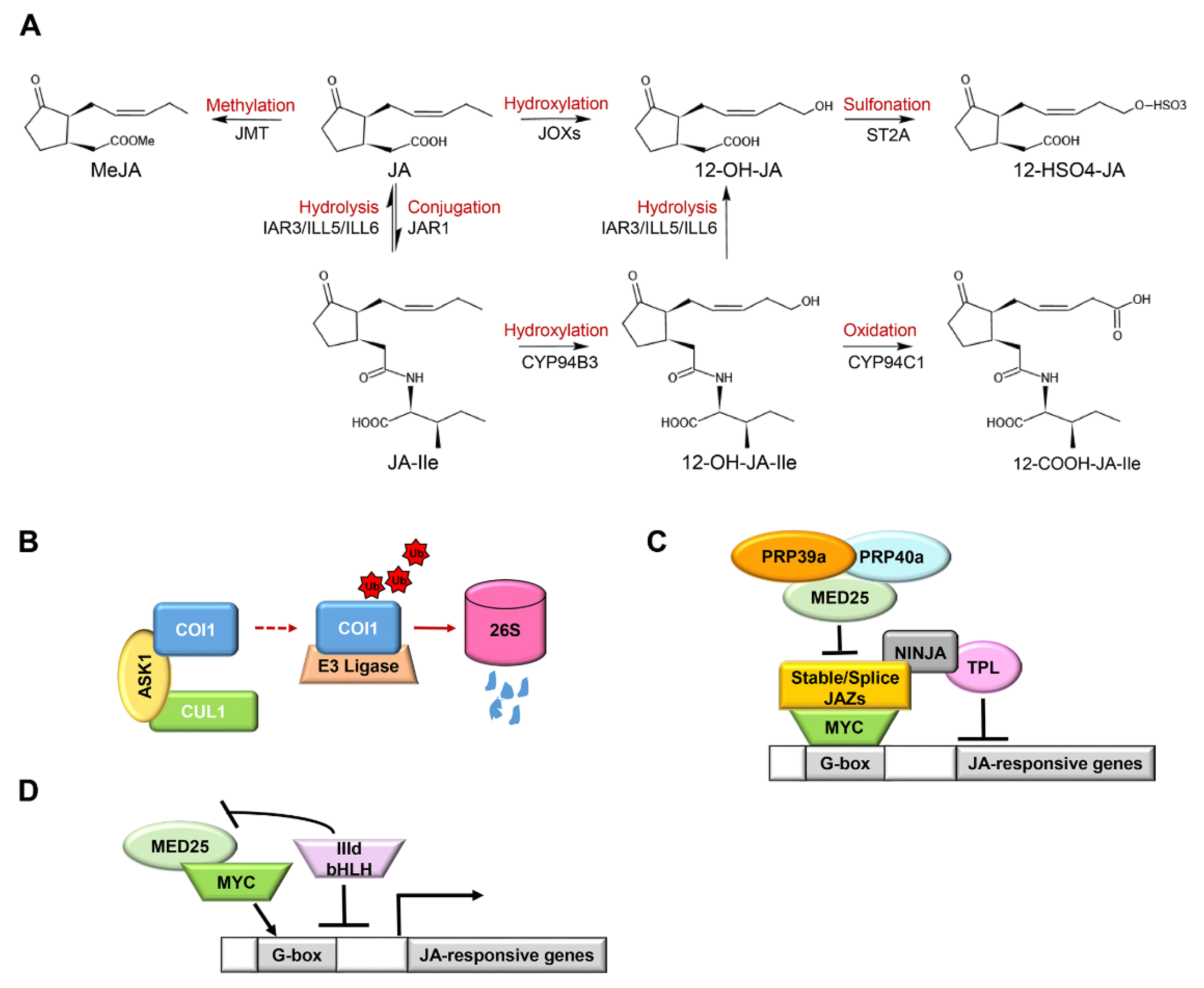
3.1. The Core JA-Ile Pathway
3.2. The Termination of JA Signaling
4. The Functions of JA in Growth and Development
4.1. Promotion of Plant Regeneration
4.2. Regulation of Reproductive Growth
4.3. Actions of JA in Vegetative Growth
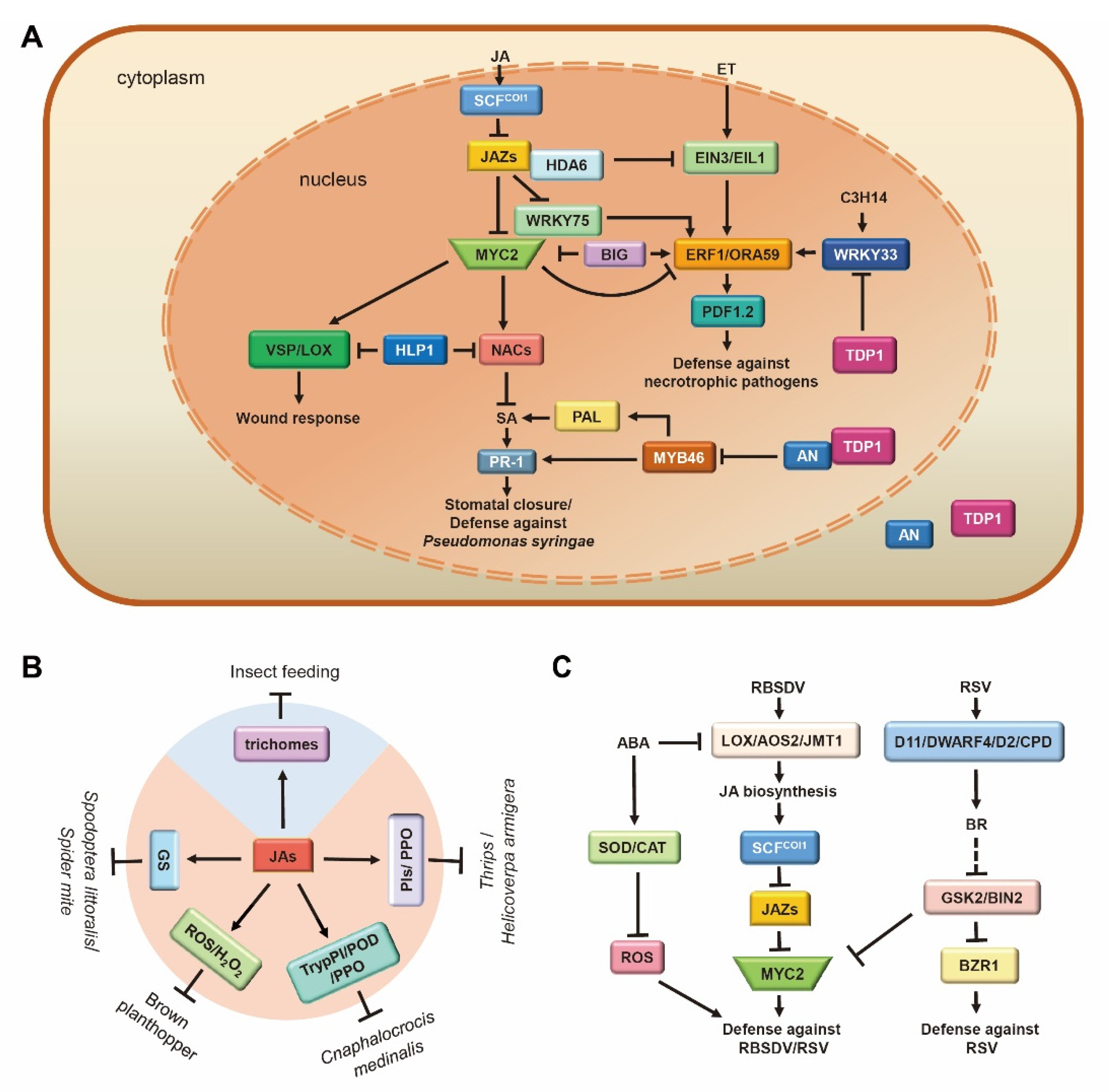
5. Role of JA during Plant Defense Responses
5.1. JA Mediates Plant Defense against Pathogens
5.2. JA Acts as a Double Agent in Plant Defense against Herbivorous Insects
5.3. JA Plays Vital Roles in the Arms Race between Plants and Viruses
5.4. JA Regulates Plant Tolerance against Abiotic Stresses
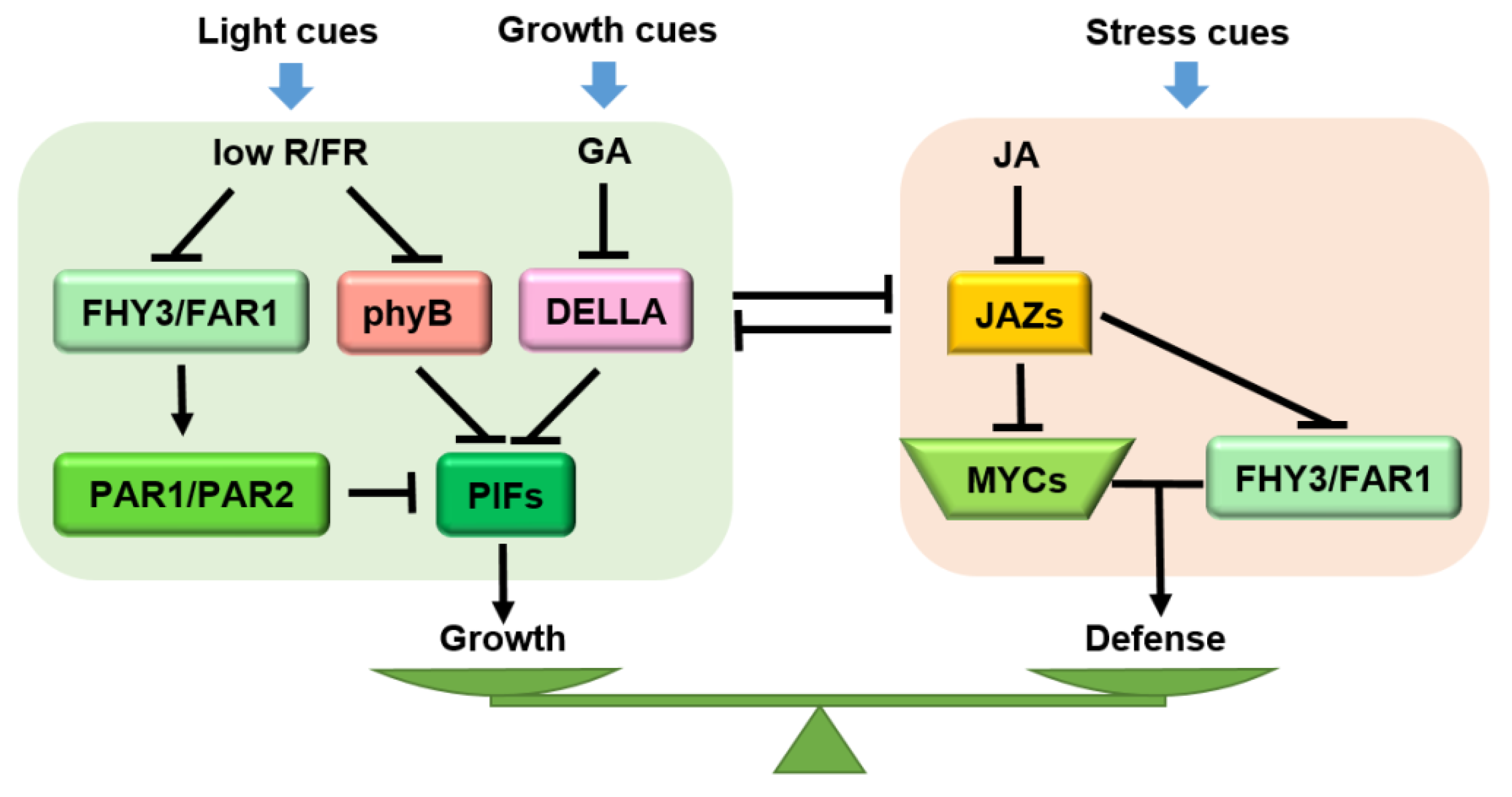
6. JA Mediates the Trade-Offs between Growth and Defense
6.1. Interactions among JA Core Components
6.2. Crosstalk between JA and Other Phytohormones
6.3. Crosstalk between JA and Phytochrome Signaling Pathway
7. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ueda, J.; Kato, J. Isolation and Identification of a Senescence-promoting Substance from Wormwood (Artemisia absinthium L.). Plant Physiol. 1980, 66, 246–249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dathe, W.; Preiss, A.; Schade, W.; Sembdner, G.; Schreiber, K. Endogenous plant hormones of the broad bean, Vicia faba L. (-)-jasmonic acid, a plant growth inhibitor in pericarp. Planta 1981, 153, 530–535. [Google Scholar] [CrossRef] [PubMed]
- Staswick, P.E. Novel Regulation of Vegetative Storage Protein Genes. Plant Cell. 1990, 2, 1–6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kashihara, K.; Onohata, T.; Yariuchi, R.; Tanaka, S.; Akimitsu, K.; Gomi, K. The overexpression of OsSRO1a, which encodes an OsNINJA1- and OsMYC2-interacting protein, negatively affects OsMYC2-mediated jasmonate signaling in rice. Plant Cell Rep. 2020, 39, 489–500. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.-T.; Xu, P.; Zhao, P.-X.; Liu, R.; Yu, L.-H.; Xiang, C.-B. Arabidopsis ERF109 mediates cross-talk between jasmonic acid and auxin biosynthesis during lateral root formation. Nat. Commun. 2014, 5, 5833. [Google Scholar] [CrossRef]
- Thines, B.; Mandaokar, A.; Browse, J. Characterizing Jasmonate Regulation of Male Fertility in Arabidopsis. In Jasmonate Signaling; Springer Nature: Cham, Switzerland, 2013; Volume 1011, pp. 13–23. [Google Scholar] [CrossRef]
- He, Y.; Fukushige, H.; Hildebrand, D.F.; Gan, S. Evidence Supporting a Role of Jasmonic Acid in Arabidopsis Leaf Senescence. Plant Physiol. 2002, 128, 876–884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wasternack, C.; Song, S. Jasmonates: Biosynthesis, metabolism, and signaling by proteins activating and repressing transcription. J. Exp. Bot. 2017, 68, 1303–1321. [Google Scholar] [CrossRef]
- Liu, Y.; Du, M.; Deng, L.; Shen, J.; Fang, M.; Chen, Q.; Lu, Y.; Wang, Q.; Li, C.; Zhai, Q. MYC2 Regulates the Termination of Jasmonate Signaling via an Autoregulatory Negative Feedback Loop. Plant Cell 2019, 31, 106–127. [Google Scholar] [CrossRef] [Green Version]
- Fonseca, S.; Calvo, P.F.; Fernández, G.M.; Diez-Diaz, M.; Gimenez-Ibanez, S.; López-Vidriero, I.; Godoy, M.; Fernández-Barbero, G.; Van Leene, J.; De Jaeger, G.; et al. bHLH003, bHLH013 and bHLH017 Are New Targets of JAZ Repressors Negatively Regulating JA Responses. PLoS ONE 2014, 9, e86182. [Google Scholar] [CrossRef] [Green Version]
- Ballaré, C.L.; Austin, A.T. Recalculating growth and defense strategies under competition: Key roles of photoreceptors and jasmonates. J. Exp. Bot. 2019, 70, 3425–3434. [Google Scholar] [CrossRef]
- Major, I.T.; Yoshida, Y.; Campos, M.L.; Kapali, G.; Xin, X.F.; Sugimoto, K.; de Oliveira Ferreira, D.; He, S.Y.; Howe, G.A. Regulation of growth-defense balance by the Jasmonate Zim-Domain (Jaz)-Myc transcriptional module. New Phytol. 2017, 215, 1533–1547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campos, M.L.; Yoshida, Y.; Major, I.T.; de Ferreira, D.O.; Weraduwage, S.M.; Froehlich, J.E.; Johnson, B.F.; Kramer, D.M.; Jander, G.; Sharkey, T.D.; et al. Rewiring of jasmonate and phytochrome B signalling uncouples plant growth-defense tradeoffs. Nat. Commun. 2016, 7, 12570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Major, I.T.; Guo, Q.; Zhai, J.; Kapali, G.; Kramer, D.M.; Howe, G.A. A Phytochrome B-Independent Pathway Restricts Growth at High Levels of Jasmonate Defense. Plant Physiol. 2020, 183, 733–749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, F.; Yang, C.; Cao, J.; Chen, H.; Pang, J.; Zhao, Q.; Wang, Z.; Fu, Z.Q.; Liu, J. A bHLH transcription activator regulates defense signaling by nucleo-cytosolic trafficking in rice. J. Integr. Plant Biol. 2020, 62, 1552–1573. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.-X.; Ge, S.; He, J.; Li, S.; Hao, Y.; Du, H.; Liu, Z.; Cheng, R.; Feng, Y.-Q.; Xiong, L.; et al. BIG regulates stomatal immunity and jasmonate production in Arabidopsis. New Phytol. 2018, 222, 335–348. [Google Scholar] [CrossRef] [PubMed]
- Yimer, H.Z.; Nahar, K.; Kyndt, T.; Haeck, A.; Van Meulebroek, L.; Vanhaecke, L.; Demeestere, K.; Hofte, M.; Gheysen, G. Gibberellin antagonizes jasmonate-induced defense against Meloidogyne graminicola in rice. New Phytol. 2018, 218, 646–660. [Google Scholar] [CrossRef] [Green Version]
- Bonaventure, G.; Schuck, S.; Baldwin, I.T. Revealing complexity and specificity in the activation of lipase-mediated oxylipin biosynthesis: A specific role of the Nicotiana attenuata GLA1 lipase in the activation of jasmonic acid biosynthesis in leaves and roots. Plant Cell Environ. 2011, 34, 1507–1520. [Google Scholar] [CrossRef]
- Chini, A.; Monte, I.; Zamarreño, A.M.; Hamberg, M.; Lassueur, S.; Reymond, P.; Weiss, S.; Stintzi, A.; Schaller, A.; Porzel, A.; et al. An OPR3-independent pathway uses 4,5-didehydrojasmonate for jasmonate synthesis. Nat. Chem. Biol. 2018, 14, 171–178. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, P.; Rasool, S.; Gul, A.; Sheikh, S.A.; Akram, N.A.; Ashraf, M.; Kazi, A.M.; Gucel, S. Jasmonates: Multifunctional Roles in Stress Tolerance. Front. Plant Sci. 2016, 7, 813. [Google Scholar] [CrossRef] [Green Version]
- Ellinger, D.; Stingl, N.; Kubigsteltig, I.I.; Bals, T.; Juenger, M.; Pollmann, S.; Berger, S.; Schuenemann, D.; Mueller, M.J. DONGLE and DEFECTIVE IN ANTHER DEHISCENCE1 lipases are not essential for wound- and pathogen-induced jasmonate biosynthesis: Redundant lipases contribute to jasmonate formation. Plant Physiol. 2010, 153, 114–127. [Google Scholar] [CrossRef] [Green Version]
- Guan, L.; Denkert, N.; Eisa, A.; Lehmann, M.; Sjuts, I.; Weiberg, A.; Soll, J.; Meinecke, M.; Schwenkert, S. JASSY, a chloroplast outer membrane protein required for jasmonate biosynthesis. Proc. Natl. Acad. Sci. USA 2019, 116, 10568–10575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Theodoulou, F.L.; Job, K.; Slocombe, S.P.; Footitt, S.; Holdsworth, M.; Baker, A.; Larson, T.R.; Graham, I.A. Jasmonic Acid Levels Are Reduced in COMATOSE ATP-Binding Cassette Transporter Mutants. Implications for Transport of Jasmonate Precursors into Peroxisomes. Plant Physiol. 2005, 137, 835–840. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheard, L.B.; Tan, X.; Mao, H.; Withers, J.; Ben-Nissan, G.; Hinds, T.R.; Kobayashi, Y.; Hsu, F.-F.; Sharon, M.; Browse, J.; et al. Jasmonate perception by inositol-phosphate-potentiated COI1–JAZ co-receptor. Nature 2010, 468, 400–405. [Google Scholar] [CrossRef] [PubMed]
- Schaller, A.; Stintzi, A. Enzymes in jasmonate biosynthesis-structure, function, regulation. Phytochemistry 2009, 70, 1532–1538. [Google Scholar] [CrossRef]
- Howe, G.A. Plant hormones: Metabolic end run to jasmonate. Nat. Chem. Biol. 2018, 14, 109–110. [Google Scholar] [CrossRef]
- Xu, L.; Liu, F.; Lechner, E.; Genschik, P.; Crosby, W.L.; Ma, H.; Peng, W.; Huang, D.; Xie, D. The SCF(COI1) ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell. 2002, 14, 1919–1935. [Google Scholar] [CrossRef] [Green Version]
- Devoto, A.; Nieto-Rostro, M.; Xie, D.; Ellis, C.; Harmston, R.; Patrick, E.; Davis, J.; Sherratt, L.; Coleman, M.; Turner, J.G. COI1 links jasmonate signalling and fertility to the SCF ubiquitin-ligase complex in Arabidopsis. Plant J. 2002, 32, 457–466. [Google Scholar] [CrossRef] [Green Version]
- Thines, B.; Katsir, L.; Melotto, M.; Niu, Y.; Mandaokar, A.; Liu, G.; Nomura, K.; He, S.Y.; Howe, G.A.; Browse, J. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 2007, 448, 661–665. [Google Scholar] [CrossRef]
- Xie, D.-X.; Feys, B.F.; James, S.; Nieto-Rostro, M.; Turner, J.G. COI1: An Arabidopsis Gene Required for Jasmonate-Regulated Defense and Fertility. Science 1998, 280, 1091–1094. [Google Scholar] [CrossRef]
- Howe, G.A.; Major, I.T.; Koo, A.J. Modularity in Jasmonate Signaling for Multistress Resilience. Annu. Rev. Plant Biol. 2018, 69, 387–415. [Google Scholar] [CrossRef] [Green Version]
- Fernandez-Calvo, P.; Chini, A.; Fernandez-Barbero, G.; Chico, J.M.; Gimenez-Ibanez, S.; Geerinck, J.; Eeckhout, D.; Schweizer, F.; Godoy, M.; Franco-Zorrilla, J.M.; et al. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell. 2011, 23, 701–715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chini, A.; Fonseca, S.; Fernández, G.; Adie, B.; Chico, J.M.; Lorenzo, O.; García-Casado, G.; López-Vidriero, I.; Lozano, F.M.; Ponce, M.R.; et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 2007, 448, 666–671. [Google Scholar] [CrossRef] [PubMed]
- Melotto, M.; Mecey, C.; Niu, Y.; Chung, H.S.; Katsir, L.; Yao, J.; Zeng, W.; Thines, B.; Staswick, P.; Browse, J.; et al. A critical role of two positively charged amino acids in the Jas motif of Arabidopsis JAZ proteins in mediating coronatine- and jasmonoyl isoleucine-dependent interactions with the COI1 F-box protein. Plant J. 2008, 55, 979–988. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, Y.; Stolz, S.; Chételat, A.; Reymond, P.; Pagni, M.; Dubugnon, L.; Farmer, E.E. A Downstream Mediator in the Growth Repression Limb of the Jasmonate Pathway. Plant Cell 2007, 19, 2470–2483. [Google Scholar] [CrossRef] [Green Version]
- Shyu, C.; Figueroa, P.; DePew, C.L.; Cooke, T.F.; Sheard, L.B.; Moreno, J.; Katsir, L.; Zheng, N.; Browse, J.; Howe, G.A. JAZ8 Lacks a Canonical Degron and Has an EAR Motif That Mediates Transcriptional Repression of Jasmonate Responses in Arabidopsis. Plant Cell 2012, 24, 536–550. [Google Scholar] [CrossRef] [Green Version]
- Pauwels, L.; Barbero, G.F.; Geerinck, J.; Tilleman, S.; Grunewald, W.; Pérez, A.C.; Chico, J.M.; Bossche, R.V.; Sewell, J.; Gil, E.; et al. NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 2010, 464, 788–791. [Google Scholar] [CrossRef] [Green Version]
- Ke, J.; Ma, H.; Gu, X.; Thelen, A.; Brunzelle, J.S.; Li, J.; Xu, H.E.; Melcher, K. Structural basis for recognition of diverse transcriptional repressors by the TOPLESS family of corepressors. Sci. Adv. 2015, 1, e1500107. [Google Scholar] [CrossRef] [Green Version]
- Kazan, K.; Manners, J.M. MYC2: The Master in Action. Mol. Plant 2013, 6, 686–703. [Google Scholar] [CrossRef] [Green Version]
- Monte, I.; Ishida, S.; Zamarreño, A.M.; Hamberg, M.; Franco-Zorrilla, J.M.; García-Casado, G.; Gouhier-Darimont, C.; Reymond, P.; Takahashi, K.; García-Mina, J.M.; et al. Ligand-receptor co-evolution shaped the jasmonate pathway in land plants. Nat. Chem. Biol. 2018, 14, 480–488. [Google Scholar] [CrossRef] [Green Version]
- Howe, G.A.; Yoshida, Y. Evolutionary Origin of JAZ Proteins and Jasmonate Signaling. Mol. Plant 2019, 12, 153–155. [Google Scholar] [CrossRef] [Green Version]
- Monte, I.; Franco-Zorrilla, J.M.; García-Casado, G.; Zamarreño, A.M.; García-Mina, J.M.; Nishihama, R.; Kohchi, T.; Solano, R. A Single JAZ Repressor Controls the Jasmonate Pathway in Marchantia polymorpha. Mol. Plant 2018, 12, 185–198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, Q.; Yoshida, Y.; Major, I.T.; Wang, K.; Sugimoto, K.; Kapali, G.; Havko, N.E.; Benning, C.; Howe, G.A. JAZ repressors of metabolic defense promote growth and reproductive fitness in Arabidopsis. Proc. Natl. Acad. Sci. USA 2018, 115, E10768–E10777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schluttenhofer, C. Origin and evolution of jasmonate signaling. Plant Sci. 2020, 298, 110542. [Google Scholar] [CrossRef] [PubMed]
- Moreno, J.; Shyu, C.; Campos, M.; Patel, L.C.; Chung, H.S.; Yao, J.; He, S.Y.; Howe, G.A. Negative Feedback Control of Jasmonate Signaling by an Alternative Splice Variant of JAZ10. Plant Physiol. 2013, 162, 1006–1017. [Google Scholar] [CrossRef] [Green Version]
- McConn, M.; Browse, J. The critical requirement for linolenic acid is pollen development, not photosynthesis, in an Arabidopsis mutant. Plant Cell. 1996, 8, 403–416. [Google Scholar] [CrossRef]
- Stintzi, A.; Weber, H.; Reymond, P.; Browse, J.; Farmer, E.E. Plant defense in the absence of jasmonic acid: The role of cyclopentenones. Proc. Natl. Acad. Sci. USA 2001, 98, 12837–12842. [Google Scholar] [CrossRef] [Green Version]
- Stintzi, A.; Browse, J. The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc. Natl. Acad. Sci. USA 2000, 97, 10625–10630. [Google Scholar] [CrossRef] [Green Version]
- Staswick, P.E.; Yuen, G.Y.; Lehman, C.C. Jasmonate signaling mutants of Arabidopsis are susceptible to the soil fungus Pythium irregulare. Plant J. 1998, 15, 747–754. [Google Scholar] [CrossRef]
- Staswick, P.E.; Su, W.; Howell, S.H. Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proc. Natl. Acad. Sci. USA 1992, 89, 6837–6840. [Google Scholar] [CrossRef] [Green Version]
- Ishiguro, S.; Kawai-Oda, A.; Ueda, J.; Nishida, I.; Okada, K. The DEFECTIVE IN ANTHER DEHISCIENCE gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell. 2001, 13, 2191–2209. [Google Scholar] [CrossRef] [Green Version]
- Sanders, P.M.; Lee, P.Y.; Biesgen, C.; Boone, J.D.; Beals, T.P.; Weiler, E.W.; Goldberg, R.B. The arabidopsis DELAYED DEHISCENCE1 gene encodes an enzyme in the jasmonic acid synthesis pathway. Plant Cell. 2000, 12, 1041–1061. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, J.-H.; Halitschke, R.; Kim, H.B.; Baldwin, I.; Feldmann, K.A.; Feyereisen, R. A knock-out mutation in allene oxide synthase results in male sterility and defective wound signal transduction in Arabidopsis due to a block in jasmonic acid biosynthesis. Plant J. 2002, 31, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Schilmiller, A.; Koo, A.; Howe, G.A. Functional Diversification of Acyl-Coenzyme A Oxidases in Jasmonic Acid Biosynthesis and Action. Plant Physiol. 2006, 143, 812–824. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caldelari, D.; Wang, G.; Farmer, E.E.; Dong, X. Arabidopsis lox3 lox4 double mutants are male sterile and defective in global proliferative arrest. Plant Mol. Biol. 2010, 75, 25–33. [Google Scholar] [CrossRef]
- Delker, C.; Zolman, B.K.; Miersch, O.; Wasternack, C. Jasmonate biosynthesis in Arabidopsis thaliana requires peroxisomal beta-oxidation enzymes--additional proof by properties of pex6 and aim1. Phytochemistry. 2007, 68, 1642–1650. [Google Scholar] [CrossRef]
- Richmond, T.A.; Bleecker, A.B. A defect in beta-oxidation causes abnormal inflorescence development in Arabidopsis. Plant Cell. 1999, 11, 1911–1924. [Google Scholar] [PubMed] [Green Version]
- Wiszniewski, A.A.; Bussell, J.D.; Long, R.L.; Smith, S.M. Knockout of the two evolutionarily conserved peroxisomal 3-ketoacyl-CoA thiolases in Arabidopsis recapitulates the abnormal inflorescence meristem 1 phenotype. J. Exp. Bot. 2014, 65, 6723–6733. [Google Scholar] [CrossRef] [Green Version]
- Heitz, T.; Widemann, E.; Lugan, R.; Miesch, L.; Ullmann, P.; Désaubry, L.; Holder, E.; Grausem, B.; Kandel, S.; Miesch, M.; et al. Cytochromes P450 CYP94C1 and CYP94B3 Catalyze Two Successive Oxidation Steps of Plant Hormone Jasmonoyl-isoleucine for Catabolic Turnover. J. Biol. Chem. 2012, 287, 6296–6306. [Google Scholar] [CrossRef] [Green Version]
- Kloek, A.P.; Verbsky, M.L.; Sharma, S.B.; Schoelz, J.E.; Vogel, J.; Klessig, D.F.; Kunkel, B.N. Resistance to Pseudomonas syringae conferred by an Arabidopsis thaliana coronatine-insensitive (coi1) mutation occurs through two distinct mechanisms. Plant J. 2001, 26, 509–522. [Google Scholar] [CrossRef]
- Feys, B.; Benedetti, C.E.; Penfold, C.N.; Turner, J.G. Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 1994, 6, 751–759. [Google Scholar] [CrossRef] [Green Version]
- Castillo, M.C.; Leon, J. Expression of the beta-oxidation gene 3-ketoacyl-CoA thiolase 2(KAT2) is required for the timely onset of natural and dark-induced leaf senescence in Arabidopsis. J. Exp. Bot. 2008, 59, 2171–2179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, F.; Deng, L.; Zhai, Q.; Zhao, J.; Chen, Q.; Li, C. Mediator Subunit MED25 Couples Alternative Splicing of JAZ Genes with Fine-Tuning of Jasmonate Signaling. Plant Cell 2019, 32, 429–448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shan, X.; Wang, J.; Chua, L.; Jiang, D.; Peng, W.; Xie, D. The Role of Arabidopsis Rubisco Activase in Jasmonate-Induced Leaf Senescence. Plant Physiol. 2010, 155, 751–764. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alonso, J.M.; Stepanova, A.N.; Leisse, T.J.; Kim, C.J.; Chen, H.; Shinn, P.; Stevenson, D.K.; Zimmerman, J.; Barajas, P.; Cheuk, R.; et al. Genome-Wide Insertional Mutagenesis of Arabidopsis thaliana. Science 2003, 301, 653–657. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.; Dotson, B.; Rey, C.; Lindsey, J.; Bleecker, A.B.; Binder, B.M.; Patterson, S.E. New Clothes for the Jasmonic Acid Receptor COI1: Delayed Abscission, Meristem Arrest and Apical Dominance. PLoS ONE 2013, 8, e60505. [Google Scholar] [CrossRef] [Green Version]
- Gimenez-Ibanez, S.; Boter, M.; Ortigosa, A.; García-Casado, G.; Chini, A.; Lewsey, M.G.; Ecker, J.; Ntoukakis, V.; Solano, R. JAZ 2 controls stomata dynamics during bacterial invasion. New Phytol. 2016, 213, 1378–1392. [Google Scholar] [CrossRef] [Green Version]
- Yang, D.-L.; Yao, J.; Mei, C.-S.; Tong, X.-H.; Zeng, L.-J.; Li, Q.; Xiao, L.-T.; Sun, T.-P.; Li, J.; Deng, X.-W.; et al. Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc. Natl. Acad. Sci. USA 2012, 109, E1192–E1200. [Google Scholar] [CrossRef] [Green Version]
- Demianski, A.J.; Chung, K.M.; Kunkel, B.N. Analysis of Arabidopsis JAZ gene expression during Pseudomonas syringae pathogenesis. Mol. Plant Pathol. 2011, 13, 46–57. [Google Scholar] [CrossRef]
- Yan, H.; Yoo, M.-J.; Koh, J.; Liu, L.; Chen, Y.; Acikgoz, D.; Wang, Q.; Chen, S. Molecular Reprogramming of Arabidopsis in Response to Perturbation of Jasmonate Signaling. J. Proteome Res. 2014, 13, 5751–5766. [Google Scholar] [CrossRef]
- Ingle, R.; Stoker, C.; Stone, W.; Adams, N.; Smith, R.; Grant, M.; Carre, I.; Roden, L.C.; Denby, K. Jasmonate signalling drives time-of-day differences in susceptibility of Arabidopsis to the fungal pathogen Botrytis cinerea. Plant J. 2015, 84, 937–948. [Google Scholar] [CrossRef] [Green Version]
- Sehr, E.M.; Agusti, J.; Lehner, R.; Farmer, E.E.; Schwarz, M.; Greb, T. Analysis of secondary growth in the Arabidopsis shoot reveals a positive role of jasmonate signalling in cambium formation. Plant J. 2010, 63, 811–822. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Zhang, Y.; Di, C.; Zhang, Q.; Zhang, K.; Wang, C.; You, Q.; Yan, H.; Dai, S.Y.; Yuan, J.S.; et al. JAZ7 negatively regulates dark-induced leaf senescence in Arabidopsis. J. Exp. Bot. 2016, 67, 751–762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Torres Zabala, M.; Zhai, B.; Jayaraman, S.; Eleftheriadou, G.; Winsbury, R.; Yang, R.; Truman, W.; Tang, S.; Smirnoff, N.; Grant, M. Novel JAZ co-operativity and unexpected JA dynamics underpin Arabidopsis defence responses to Pseudomonas syringae infection. New Phytol. 2016, 209, 1120–1134. [Google Scholar] [CrossRef] [Green Version]
- Qi, T.; Huang, H.; Song, S.; Xie, D. Regulation of Jasmonate-Mediated Stamen Development and Seed Production by a bHLH-MYB Complex in Arabidopsis. Plant Cell 2015, 27, 1620–1633. [Google Scholar] [PubMed] [Green Version]
- Schweizer, F.; Fernandez-Calvo, P.; Zander, M.; Diez-Diaz, M.; Fonseca, S.; Glauser, G.; Lewsey, M.G.; Ecker, J.R.; Solano, R.; Reymond, P. Arabidopsis basic helix-loop-helix transcription factors MYC2, MYC3, and MYC4 regulate glucosinolate biosynthesis, insect performance, and feeding behavior. Plant Cell 2013, 25, 3117–3132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhurov, V.; Navarro, M.; Bruinsma, K.A.; Arbona, V.; Santamaria, M.E.; Cazaux, M.; Wybouw, N.; Osborne, E.J.; Ens, C.; Rioja, C.; et al. Reciprocal Responses in the Interaction between Arabidopsis and the Cell-Content-Feeding Chelicerate Herbivore Spider Mite. Plant Physiol. 2013, 164, 384–399. [Google Scholar] [CrossRef] [Green Version]
- Meng, L.; Zhang, T.; Geng, S.; Scott, P.B.; Li, H.; Chen, S. Comparative proteomics and metabolomics of JAZ7-mediated drought tolerance in Arabidopsis. J. Proteom. 2019, 196, 81–91. [Google Scholar] [CrossRef]
- Chung, H.S.; Howe, G.A. A Critical Role for the TIFY Motif in Repression of Jasmonate Signaling by a Stabilized Splice Variant of the JASMONATE ZIM-Domain Protein JAZ10 inArabidopsis. Plant Cell 2009, 21, 131–145. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Wang, X.; Zu, H.; Zeng, X.; Baldwin, I.T.; Lou, Y.; Li, R. Molecular dissection of rice phytohormone signaling involved in resistance to a piercing-sucking herbivore. New Phytol. 2021, 230, 1639–1652. [Google Scholar] [CrossRef]
- Dhakarey, R.; Raorane, M.L.; Treumann, A.; Peethambaran, P.K.; Schendel, R.R.; Sahi, V.P.; Hause, B.; Bunzel, M.; Henry, A.; Kohli, A.; et al. Physiological and proteomic analysis of the rice mutant cpm2 suggests a negative regulatory role of jasmonic acid in drought tolerance. Front Plant Sci. 2017, 8, 1903. [Google Scholar] [CrossRef] [Green Version]
- Qi, J.; Li, J.; Han, X.; Li, R.; Wu, J.; Yu, H.; Hu, L.; Xiao, Y.; Lu, J.; Lou, Y. Jasmonic acid carboxyl methyltransferase regulates development and herbivory-induced defense response in rice. J. Integr. Plant Biol. 2015, 58, 564–576. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Zhang, H.; Sun, Z.; Li, J.; Hong, G.; Zhu, Q.; Zhou, X.; MacFarlane, S.; Yan, F.; Chen, J. Jasmonic acid-mediated defense suppresses brassinosteroid-mediated susceptibility to Rice black streaked dwarf virus infection in rice. New Phytol. 2016, 214, 388–399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, J.; Wu, H.; Ma, S.; Xiang, D.; Liu, R.; Xiong, L. OsJAZ1 Attenuates Drought Resistance by Regulating JA and ABA Signaling in Rice. Front. Plant Sci. 2017, 8, 2108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, L.; Tian, J.; Liu, Y.; Chen, X.; Li, S.; Persson, S.; Lu, D.; Chen, M.; Luo, Z.; Zhang, D.; et al. Ectopic expression of OsJAZ6, which interacts with OsJAZ1, alters JA signaling and spikelet development in rice. Plant J. 2021, 108, 1083–1096. [Google Scholar] [CrossRef]
- Xiao, Y.; Zhang, G.; Liu, D.; Niu, M.; Tong, H.; Chu, C. GSK2 stabilizes OFP3 to suppress brassinosteroid responses in rice. Plant J. 2020, 102, 1187–1201. [Google Scholar] [CrossRef]
- Hu, J.; Huang, J.; Xu, H.; Wang, Y.; Li, C.; Wen, P.; You, X.; Zhang, X.; Pan, G.; Li, Q.; et al. Rice stripe virus suppresses jasmonic acid-mediated resistance by hijacking brassinosteroid signaling pathway in rice. PLoS Pathog. 2020, 16, e1008801. [Google Scholar] [CrossRef]
- Yamada, S.; Kano, A.; Tamaoki, D.; Miyamoto, A.; Shishido, H.; Miyoshi, S.; Taniguchi, S.; Akimitsu, K.; Gomi, K. Involvement of OsJAZ8 in Jasmonate-Induced Resistance to Bacterial Blight in Rice. Plant Cell Physiol. 2012, 53, 2060–2072. [Google Scholar] [CrossRef] [Green Version]
- Feng, X.; Zhang, L.; Wei, X.; Zhou, Y.; Dai, Y.; Zhu, Z. OsJAZ13 Negatively Regulates Jasmonate Signaling and Activates Hypersensitive Cell Death Response in Rice. Int. J. Mol. Sci. 2020, 21, 4379. [Google Scholar] [CrossRef]
- Howe, G.A.; Lightner, J.; Browse, J.; Ryan, C.A. An octadecanoid pathway mutant (JL5) of tomato is compromised in signaling for defense against insect attack. Plant Cell 1996, 8, 2067–2077. [Google Scholar]
- Lightner, J.; Pearce, G.; Ryan, C.A.; Browse, J. Isolation of signaling mutants of tomato (Lycopersicon esculentum). Mol. Gen. Genet. MGG 1993, 241, 595–601. [Google Scholar] [CrossRef]
- Lee, G.I.; Howe, G.A. The tomato mutant spr1 is defective in systemin perception and the production of a systemic wound signal for defense gene expression. Plant J. 2003, 33, 567–576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.; Liu, G.; Xu, C.; Lee, G.I.; Bauer, P.; Ling, H.-Q.; Ganal, M.W.; Howe, G.A. The Tomato Suppressor of prosystemin-mediated responses2 Gene Encodes a Fatty Acid Desaturase Required for the Biosynthesis of Jasmonic Acid and the Production of a Systemic Wound Signal for Defense Gene Expression. Plant Cell 2003, 15, 1646–1661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, L.; Li, C.; Lee, G.I.; Howe, G.A. Distinct roles for jasmonate synthesis and action in the systemic wound response of tomato. Proc. Natl. Acad. Sci. USA 2002, 99, 6416–6421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campos, M.; Kang, J.-H.; Howe, G.A. Jasmonate-Triggered Plant Immunity. J. Chem. Ecol. 2014, 40, 657–675. [Google Scholar] [CrossRef]
- Thaler, J.S.; Owen, B.; Higgins, V.J. The Role of the Jasmonate Response in Plant Susceptibility to Diverse Pathogens with a Range of Lifestyles. Plant Physiol. 2004, 135, 530–538. [Google Scholar] [CrossRef] [Green Version]
- AbuQamar, S.; Chai, M.-F.; Luo, H.; Song, F.; Mengiste, T. Tomato Protein Kinase 1b Mediates Signaling of Plant Responses to Necrotrophic Fungi and Insect Herbivory. Plant Cell 2008, 20, 1964–1983. [Google Scholar] [CrossRef] [Green Version]
- Ortigosa, A.; Gimenez-Ibanez, S.; Leonhardt, N.; Solano, R. Design of a bacterial speck resistant tomato by CRISPR/Cas9-mediated editing of SlJAZ2. Plant Biotechnol. J. 2018, 17, 665–673. [Google Scholar] [CrossRef] [Green Version]
- Yu, X.; Chen, G.; Tang, B.; Zhang, J.; Zhou, S.; Hu, Z. The Jasmonate ZIM-domain protein gene SlJAZ2 regulates plant morphology and accelerates flower initiation in Solanum lycopersicum plants. Plant Sci. 2017, 267, 65–73. [Google Scholar] [CrossRef]
- Li, L.; Zhao, Y.; McCaig, B.C.; Wingerd, B.A.; Wang, J.; Whalon, M.E.; Pichersky, E.; Howe, G.A. The Tomato Homolog of CORONATINE-INSENSITIVE1 Is Required for the Maternal Control of Seed Maturation, Jasmonate-Signaled Defense Responses, and Glandular Trichome Development. Plant Cell 2004, 16, 126–143. [Google Scholar] [CrossRef] [Green Version]
- Widemann, E.; Miesch, L.; Lugan, R.; Holder, E.; Heinrich, C.; Aubert, Y.; Miesch, M.; Pinot, F.; Heitz, T. The Amidohydrolases IAR3 and ILL6 Contribute to Jasmonoyl-Isoleucine Hormone Turnover and Generate 12-Hydroxyjasmonic Acid Upon Wounding in Arabidopsis Leaves. J. Biol. Chem. 2013, 288, 31701–31714. [Google Scholar] [CrossRef] [Green Version]
- Wasternack, C.; Strnad, M. Jasmonate signaling in plant stress responses and development—Active and inactive compounds. New Biotechnol. 2016, 33, 604–613. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.; Herrfurth, C.; Xin, M.; Savchenko, T.; Feussner, I.; Goossens, A.; De Smet, I. Warm temperature triggers JOX and ST2A-mediated jasmonate catabolism to promote plant growth. Nat. Commun. 2021, 12, 1–8. [Google Scholar] [CrossRef]
- Fernández-Milmanda, G.L.; Crocco, C.D.; Reichelt, M.; Mazza, C.A.; Köllner, T.G.; Zhang, T.; Cargnel, M.D.; Lichy, M.Z.; Fiorucci, A.-S.; Fankhauser, C.; et al. A light-dependent molecular link between competition cues and defence responses in plants. Nat. Plants 2020, 6, 223–230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, M.; Yu, G.; Cao, C.; Liu, P. Metabolism, signaling, and transport of jasmonates. Plant Commun. 2021, 2, 100231. [Google Scholar] [CrossRef] [PubMed]
- Glauser, G.; Dubugnon, L.; Mousavi, S.A.; Rudaz, S.; Wolfender, J.-L.; Farmer, E.E. Velocity Estimates for Signal Propagation Leading to Systemic Jasmonic Acid Accumulation in Wounded Arabidopsis. J. Biol. Chem. 2009, 284, 34506–34513. [Google Scholar] [CrossRef] [Green Version]
- Yan, J.; Li, H.; Li, S.; Yao, R.; Deng, H.; Xie, Q.; Xie, D. The Arabidopsis F-Box Protein Coronatine Insensitive1 Is Stabilized by SCFCOI1 and Degraded via the 26S Proteasome Pathway. Plant Cell 2013, 25, 486–498. [Google Scholar] [CrossRef] [Green Version]
- Thireault, C.; Shyu, C.; Yoshida, Y.; St Aubin, B.; Campos, M.L.; Howe, G.A. Repression of jasmonate signaling by a non-TIFY JAZ protein in Arabidopsis. Plant J. 2015, 82, 669–679. [Google Scholar] [CrossRef]
- Nakata, M.; Mitsuda, N.; Herde, M.; Koo, A.J.; Moreno, J.E.; Suzuki, K.; Howe, G.A.; Ohme-Takagi, M. A bHLH-type transcription factor, aba-inducible bhlh-type transcription factor/ja-associated myc2-like1, acts as a repressor to negatively regulate jasmonate signaling in arabidopsis. Plant Cell. 2013, 25, 1641–1656. [Google Scholar] [CrossRef] [Green Version]
- Qi, T.; Wang, J.; Huang, H.; Liu, B.; Gao, H.; Liu, Y.; Song, S.; Xie, D. Regulation of Jasmonate-Induced Leaf Senescence by Antagonism between bHLH Subgroup IIIe and IIId Factors in Arabidopsis. Plant Cell 2015, 27, 1634–1649. [Google Scholar] [CrossRef] [Green Version]
- Zhang, G.; Liu, W.; Gu, Z.; Wu, S.; Zhou, W.; Lin, J.; Xu, L. Roles of the wound hormone jasmonate in plant regeneration. J. Exp. Bot. 2021. [Google Scholar] [CrossRef]
- Zhang, G.; Zhao, F.; Chen, L.; Pan, Y.; Sun, L.; Bao, N.; Zhang, T.; Cui, C.-X.; Qiu, Z.; Zhang, Y.; et al. Jasmonate-mediated wound signalling promotes plant regeneration. Nat. Plants 2019, 5, 491–497. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Lozano-Torres, J.L.; Blilou, I.; Zhang, X.; Zhai, Q.; Smant, G.; Li, C.; Scheres, B. A Jasmonate Signaling Network Activates Root Stem Cells and Promotes Regeneration. Cell 2019, 177, 942–956.e14. [Google Scholar] [CrossRef] [PubMed]
- Weber, H. Fatty acid-derived signals in plants. Trends Plant Sci. 2002, 7, 217–224. [Google Scholar] [CrossRef]
- Raza, A.; Charagh, S.; Zahid, Z.; Mubarik, M.S.; Javed, R.; Siddiqui, M.H.; Hasanuzzaman, M. Jasmonic acid: A key frontier in conferring abiotic stress tolerance in plants. Plant Cell Rep. 2021, 40, 1513–1541. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Christensen, S.; Isakeit, T.; Engelberth, J.; Meeley, R.; Hayward, A.; Emery, R.N.; Kolomiets, M.V. Disruption of OPR7 and OPR8 Reveals the Versatile Functions of Jasmonic Acid in Maize Development and Defense. Plant Cell 2012, 24, 1420–1436. [Google Scholar] [CrossRef] [Green Version]
- Reeves, P.H.; Ellis, C.M.; Ploense, S.E.; Wu, M.-F.; Yadav, V.; Tholl, D.; Chételat, A.; Haupt, I.; Kennerley, B.J.; Hodgens, C.; et al. A Regulatory Network for Coordinated Flower Maturation. PLoS Genet. 2012, 8, e1002506. [Google Scholar] [CrossRef] [Green Version]
- Song, S.; Qi, T.; Huang, H.; Ren, Q.; Wu, D.; Chang, C.; Peng, W.; Liu, Y.; Peng, J.; Xie, D. The Jasmonate-ZIM Domain Proteins Interact with the R2R3-MYB Transcription Factors MYB21 and MYB24 to Affect Jasmonate-Regulated Stamen Development in Arabidopsis. Plant Cell 2011, 23, 1000–1013. [Google Scholar] [CrossRef] [Green Version]
- Zhai, Q.; Zhang, X.; Wu, F.; Feng, H.; Deng, L.; Xu, L.; Zhang, M.; Wang, Q.; Li, C. Transcriptional Mechanism of Jasmonate Receptor COI1-Mediated Delay of Flowering Time in Arabidopsis. Plant Cell 2015, 27, 2814–2828. [Google Scholar] [CrossRef] [Green Version]
- Xu, Q.; Truong, T.T.; Barrero, J.M.; Jacobsen, J.V.; Hocart, C.H.; Gubler, F. A role for jasmonates in the release of dormancy by cold stratification in wheat. J. Exp. Bot. 2016, 67, 3497–3508. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Hou, Y.; Qiu, J.; Wang, H.; Wang, S.; Tang, L.; Tong, X.; Zhang, J. Abscisic acid promotes jasmonic acid biosynthesis via a ‘SAPK10-bZIP72-AOC’ pathway to synergistically inhibit seed germination in rice (Oryza sativa). New Phytol. 2020, 228, 1336–1353. [Google Scholar] [CrossRef]
- Zhuo, M.; Sakuraba, Y.; Yanagisawa, S. A Jasmonate-activated MYC2-Dof2.1-MYC2 transcriptional loop promotes leaf senescence in Arabidopsis. Plant Cell 2020, 32, 242–262. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, Y.; Wei, H.; Li, N.; Tian, W.; Chong, K.; Wang, L. Circadian Evening Complex Represses Jasmonate-Induced Leaf Senescence in Arabidopsis. Mol. Plant 2018, 11, 326–337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Z.; Wu, Y.; Yang, Y.; Du, M.; Zhang, X.; Guo, Y.; Li, C.; Zhou, J.-M. An Arabidopsis Plasma Membrane Proton ATPase Modulates JA Signaling and Is Exploited by the Pseudomonas syringae Effector Protein AvrB for Stomatal Invasion. Plant Cell 2015, 27, 2032–2041. [Google Scholar] [CrossRef] [Green Version]
- Hua, B.; Chang, J.; Xu, Z.; Han, X.; Xu, M.; Yang, M.; Yang, C.; Ye, Z.; Wu, S. HOMEODOMAIN PROTEIN8 mediates jasmonate-triggered trichome elongation in tomato. New Phytol. 2021, 230, 1063–1077. [Google Scholar] [CrossRef] [PubMed]
- Yan, T.; Chen, M.; Shen, Q.; Li, L.; Fu, X.; Pan, Q.; Tang, Y.; Shi, P.; Lv, Z.; Jiang, W.; et al. HOMEODOMAIN PROTEIN 1 is required for jasmonate-mediated glandular trichome initiation in Artemisia annua. New Phytol. 2016, 213, 1145–1155. [Google Scholar] [CrossRef] [PubMed]
- Qi, T.; Huang, H.; Wu, D.; Yan, J.; Qi, Y.; Song, S.; Xie, D. Arabidopsis DELLA and JAZ proteins bind the WD-repeat/bHLH/MYB complex to modulate gibberellin and jasmonate signaling synergy. Plant Cell 2014, 26, 1118–1133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Q.; Sun, J.; Zhai, Q.; Zhou, W.; Qi, L.; Xu, L.; Wang, B.; Chen, R.; Jiang, H.; Qi, J.; et al. The basic helix-loop-helix transcription factor MYC2 directly represses PLETHORA expression during jasmonate-mediated modulation of the root stem cell niche in Arabidopsis. Plant Cell 2011, 23, 3335–3352. [Google Scholar] [CrossRef] [Green Version]
- Hentrich, M.; Böttcher, C.; Düchting, P.; Cheng, Y.; Zhao, Y.; Berkowitz, O.; Masle, J.; Medina, J.; Pollmann, S. The jasmonic acid signaling pathway is linked to auxin homeostasis through the modulation ofYUCCA8andYUCCA9gene expression. Plant J. 2013, 74, 626–637. [Google Scholar] [CrossRef] [Green Version]
- Taki, N.; Sasaki-Sekimoto, Y.; Obayashi, T.; Kikuta, A.; Kobayashi, K.; Ainai, T.; Yagi, K.; Sakurai, N.; Suzuki, H.; Masuda, T.; et al. 12-Oxo-Phytodienoic Acid Triggers Expression of a Distinct Set of Genes and Plays a Role in Wound-Induced Gene Expression in Arabidopsis. Plant Physiol. 2005, 139, 1268–1283. [Google Scholar] [CrossRef] [Green Version]
- Qi, L.; Yan, J.; Li, Y.; Jiang, H.; Sun, J.; Chen, Q.; Li, H.; Chu, J.; Yan, C.; Sun, X.; et al. Arabidopsis thaliana plants differentially modulate auxin biosynthesis and transport during defense responses to the necrotrophic pathogen Alternaria brassicicola. New Phytol. 2012, 195, 872–882. [Google Scholar] [CrossRef]
- Yan, C.; Xie, D. Jasmonate in plant defence: Sentinel or double agent? Plant Biotechnol. J. 2015, 13, 1233–1240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomma, B.P.H.J.; Eggermont, K.; Penninckx, I.A.M.A.; Mauch-Mani, B.; Vogelsang, R.; Cammue, B.P.A.; Broekaert, W.F. Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc. Natl. Acad. Sci. USA 1998, 95, 15107–15111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Y.; Jiang, Y.; Han, X.; Wang, H.; Pan, J.; Yu, D. Jasmonate regulates leaf senescence and tolerance to cold stress: Crosstalk with other phytohormones. J. Exp. Bot. 2017, 68, 1361–1369. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhang, L.; Xiang, S.; Chen, Y.; Zhang, H.; Yu, D. The transcription factor WRKY75 positively regulates jasmonate-mediated plant defense to necrotrophic fungal pathogens. J. Exp. Bot. 2020, 72, 1473–1489. [Google Scholar] [CrossRef] [PubMed]
- Lv, T.; Li, X.; Fan, T.; Luo, H.; Xie, C.; Zhou, Y.; Tian, C.-E. The Calmodulin-Binding Protein IQM1 Interacts with CATALASE2 to Affect Pathogen Defense. Plant Physiol. 2019, 181, 1314–1327. [Google Scholar] [CrossRef] [PubMed]
- Ren, H.; Bai, M.; Sun, J.; Liu, J.; Ren, M.; Dong, Y.; Wang, N.; Ning, G.; Wang, C. RcMYB84 and RcMYB123 mediate jasmonate-induced defense responses against Botrytis cinerea in rose (Rosa chinensis). Plant J. 2020, 103, 1839–1849. [Google Scholar] [CrossRef] [PubMed]
- Dombrecht, B.; Xue, G.P.; Sprague, S.J.; Kirkegaard, J.A.; Ross, J.J.; Reid, J.B.; Fitt, G.P.; Sewelam, N.; Schenk, P.M.; Manners, J.M.; et al. MYC2 Differentially Modulates Diverse Jasmonate-Dependent Functions in Arabidopsis. Plant Cell 2007, 19, 2225–2245. [Google Scholar] [CrossRef] [Green Version]
- Lorenzo, O.; Chico, J.M.; Saénchez-Serrano, J.J.; Solano, R. JASMONATE-INSENSITIVE1 Encodes a MYC Transcription Factor Essential to Discriminate between Different Jasmonate-Regulated Defense Responses in Arabidopsis. Plant Cell 2004, 16, 1938–1950. [Google Scholar] [CrossRef] [Green Version]
- Du, M.; Zhao, J.; Tzeng, D.T.; Liu, Y.; Deng, L.; Yang, T.; Zhai, Q.; Wu, F.; Huang, Z.; Zhou, M.; et al. MYC2 Orchestrates a Hierarchical Transcriptional Cascade That Regulates Jasmonate-Mediated Plant Immunity in Tomato. Plant Cell 2017, 29, 1883–1906. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Z.; An, F.; Feng, Y.; Li, P.; Xue, L.; Mu, A.; Jiang, Z.; Kim, J.M.; To, T.K.; Li, W.; et al. Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 12539–12544. [Google Scholar] [CrossRef] [Green Version]
- Pre, M.; Atallah, M.; Champion, A.; De Vos, M.; Pieterse, C.M.; Memelink, J. The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol. 2008, 147, 1347–1357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, D.; Xu, H.; Huang, J.; Kong, Y.; AbuQamar, S.; Yu, D.; Liu, S.; Zhou, G.; Chai, G. The Arabidopsis CCCH protein C3H14 contributes to basal defense against Botrytis cinerea mainly through the WRKY33-dependent pathway. Plant Cell Environ. 2020, 43, 1792–1806. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Prado, J.S.; Latrasse, D.; Rodriguez-Granados, N.Y.; Huang, Y.; Manza-Mianza, D.; Brik-Chaouche, R.; Jaouannet, M.; Citerne, S.; Bendahmane, A.; Hirt, H.; et al. The Polycomb protein LHP 1 regulates Arabidopsis thaliana stress responses through the repression of the MYC 2-dependent branch of immunity. Plant J. 2019, 100, 1118–1131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, M.; Zhang, J.; Yao, T.; Bryan, A.C.; Pu, Y.; Labbé, J.; Pelletier, D.A.; Engle, N.; Morrell-Falvey, J.L.; Schmutz, J.; et al. Arabidopsis C-terminal binding protein ANGUSTIFOLIA modulates transcriptional co-regulation of MYB46 and WRKY33. New Phytol. 2020, 228, 1627–1639. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, Q.; Zhai, H.; Gao, S.; Yang, L.; Wang, Z.; Xu, Y.; Huo, J.; Ren, Z.; Zhao, N.; et al. IbBBX24 Promotes the Jasmonic Acid Pathway and Enhances Fusarium Wilt Resistance in Sweet Potato. Plant Cell 2020, 32, 1102–1123. [Google Scholar] [CrossRef]
- Hou, Y.; Wang, Y.; Tang, L.; Tong, X.; Wang, L.; Liu, L.; Huang, S.; Zhang, J. SAPK10-Mediated Phosphorylation on WRKY72 Releases Its Suppression on Jasmonic Acid Biosynthesis and Bacterial Blight Resistance. iScience 2019, 16, 499–510. [Google Scholar] [CrossRef] [Green Version]
- Boter, M.; Ruíz-Rivero, O.; Abdeen, A.; Prat, S. Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Genes Dev. 2004, 18, 1577–1591. [Google Scholar] [CrossRef] [Green Version]
- Falk, K.L.; Kastner, J.; Bodenhausen, N.; Schramm, K.; Paetz, C.; Vassao, D.G.; Reichelt, M.; von Knorre, D.; Bergelson, J.; Erb, M.; et al. The role of glucosinolates and the jasmonic acid pathway in resistance of Arabidopsis thaliana against molluscan herbivores. Mol. Ecol. 2014, 23, 1188–1203. [Google Scholar] [CrossRef] [Green Version]
- Mikkelsen, M.D.; Petersen, B.L.; Glawischnig, E.; Jensen, A.B.; Andreasson, E.; Halkier, B. Modulation of CYP79 Genes and Glucosinolate Profiles in Arabidopsis by Defense Signaling Pathways. Plant Physiol. 2003, 131, 298–308. [Google Scholar] [CrossRef] [Green Version]
- Reymond, P.; Bodenhausen, N.; Van Poecke, R.M.P.; Krishnamurthy, V.; Dicke, M.; Farmer, E.E. A conserved transcript pattern in response to a specialist and a generalist herbivore. Plant Cell. 2004, 16, 3132–3147. [Google Scholar] [CrossRef] [Green Version]
- Thaler, J.S.; Stout, M.J.; Karban, R.; Duffey, S.S. Jasmonate-mediated induced plant resistance affects a community of herbivores. Ecol. Èntomol. 2001, 26, 312–324. [Google Scholar] [CrossRef]
- Peiffer, M.; Tooker, J.; Luthe, D.S.; Felton, G.W. Plants on early alert: Glandular trichomes as sensors for insect herbivores. New Phytol. 2009, 184, 644–656. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.-H.; McRoberts, J.; Shi, F.; Moreno, J.; Jones, A.D.; Howe, G.A. The Flavonoid Biosynthetic Enzyme Chalcone Isomerase Modulates Terpenoid Production in Glandular Trichomes of Tomato. Plant Physiol. 2014, 164, 1161–1174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pérez-Alonso, M.-M.; Sánchez-Parra, B.; Ortiz-García, P.; Santamaría, M.E.; Díaz, I.; Pollmann, S. Jasmonic Acid-Dependent MYC Transcription Factors Bind to a Tandem G-Box Motif in the YUCCA8 and YUCCA9 Promoters to Regulate Biotic Stress Responses. Int. J. Mol. Sci. 2021, 22, 9768. [Google Scholar] [CrossRef] [PubMed]
- Machado, R.A.R.; Robert, C.; Arce, C.C.M.; Ferrieri, A.P.; Xu, S.; Jimenez-Aleman, G.; Baldwin, I.T.; Erb, M. Auxin Is Rapidly Induced by Herbivore Attack and Regulates a Subset of Systemic, Jasmonate-Dependent Defenses. Plant Physiol. 2016, 172, 521–532. [Google Scholar] [CrossRef] [Green Version]
- Zhang, P.-J.; Wei, J.-N.; Zhao, C.; Zhang, Y.-F.; Li, C.-Y.; Liu, S.-S.; Dicke, M.; Yu, X.-P.; Turlings, T.C.J. Airborne host–plant manipulation by whiteflies via an inducible blend of plant volatiles. Proc. Natl. Acad. Sci. USA 2019, 116, 7387–7396. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.-Y.; Liu, Y.-Q.; Song, W.-M.; Chen, D.-Y.; Chen, F.-Y.; Chen, X.-Y.; Chen, Z.-W.; Ge, S.-X.; Wang, C.-Z.; Zhan, S.; et al. An effector from cotton bollworm oral secretion impairs host plant defense signaling. Proc. Natl. Acad. Sci. USA 2019, 116, 14331–14338. [Google Scholar] [CrossRef] [Green Version]
- Pan, G.; Liu, Y.; Ji, L.; Zhang, X.; He, J.; Huang, J.; Qiu, Z.; Liu, D.; Sun, Z.; Xu, T.; et al. Brassinosteroids mediate susceptibility to brown planthopper by integrating with the salicylic acid and jasmonic acid pathways in rice. J. Exp. Bot. 2018, 69, 4433–4442. [Google Scholar] [CrossRef]
- Xie, K.; Li, L.; Zhang, H.; Wang, R.; Tan, X.; He, Y.; Hong, G.; Li, J.; Ming, F.; Yao, X.; et al. Abscisic acid negatively modulates plant defence against rice black-streaked dwarf virus infection by suppressing the jasmonate pathway and regulating reactive oxygen species levels in rice. Plant Cell Environ. 2018, 41, 2504–2514. [Google Scholar] [CrossRef]
- Li, L.; Zhang, H.; Chen, C.; Huang, H.; Tan, X.; Wei, Z.; Li, J.; Yan, F.; Zhang, C.; Chen, J.; et al. A class of independently evolved transcriptional repressors in plant RNA viruses facilitates viral infection and vector feeding. Proc. Natl. Acad. Sci. USA 2021, 118. [Google Scholar] [CrossRef]
- He, L.; Chen, X.; Yang, J.; Zhang, T.; Li, J.; Zhang, S.; Zhong, K.; Zhang, H.; Chen, J. Rice black-streaked dwarf virus -encoded P5-1 regulates the ubiquitination activity of SCF E3 ligases and inhibits jasmonate signaling to benefit its infection in rice. New Phytol. 2019, 225, 896–912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, K.; Huang, H.; Zheng, H.; Ji, M.; Yuan, Q.; Cui, W.; Zhang, H.; Peng, J.; Lu, Y.; Rao, S.; et al. Rice stripe virus coat protein induces the accumulation of jasmonic acid, activating plant defence against the virus while also attracting its vector to feed. Mol. Plant Pathol. 2020, 21, 1647–1653. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Abellan, J.O.; Fernandez-Garcia, N.; Lopez-Berenguer, C.; Egea, I.; Flores, F.B.; Angosto, T.; Capel, J.; Lozano, R.; Pineda, B.; Moreno, V.; et al. The tomato res mutant which accumulates JA in roots in non-stressed conditions restores cell structure alterations under salinity. Physiol Plantarum. 2015, 155, 296–314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seo, J.-S.; Joo, J.; Kim, M.-J.; Kim, Y.-K.; Nahm, B.H.; Song, S.I.; Cheong, J.-J.; Lee, J.S.; Kim, J.-K.; Choi, Y.D. OsbHLH148, a basic helix-loop-helix protein, interacts with OsJAZ proteins in a jasmonate signaling pathway leading to drought tolerance in rice. Plant J. 2011, 65, 907–921. [Google Scholar] [CrossRef]
- Demkura, P.V.; Abdala, G.; Baldwin, I.; Ballare, C.L. Jasmonate-Dependent and -Independent Pathways Mediate Specific Effects of Solar Ultraviolet B Radiation on Leaf Phenolics and Antiherbivore Defense. Plant Physiol. 2009, 152, 1084–1095. [Google Scholar] [CrossRef] [Green Version]
- Rao, M.V.; Lee, H.; Creelman, R.A.; Mullet, J.E.; Davis, K.R. Jasmonic acid signaling modulates ozone-induced hypersensitive cell death. Plant Cell 2000, 12, 1633–1646. [Google Scholar] [CrossRef] [Green Version]
- Sivasankar, S.; Sheldrick, B.; Rothstein, S.J. Expression of Allene Oxide Synthase Determines Defense Gene Activation in Tomato. Plant Physiol. 2000, 122, 1335–1342. [Google Scholar] [CrossRef] [Green Version]
- Howe, G.A.; Schilmiller, A. Oxylipin metabolism in response to stress. Curr. Opin. Plant Biol. 2002, 5, 230–236. [Google Scholar] [CrossRef]
- Wang, X.; Li, Q.; Xie, J.; Huang, M.; Cai, J.; Zhou, Q.; Dai, T.; Jiang, D. Abscisic acid and jasmonic acid are involved in drought priming-induced tolerance to drought in wheat. Crop J. 2020, 9, 120–132. [Google Scholar] [CrossRef]
- An, J.P.; Wang, X.F.; Zhang, X.W.; You, C.X.; Hao, Y.J. Apple B-box protein BBX37 regulates jasmonic acid mediated cold tolerance through the JAZ-BBX37-ICE1-CBF pathway and undergoes MIEL1-mediated ubiquitination and degradation. New Phytol. 2021, 229, 2707–2729. [Google Scholar] [CrossRef]
- Hou, X.; Lee, L.Y.C.; Xia, K.; Yan, Y.; Yu, H. DELLAs Modulate Jasmonate Signaling via Competitive Binding to JAZs. Dev. Cell 2010, 19, 884–894. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoffmann, M.; Hentrich, M.; Pollmann, S. Auxin-Oxylipin Crosstalk: Relationship of AntagonistsF. J. Integr. Plant Biol. 2011, 53, 429–445. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Alonso, M.-M. How Auxin May Contribute to the Regulation of Plant Defense Responses against Herbivory. Austin J. Plant Biol. 2018, 4, 1019. [Google Scholar]
- Zhang, T.; Poudel, A.N.; Jewell, J.B.; Kitaoka, N.; Staswick, P.; Matsuura, H.; Koo, A.J. Hormone crosstalk in wound stress response: Wound-inducible amidohydrolases can simultaneously regulate jasmonate and auxin homeostasis in Arabidopsis thaliana. J. Exp. Bot. 2015, 67, 2107–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Risseeuw, E.P.; Daskalchuk, T.E.; Banks, T.W.; Liu, E.; Cotelesage, J.; Hellmann, H.; Estelle, M.; Somers, D.E.; Crosby, W.L. Protein interaction analysis of SCF ubiquitin E3 ligase subunits from Arabidopsis. Plant J. 2003, 34, 753–767. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, C.; Pan, J.; Peng, W.; Genschik, P.; Hobbie, L.; Hellmann, H.; Estelle, M.; Gao, B.; Peng, J.; Sun, C.; et al. Point mutations in Arabidopsis Cullin1 reveal its essential role in jasmonate response. Plant J. 2005, 42, 514–524. [Google Scholar] [CrossRef]
- Liu, Y.; Wei, H.; Ma, M.; Li, Q.; Kong, D.; Sun, J.; Ma, X.; Wang, B.; Chen, C.; Xie, Y.; et al. Arabidopsis FHY3 and FAR1 Regulate the Balance between Growth and Defense Responses under Shade Conditions. Plant Cell 2019, 31, 2089–2106. [Google Scholar] [CrossRef] [Green Version]
- Savchenko, T.V.; Rolletschek, H.; Dehesh, K. Jasmonates-Mediated Rewiring of Central Metabolism Regulates Adaptive Responses. Plant Cell Physiol. 2019, 60, 2613–2620. [Google Scholar] [CrossRef]
- Zhu, X.; Zhu, J.-K. Double Repression in Jasmonate-Mediated Plant Defense. Mol. Cell 2013, 50, 459–460. [Google Scholar] [CrossRef] [Green Version]
- Ueda, M.; Kaji, T.; Kozaki, W. Recent Advances in Plant Chemical Biology of Jasmonates. Int. J. Mol. Sci. 2020, 21, 1124. [Google Scholar] [CrossRef] [Green Version]
- Bastías, D.A.; Gianoli, E.; Gundel, P.E. Fungal endophytes can eliminate the plant growth-defence trade-off. New Phytol. 2021, 230, 2105–2113. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Guo, W.; Yang, D.; Hou, Z.; Liang, Z. Transcriptional Profiles of SmWRKY Family Genes and Their Putative Roles in the Biosynthesis of Tanshinone and Phenolic Acids in Salvia miltiorrhiza. Int. J. Mol. Sci. 2018, 19, 1593. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Species | Name | Description | Alteration in JA Responses | Ref. |
|---|---|---|---|---|
| At | fad3-2fad7-2fad8 | Cross between fad3-2, fad7-2 and fad8 | No JA produced; Male sterile; Hypersensitive to Alternaria brassiccola | [46,47] |
| opr3 | T-DNA insertion mutant | No JA produced; Defective anther and pollen development, male sterile; Enlarged petals; Resistant to A. brassicicola infection | [47,48] | |
| jar1-1 | / | JA-insensitive phenotype; Defect in JA-Ile synthesis; Increased susceptibility to P. irregular | [49,50] | |
| jassy | T-DNA insertion mutant | Defective in OPDA transportation; Reduced cold tolerance; Increased susceptibility to B. cinerea | [22] | |
| dad1 | T-DNA insertion mutant | Defective in JA biosynthesis; Male sterile | [51] | |
| dde1 | T-DNA insertion mutant | Defective in JA biosynthesis; Male sterile | [52] | |
| aos | T-DNA knockout mutant | No JA produced; Male sterile | [53] | |
| acx-1acx-5 | Cross between acx-1 and acx-5 | Poor pollen viability; Increased susceptibility to Trichoplusia ni larvae and Frankliniella occidentalis; Remain resistant to the A. brassicicola | [54] | |
| lox3lox4 | T-DNA insertion mutant | Defective in JA biosynthesis; Male sterile | [55] | |
| aim1 | T-DNA insertion mutant | Defective in wound-induced formation of JA; Defective in floral development | [56,57] | |
| pex6 | / | Defective in wound-induced formation of JA; Increased OPDA level | [56] | |
| kat2kat5 | Knockout mutant | Growth defect; Male sterile; Phenotype similar to aim1 | [58] | |
| cyp94c1−1 | T-DNA insertion mutant | Decreased 12COOH-JA-Ile accumulation | [59] | |
| cyp94b3cyp94c1 | / | Increased JA-Ile accumulation; JA-insensitive phenotype; Sensitive to exogenous JA | [59] | |
| coi1-1 | EMS mutagenized, W467 * nonsense mutation | JA insensitivity; Male sterile; Increased susceptibility to fungal pathogens and Erwinea carotovora; Dark-induced senescence | [60,61,62] | |
| coi1-2 | EMS mutagenized, L245F missense mutation | Reduced JA insensitivity; Partial fertility | [27,63,64] | |
| coi1-8 | EMS mutagenized, G543L missense mutation | Reduced JA insensitivity; Insensitivity to JA-inhibitory root elongation; Partial fertility | [27] | |
| coi1-16 | / | Fertility in a temperature-sensitive manner | [28,65] | |
| coi1-20 | EMS mutagenized | Male sterility; Resistant to P. syringae | [60] | |
| coi1-21 | T-DNA in F-box | Male sterility | [65,66] | |
| coi1-30 | / | Enhanced resistance to P. syringae; Longer hypocotyls and petioles under low-intensity light conditions; Early flowering | [67,68] | |
| coi1-37 | T-DNA insertion line, a 1537 bp deletion in the promoter, and the first exon | Male sterility; Leaf epinasty; Dark green leaves; Strong apical dominance; Enhanced meristem longevity | [66] | |
| jaz1-1 | Loss-of-function | Normal JA responses | [69] | |
| jaz2-1 | T-DNA insertion at the fourth intron | JA insensitivity | [70] | |
| jaz2-3 | Transposon insertion leads to JAZ2 knockout | Partially impaired in stomatal closing; More susceptible to P. syringae | [67] | |
| jaz2∆jas | T-DNA insertion in the third exon; Lack the Jas domain | Resistant to P. syringae and necrotrophs | [67] | |
| jaz5-1 | / | Normal JA responses | [69] | |
| jaz6-1 | / | Short filament; Delayed anther dehiscence; Unviable pollen grains | [71] | |
| jaz7 | T-DNA insertion at the promoter, overexpression | JA sensitivity; Significantly short roots; Reduced weight; Enhanced defense | [36,70] | |
| jaz7-1 | T-DNA insertion at 384 bp from the 5’-UTR, Loss-of-function | Week regulation of cambium initiation; Dark-induced leaf senescence hypersensitive | [43,72,73] | |
| jaz9-1 | Loss-of-function | Partial GA insensitivity; Normal JA phenotype | [68] | |
| jaz9-3 | Loss-of-function | Partial GA insensitivity | [68] | |
| jaz10-1 | Open reading frame was disrupted, Loss-of-function | JA-hypersensitive; Enhanced susceptibility to P. syringae infection; Enhanced cambium initiation | [69,72,74] | |
| jaz10-2 | A weak allele, Loss-of-function unclear | Weak regulation of cambium initiation | [72] | |
| jazQ | T-DNA insertion mutations in 5 JAZ genes (JAZ1/3/4/9/10) | JA-hypersensitive root growth; Enhanced susceptibility to P. syringae; Heightened resistance to Trichoplusia ni | [13] | |
| jazD | T-DNA insertion mutations in 10 JAZ genes (JAZ1-7, -9, -10, -13) | Resistant to insect herbivores and fungal pathogens; Slow vegetative growth; Poor fertility | [43] | |
| myc2-3 | / | Reduced formation of interfascicular cambium | [72] | |
| myc3-1 | MYC3 knockout | Enhanced resistance to P. syringae and S. littoralis larvae | [32] | |
| myc4-1 | MYC4 knockout | Enhanced resistance to P. syringae and S. littoralis larvae | [32] | |
| myc5 | / | Normal development in flower and stamen | [75] | |
| myc1/3/4 | / | Hypersensitive to S. littoralis and spider mite; reduced JA-mediated root inhibition | [76,77] | |
| myc2/3/4/5 | / | Short filament; Delayed anther dehiscence; Unviable pollen grains | [75] | |
| OE JAZ1∆3A | Lack residues 202-228 | JA-insensitive phenotypes; Male sterility | [29] | |
| OE JAZ8 | / | JA-insensitive root growth; Vulnerability to herbivore attack | [36] | |
| OE JAZ7 | / | Enhanced drought tolerance | [78] | |
| OE JAZ9 | / | Longer hypocotyls and petioles under low-in-tensity light condition; Early flowering | [29] | |
| OE JAZ10.4 | Lack the Jas domain | JA-insensitive; Resistant to JA-induced degradation | [79] | |
| Os | aoc-2 | T-DNA insertion mutant | Decreased JA accumulation; Susceptible to BPH attack | [80] |
| cpm2 | An 11 bp deletion within the first exon of AOC | Enhanced adaptability to drought; Male sterile; Strong root systems | [81] | |
| OE JMT | / | Increased MeJA accumulation; Reduced height and yield; Increased resistance to BPH nymphs | [82] | |
| coi1-13 | RNAi line | JA-insensitive; Increased plant height; More susceptible to virus infection | [68,83] | |
| coi1-18 | RNAi line | JA-insensitive; Increased plant height | [68] | |
| jaz1 | T-DNA insertion mutant | Increased drought tolerance | [84] | |
| jaz6 | / | Normal JA responses | [85] | |
| myc2 | Loss-of-function | Reduced JA-mediate RSV defense response | [86,87] | |
| OE JAZ1 | / | More sensitive to drought stress | [84] | |
| OE JAZ6 | / | JA-insensitive phenotype; Abnormal spikelet development; Weak root inhibition | [85] | |
| OE JAZ8∆C | Lack the Jas domain | JA-insensitive phenotype; Negatively regulated the JA-induced resistance to Xoo | [88] | |
| OEJAZ13a | Lack an intron | JA-insensitive root growth; Developed lesion mimics in the sheath and tillers | [89] | |
| Sl | def1 | / | Decreased JA accumulation; Increased susceptibility to Manduca sexta | [90,91] |
| spr1 | / | Decreased JA accumulation; Defective in wound signal-mediated PI expression | [92] | |
| spr2 | / | Defective in JA biosynthesis; Increased susceptibility to tobacco hornworm larvae | [93,94] | |
| jai1 | A 525 bp downstream intron-1 sequence deletion | Reduced pollen viability; Abnormal development of glandular trichomes; Increased susceptibility to two-spotted spider mites, B. cinerea, Pythium, and Fusarium | [95,96,97] | |
| JAZ2Δjas | Lack the Jas domain | Inhibited stomatal reopening by COR and enhanced resistance to P. syringae; Remain resistant to the B. cinerea | [98] | |
| OE JAZ2 | / | Quicker leaf initiation; Reduced plant height; Decreased trichomes; Earlier lateral bud emergence; Advanced flowering transition | [99] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, C.; Xu, M.; Cai, X.; Han, Z.; Si, J.; Chen, D. Jasmonate Signaling Pathway Modulates Plant Defense, Growth, and Their Trade-Offs. Int. J. Mol. Sci. 2022, 23, 3945. https://doi.org/10.3390/ijms23073945
Li C, Xu M, Cai X, Han Z, Si J, Chen D. Jasmonate Signaling Pathway Modulates Plant Defense, Growth, and Their Trade-Offs. International Journal of Molecular Sciences. 2022; 23(7):3945. https://doi.org/10.3390/ijms23073945
Chicago/Turabian StyleLi, Cong, Mengxi Xu, Xiang Cai, Zhigang Han, Jinping Si, and Donghong Chen. 2022. "Jasmonate Signaling Pathway Modulates Plant Defense, Growth, and Their Trade-Offs" International Journal of Molecular Sciences 23, no. 7: 3945. https://doi.org/10.3390/ijms23073945
APA StyleLi, C., Xu, M., Cai, X., Han, Z., Si, J., & Chen, D. (2022). Jasmonate Signaling Pathway Modulates Plant Defense, Growth, and Their Trade-Offs. International Journal of Molecular Sciences, 23(7), 3945. https://doi.org/10.3390/ijms23073945






