Profiling Blood Serum Extracellular Vesicles in Plaque Psoriasis and Psoriatic Arthritis Patients Reveals Potential Disease Biomarkers
Abstract
:1. Introduction
2. Results
2.1. Purification of Extracellular Vesicles
2.2. Size and ZP Profile of Purified EV Samples
2.3. EV Array Phenotyping
2.4. Small RNA Profile of EVs
2.5. Differentially Enriched miRNAs
3. Discussion
4. Materials and Methods
4.1. Patient Groups and Blood Serum Samples
4.2. Purification of Extracellular Vesicles
4.3. Nanoparticle Tracking Analysis
4.4. Western Blot Analysis
4.5. Transmission Electron Microscopy
4.6. Multiplexed Phenotyping of EVs by EV Array
4.7. Small RNA Sequencing
4.8. Statistical Analysis
4.8.1. NTA Data
4.8.2. EV Array Data
4.8.3. Small RNA Sequencing Data
4.8.4. Data Visualization
4.9. Experimental Design
4.9.1. Investigating the Physical Characteristics of Blood Serum-Derived EVs
4.9.2. EV miRNA and Surface Proteome Profiling of Patient and Control Groups
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ocampo, D.V.; Gladman, D. Psoriatic arthritis. F1000Res 2019, 8, 1665. [Google Scholar] [CrossRef]
- Gudu, T.; Gossec, L. Quality of life in psoriatic arthritis. Expert Rev. Clin. Immunol. 2018, 14, 405–417. [Google Scholar] [CrossRef]
- Coates, L.C.; Helliwell, P.S. Psoriatic arthritis: State of the art review. Clin. Med. 2017, 17, 65–70. [Google Scholar] [CrossRef]
- Haroon, M.; Gallagher, P.; FitzGerald, O. Diagnostic delay of more than 6 months contributes to poor radiographic and functional outcome in psoriatic arthritis. Ann. Rheum. Dis. 2015, 74, 1045–1050. [Google Scholar] [CrossRef]
- Villani, A.P.; Rouzaud, M.; Sevrain, M.; Barnetche, T.; Paul, C.; Richard, M.A.; Beylot-Barry, M.; Misery, L.; Joly, P.; Le Maitre, M.; et al. Prevalence of undiagnosed psoriatic arthritis among psoriasis patients: Systematic review and meta-analysis. J. Am. Acad. Dermatol. 2015, 73, 242–248. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, C.E.; Barker, J.N. Pathogenesis and clinical features of psoriasis. Lancet 2007, 370, 263–271. [Google Scholar] [CrossRef]
- Dastoli, S.; Nisticò, S.P.; Morrone, P.; Patruno, C.; Leo, A.; Citraro, R.; Gallelli, L.; Russo, E.; De Sarro, G.; Bennardo, L. Colchicine in managing skin conditions: A systematic review. Pharmaceutics 2022, 14, 294. [Google Scholar] [CrossRef] [PubMed]
- Iannone, L.F.; Bennardo, L.; Palleria, C.; Roberti, R.; De Sarro, C.; Naturale, M.D.; Dastoli, S.; Donato, L.; Manti, A.; Valenti, G.; et al. Safety profile of biologic drugs for psoriasis in clinical practice: An Italian prospective pharmacovigilance study. PLoS ONE 2020, 15, e0241575. [Google Scholar]
- Christophers, E. Psoriasis—Epidemiology and clinical spectrum. Clin. Exp. Dermatol. 2001, 26, 314–320. [Google Scholar] [CrossRef]
- Mc Ardle, A.; Flatley, B.; Pennington, S.R.; FitzGerald, O. Early biomarkers of joint damage in rheumatoid and psoriatic arthritis. Arthritis Res. Ther. 2015, 17, 141. [Google Scholar] [CrossRef] [Green Version]
- Generali, E.; Scirè, C.A.; Favalli, E.G.; Selmi, C. Biomarkers in psoriatic arthritis: A systematic literature review. Expert Rev. Clin. Immunol. 2016, 12, 651–660. [Google Scholar] [CrossRef] [Green Version]
- Selleck, M.J.; Senthil, M.; Wall, N.R. Making meaningful clinical use of biomarkers. Biomark. Insights 2017, 12, 1177271917715236. [Google Scholar] [CrossRef] [Green Version]
- Jiang, S.; Hinchliffe, T.E.; Wu, T. Biomarkers of an autoimmune skin disease—Psoriasis. Genom. Proteom. Bioinform. 2015, 13, 224–233. [Google Scholar] [CrossRef] [Green Version]
- Punzi, L.; Poswiadek, M.; Oliviero, F.; Lonigro, A.; Modesti, V.; Ramonda, R.; Todesco, S. Laboratory findings in psoriatic arthritis. Reumatismo 2011, 59 (Suppl. S1), 52–55. [Google Scholar] [CrossRef]
- Sokolova, M.V.; Simon, D.; Nas, K.; Zaiss, M.M.; Luo, Y.; Zhao, Y.; Rech, J.; Schett, G. A set of serum markers detecting systemic inflammation in psoriatic skin, entheseal, and joint disease in the absence of C-reactive protein and its link to clinical disease manifestations. Arthritis Res. Ther. 2020, 22, 26. [Google Scholar] [CrossRef] [Green Version]
- Tamagawa-Mineoka, R.; Katoh, N.; Kishimoto, S. Platelet activation in patients with psoriasis: Increased plasma levels of platelet-derived microparticles and soluble P-selectin. J. Am. Acad. Dermatol. 2010, 62, 621–626. [Google Scholar] [CrossRef]
- Pamuk, G.E.; Pamuk, N.; Orüm, H.; Arican, O.; Turgut, B.; Demir, M. Elevated platelet-monocyte complexes in patients with psoriatic arthritis. Platelets 2009, 20, 493–497. [Google Scholar] [CrossRef]
- Arican, O.; Aral, M.; Sasmaz, S.; Ciragil, P. Serum levels of TNF-alpha, IFN-gamma, IL-6, IL-8, IL-12, IL-17, and IL-18 in patients with active psoriasis and correlation with disease severity. Mediators Inflamm. 2005, 2005, 273–279. [Google Scholar] [CrossRef] [Green Version]
- Bosè, F.; Capsoni, F.; Molteni, S.; Raeli, L.; Diani, M.; Altomare, A.; Garavaglia, M.; Garutti, C.; Frigerio, E.; Banfi, G.; et al. Differential expression of interleukin-2 by anti-CD3-stimulated peripheral blood mononuclear cells in patients with psoriatic arthritis and patients with cutaneous psoriasis. Clin. Exp. Dermatol. 2014, 39, 385–390. [Google Scholar] [CrossRef]
- Alenius, G.-M.; Eriksson, C.; Rantapää Dahlqvist, S. Interleukin-6 and soluble interleukin-2 receptor alpha-markers of inflammation in patients with psoriatic arthritis? Clin. Exp. Rheumatol. 2009, 27, 120–123. [Google Scholar]
- Xiao, S.; Liu, X.; Wang, X.; Lv, H.; Zhao, J.; Guo, X.; Xian, F.; Ji, Y.; Zhang, G. Plasma MicroRNA expression profiles in psoriasis. J. Immunol. Res. 2020, 2020, 1561278. [Google Scholar] [CrossRef] [Green Version]
- Wade, S.M.; McGarry, T.; Wade, S.C.; Fearon, U.; Veale, D.J. Serum MicroRNA signature as a diagnostic and therapeutic marker in patients with psoriatic arthritis. J. Rheumatol. 2020, 47, 1760–1767. [Google Scholar] [CrossRef] [PubMed]
- Alberro, A.; Iparraguirre, L.; Fernandes, A.; Otaegui, D. Extracellular vesicles in blood: Sources, effects, and applications. Int. J. Mol. Sci. 2021, 22, 8163. [Google Scholar] [CrossRef]
- Palviainen, M.; Saraswat, M.; Varga, Z.; Kitka, D.; Neuvonen, M.; Puhka, M.; Joenväärä, S.; Renkonen, R.; Nieuwland, R.; Takatalo, M.; et al. Extracellular vesicles from human plasma and serum are carriers of extravesicular cargo-Implications for biomarker discovery. PLoS ONE 2020, 15, e0236439. [Google Scholar] [CrossRef]
- El Andaloussi, S.; Mäger, I.; Breakefield, X.O.; Wood MJ, A. Extracellular vesicles: Biology and emerging therapeutic opportunities. Nat. Rev. Drug. Discov. 2013, 12, 347–357. [Google Scholar] [CrossRef]
- He, X.; Park, S.; Chen, Y.; Lee, H. Extracellular vesicle-associated miRNAs as a biomarker for lung cancer in liquid biopsy. Front. Mol. Biosci. 2021, 8, 630718. [Google Scholar] [CrossRef]
- Probert, C.; Dottorini, T.; Speakman, A.; Hunt, S.; Nafee, T.; Fazeli, A.; Wood, S.; Brown, J.E.; James, V. Communication of prostate cancer cells with bone cells via extracellular vesicle RNA; a potential mechanism of metastasis. Oncogene 2019, 38, 1751–1763. [Google Scholar] [CrossRef]
- Mørk, M.; Andreasen, J.J.; Rasmussen, L.H.; Lip, G.Y.; Pedersen, S.; Bæk, R.; Jørgensen, M.M.; Kristensen, S.R. Elevated blood plasma levels of tissue factor-bearing extracellular vesicles in patients with atrial fibrillation. Thromb. Res. 2019, 173, 141–150. [Google Scholar] [CrossRef]
- Gidlöf, O.; Evander, M.; Rezeli, M.; Marko-Varga, G.; Laurell, T.; Erlinge, D. Proteomic profiling of extracellular vesicles reveals additional diagnostic biomarkers for myocardial infarction compared to plasma alone. Sci. Rep. 2019, 9, 8991. [Google Scholar] [CrossRef] [Green Version]
- Badhwar, A.; Haqqani, A.S. Biomarker potential of brain-secreted extracellular vesicles in blood in Alzheimer’s disease. Alzheimers Dement. 2020, 12, e12001. [Google Scholar] [CrossRef]
- Jiang, M.; Fang, H.; Shao, S.; Dang, E.; Zhang, J.; Qiao, P.; Yang, A.; Wang, G. Keratinocyte exosomes activate neutrophils and enhance skin inflammation in psoriasis. FASEB J. 2019, 33, 13241–13253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mangino, G.; Iuliano, M.; Carlomagno, S.; Bernardini, N.; Rosa, P.; Chiantore, M.V.; Skroza, N.; Calogero, A.; Potenza, C.; Romeo, G. Interleukin-17A affects extracellular vesicles release and cargo in human keratinocytes. Exp. Dermatol. 2019, 2, 1066–1073. [Google Scholar] [CrossRef] [PubMed]
- Marton, N.; Kovács, O.T.; Baricza, E.; Kittel, Á.; Győri, D.; Mócsai, A.; Meier FM, P.; Goodyear, C.S.; McInnes, I.B.; Buzás, E.I.; et al. Extracellular vesicles regulate the human osteoclastogenesis: Divergent roles in discrete inflammatory arthropathies. Cell. Molec. Life Sci. 2017, 74, 3599–3611. [Google Scholar] [CrossRef] [PubMed]
- Andreu, Z.; Rivas, E.; Sanguino-Pascual, A.; Lamana, A.; Marazuela, M.; González-Alvaro, I.; Sánchez-Madrid, F.; de la Fuente, H.; Yáñez-Mó, M. Comparative analysis of EV isolation procedures for miRNAs detection in serum samples. J. Extracell. Vesicles 2016, 5, 31655. [Google Scholar] [CrossRef]
- Endzeliņš, E.; Berger, A.; Melne, V.; Bajo-Santos, C.; Soboļevska, K.; Ābols, A.; Rodriguez, M.; Šantare, D.; Rudņickiha, A.; Lietuvietis, V.; et al. Detection of circulating miRNAs: Comparative analysis of extracellular vesicle-incorporated miRNAs and cell-free miRNAs in whole plasma of prostate cancer patients. BMC Cancer 2017, 17, 730. [Google Scholar] [CrossRef]
- Ramshani, Z.; Zhang, C.; Richards, K.; Chen, L.; Xu, G.; Stiles, B.L.; Hill, R.; Senapati, S.; Go, D.B.; Chang, H.-C. Extracellular vesicle microRNA quantification from plasma using an integrated microfluidic device. Commun. Biol. 2019, 2, 189. [Google Scholar] [CrossRef]
- Karimi, N.; Cvjetkovic, A.; Jang, S.C.; Crescitelli, R.; Hosseinpour Feizi, M.A.; Nieuwland, R.; Lötvall, J.; Lässer, C. Detailed analysis of the plasma extracellular vesicle proteome after separation from lipoproteins. Cell. Molec. Life Sci. CMLS 2018, 75, 2873–2886. [Google Scholar] [CrossRef] [Green Version]
- Yuana, Y.; Levels, J.; Grootemaat, A.; Sturk, A.; Nieuwland, R. Co-isolation of extracellular vesicles and high-density lipoproteins using density gradient ultracentrifugation. J. Extracell. Vesicles 2014, 3, 23262. [Google Scholar] [CrossRef]
- Berumen Sánchez, G.; Bunn, K.E.; Pua, H.H.; Rafat, M. Extracellular vesicles: Mediators of intercellular communication in tissue injury and disease. Cell. Commun. Signal. 2021, 19, 104. [Google Scholar] [CrossRef]
- Pitt, J.M.; Kroemer, G.; Zitvogel, L. Extracellular vesicles: Masters of intercellular communication and potential clinical interventions. J. Clin. Invest. 2016, 126, 1139–1143. [Google Scholar] [CrossRef] [Green Version]
- Pelosi, A.; Lunardi, C.; Fiore, P.F.; Tinazzi, E.; Patuzzo, G.; Argentino, G.; Moretta, F.; Puccetti, A.; Dolcino, M. MicroRNA expression profiling in psoriatic arthritis. Biomed. Res. Int. 2018, 2018, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Koga, Y.; Jinnin, M.; Ichihara, A.; Fujisawa, A.; Moriya, C.; Sakai, K.; Fukushima, S.; Inoue, Y.; Ihn, H. Analysis of expression pattern of serum microRNA levels in patients with psoriasis. J. Dermatol. Sci. 2014, 74, 170–171. [Google Scholar] [CrossRef]
- Lerman, G.; Avivi, C.; Mardoukh, C.; Barzilai, A.; Tessone, A.; Gradus, B.; Pavlotsky, F.; Barshack, I.; Polak-Charcon, S.; Orenstein, A. MiRNA expression in psoriatic skin: Reciprocal regulation of hsa-miR-99a and IGF-1R. PLoS ONE 2011, 6, e20916. [Google Scholar] [CrossRef]
- Liew, W.C.; Sundaram, G.M.; Quah, S.; Lum, G.G.; Tan, J.S.L.; Ramalingam, R.; Common, J.E.A.; Tang, M.B.Y.; Lane, E.B.; Thng, S.T.G.; et al. Belinostat resolves skin barrier defects in atopic dermatitis by targeting the dysregulated miR-335:SOX6 axis. J. Allergy Clin. Immunol. 2020, 146, 606–620.e12. [Google Scholar] [CrossRef] [Green Version]
- Tijsen, A.J.; Creemers, E.E.; Moerland, P.D.; de Windt, L.J.; van der Wal, A.C.; Kok, W.E.; Pinto, Y.M. MiR423-5p as a circulating biomarker for heart failure. Circ. Res. 2010, 106, 1035–1039. [Google Scholar] [CrossRef]
- Rizzacasa, B.; Morini, E.; Mango, R.; Vancheri, C.; Budassi, S.; Massaro, G.; Maletta, S.; Macrini, M.; D’Annibale, S.; Romeo, F.; et al. MiR-423 is differentially expressed in patients with stable and unstable coronary artery disease: A pilot study. PLoS ONE 2019, 14, e0216363. [Google Scholar] [CrossRef] [Green Version]
- Fourie, N.H.; Peace, R.M.; Abey, S.K.; Sherwin, L.B.; Rahim-Williams, B.; Smyser, P.A.; Wiley, J.W.; Henderson, W.A. Elevated circulating miR-150 and miR-342-3p in patients with irritable bowel syndrome. Exp. Mol. Pathol. 2014, 96, 422–425. [Google Scholar] [CrossRef] [Green Version]
- Hu, G.; Zhang, N.; Li, J.; Wang, J.; Wu, W.; Li, J.; Tong, W.; Zhao, X.; Dai, L.; Zhang, X. Tumor necrosis factor receptor associated factor 3 modulates cartilage degradation through suppression of Interleukin 17 signaling. Am. J. Pathol. 2020, 190, 1701–1712. [Google Scholar] [CrossRef]
- Liu, Z.; Chen, S.; Yang, Y.; Lu, S.; Zhao, X.; Hu, B.; Pei, H. MicroRNA-671-3p regulates the development of knee osteoarthritis by targeting TRAF3 in chondrocytes. Mol. Med. Rep. 2019, 20, 2843–2850. [Google Scholar] [CrossRef]
- Ntoumou, E.; Tzetis, M.; Braoudaki, M.; Lambrou, G.; Poulou, M.; Malizos, K.; Stefanou, N.; Anastasopoulou, L.; Tsezou, A. Serum microRNA array analysis identifies miR-140-3p, miR-33b-3p and miR-671-3p as potential osteoarthritis biomarkers involved in metabolic processes. Clin. Epigenetics 2017, 9, 127. [Google Scholar] [CrossRef] [Green Version]
- Costa, V.; De Fine, M.; Carina, V.; Conigliaro, A.; Raimondi, L.; De Luca, A.; Bellavia, D.; Salamanna, F.; Alessandro, R.; Pignatti, G.; et al. How miR-31-5p and miR-33a-5p regulates SP1/CX43 expression in osteoarthritis disease: Preliminary insights. Int. J. Mol. Sci. 2021, 22, 2471. [Google Scholar] [CrossRef]
- Huang, Z.; Xing, S.; Liu, M.; Deng, W.; Wang, Y.; Huang, Z.; Huang, Y.; Huang, X.; Wu, C.; Guo, X.; et al. MiR-26a-5p enhances cells proliferation, invasion, and apoptosis resistance of fibroblast-like synoviocytes in rheumatoid arthritis by regulating PTEN/PI3K/AKT pathway. Biosci. Rep. 2019, 39, 20182192. [Google Scholar] [CrossRef] [Green Version]
- Guo, T.; Ding, H.; Jiang, H.; Bao, N.; Zhou, L.; Zhao, J. miR-338-5p regulates the viability, proliferation, apoptosis and migration of rheumatoid arthritis fibroblast-like synoviocytes by targeting NFAT5. Cell. Physiol. Biochem. 2018, 49, 899–910. [Google Scholar] [CrossRef]
- Hussain, N.; Zhu, W.; Jiang, C.; Xu, J.; Geng, M.; Wu, X.; Hussain, S.; Wang, B.; Rajoka, M.S.R.; Li, Y.; et al. Down-regulation of miR-10a-5p promotes proliferation and restricts apoptosis via targeting T-box transcription factor 5 in inflamed synoviocytes. Biosci. Rep. 2018, 38, 20180003. [Google Scholar] [CrossRef] [Green Version]
- Song, A.-F.; Kang, L.; Wang, Y.-F.; Wang, M. MiR-34a-5p inhibits fibroblast-like synoviocytes proliferation via XBP1. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 11675–11682. [Google Scholar]
- Li, H.-Z.; Xu, X.-H.; Lin, N.; Wang, D.-W.; Lin, Y.-M.; Su, Z.-Z.; Lu, H.-D. Overexpression of miR-10a-5p facilitates the progression of osteoarthritis. Aging 2020, 12, 5948–5976. [Google Scholar] [CrossRef]
- Endisha, H.; Datta, P.; Sharma, A.; Nakamura, S.; Rossomacha, E.; Younan, C.; Ali, S.A.; Tavallaee, G.; Lively, S.; Potla, P.; et al. MicroRNA-34a-5p promotes joint destruction during osteoarthritis. Arthritis Rheumatol. 2021, 73, 426–439. [Google Scholar] [CrossRef]
- Rousseau, J.-C.; Millet, M.; Croset, M.; Sornay-Rendu, E.; Borel, O.; Chapurlat, R. Association of circulating microRNAs with prevalent and incident knee osteoarthritis in women: The OFELY study. Arthritis Res. Ther. 2020, 22, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohammed, A.; Alshamarri, T.; Adeyeye, T.; Lazariu, V.; McNutt, L.-A.; Carpenter, D.O. A comparison of risk factors for osteo- and rheumatoid arthritis using NHANES data. Prev. Med. Rep. 2020, 20, 101242. [Google Scholar] [CrossRef] [PubMed]
- Prinz, J.C. Human leukocyte antigen-Class I alleles and the autoreactive T cell response in psoriasis pathogenesis. Front. Immunol. 2018, 9, 954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, L.; Guo, W.; Zhang, S.; Wang, G. Ubiquitination-proteasome system: A new player in the pathogenesis of psoriasis and clinical implications. J. Dermatol. Sci. 2018, 89, 219–225. [Google Scholar] [CrossRef] [Green Version]
- Sun, Z.-H.; Liu, Y.; Liu, J.; Xu, D.; Li, X.; Meng, X.; Ma, T.; Huang, C.; Li, J. MeCP2 regulates PTCH1 expression through DNA methylation in rheumatoid arthritis. Inflammation 2017, 40, 1497–1508. [Google Scholar] [CrossRef]
- Miao, C.; Huang, C.; Huang, Y.; Yang, Y.; He, X.; Zhang, L.; Lv, X.-W.; Jin, Y.; Li, J. MeCP2 modulates the canonical Wnt pathway activation by targeting SFRP4 in rheumatoid arthritis fibroblast-like synoviocytes in rats. Cell. Signal. 2013, 25, 598–608. [Google Scholar] [CrossRef]
- Gibbs, J.E.; Ray, D.W. The role of the circadian clock in rheumatoid arthritis. Arthritis Res. Ther. 2013, 15, 205. [Google Scholar] [CrossRef] [Green Version]
- Ando, N.; Nakamura, Y.; Aoki, R.; Ishimaru, K.; Ogawa, H.; Okumura, K.; Shibata, S.; Shimada, S.; Nakao, A. Circadian gene clock regulates psoriasis-like skin inflammation in mice. J. Investig. Dermatol. 2015, 135, 3001–3008. [Google Scholar] [CrossRef] [Green Version]
- Bain, K.A.; Milling, S. T cell addiction: Can pathogenic T cells be controlled using dopamine receptors? Immunology 2019, 158, 151–152. [Google Scholar] [CrossRef]
- Capellino, S. Dopaminergic agents in rheumatoid arthritis. J. Neuroimmune. Pharmacol. 2020, 15, 48–56. [Google Scholar] [CrossRef] [Green Version]
- Van Nie, L.; Salinas-Tejedor, L.; Dychus, N.; Fasbender, F.; Hülser, M.-L.; Cutolo, M.; Rehart, S.; Neumann, E.; Müller-Ladner, U.; Capellino, S. Dopamine induces in vitro migration of synovial fibroblast from patients with rheumatoid arthritis. Sci. Rep. 2020, 10, 11928. [Google Scholar] [CrossRef]
- Pasquali, L.; Svedbom, A.; Srivastava, A.; Rosén, E.; Lindqvist, U.; Ståhle, M.; Pivarcsi, A.; Sonkoly, E. Circulating microRNAs in extracellular vesicles as potential biomarkers for psoriatic arthritis in patients with psoriasis. J. Eur. Acad. Dermatol. Venereol. 2020, 34, 1248–1256. [Google Scholar] [CrossRef]
- Reimann, E.; Lättekivi, F.; Keermann, M.; Abram, K.; Kõks, S.; Kingo, K.; Fazeli, A. Multicomponent biomarker approach improves the accuracy of diagnostic biomarkers for psoriasis vulgaris. Acta Derm. Venereol. 2019, 99, 1258–1265. [Google Scholar] [CrossRef]
- Helwa, I.; Cai, J.; Drewry, M.D.; Zimmerman, A.; Dinkins, M.B.; Khaled, M.L.; Seremwe, M.; Dismuke, W.M.; Bieberich, E.; Stamer, W.D.; et al. A comparative study of serum exosome isolation using differential ultracentrifugation and three commercial reagents. PLoS ONE 2017, 12, e0170628. [Google Scholar]
- Konoshenko, M.; Yu Lekchnov, E.A.; Vlassov, A.V.; Laktionov, P.P. Isolation of extracellular vesicles: General methodologies and latest trends. Biomed. Res. Int. 2018, 2018, 8545347. [Google Scholar] [CrossRef] [PubMed]
- Buschmann, D.; Kirchner, B.; Hermann, S.; Märte, M.; Wurmser, C.; Brandes, F.; Kotschote, S.; Bonin, M.; Steinlein, O.K.; Pfaffl, M.W.; et al. Evaluation of serum extracellular vesicle isolation methods for profiling miRNAs by next-generation sequencing. J. Extracell. Vesicles 2018, 7, 1481321. [Google Scholar] [CrossRef] [PubMed]
- Sódar, B.W.; Kittel, Á.; Pálóczi, K.; Vukman, K.V.; Osteikoetxea, X.; Szabó-Taylor, K.; Németh, A.; Sperlágh, B.; Baranyai, T.; Giricz, Z.; et al. Low-density lipoprotein mimics blood plasma-derived exosomes and microvesicles during isolation and detection. Sci. Rep. 2016, 6, 24316. [Google Scholar] [CrossRef] [Green Version]
- Brennan, K.; Martin, K.; FitzGerald, S.P.; O’Sullivan, J.; Wu, Y.; Blanco, A.; Richardson, C.; Mc Gee, M.M.A. A comparison of methods for the isolation and separation of extracellular vesicles from protein and lipid particles in human serum. Sci. Rep. 2020, 10, 1039. [Google Scholar] [CrossRef] [Green Version]
- Onódi, Z.; Pelyhe, C.; Terézia Nagy, C.; Brenner, G.B.; Almási, L.; Kittel, Á.; Manček-Keber, M.; Ferdinandy, P.; Buzás, E.I.; Giricz, Z. Isolation of high-purity extracellular vesicles by the combination of iodixanol density gradient ultracentrifugation and bind-elute chromatography from blood plasma. Front. Physiol. 2018, 9, 1479. [Google Scholar] [CrossRef] [Green Version]
- Vogel, R.; Coumans, F.A.W.; Maltesen, R.G.; Böing, A.N.; Bonnington, K.E.; Broekman, M.L.; Broom, M.F.; Buzás, E.I.; Christiansen, G.; Hajji, N.; et al. A standardized method to determine the concentration of extracellular vesicles using tunable resistive pulse sensing. J. Extracell. Vesicles 2016, 5, 31242. [Google Scholar] [CrossRef]
- Navajas, R.; Corrales, F.J.; Paradela, A. Serum exosome isolation by size-exclusion chromatography for the discovery and validation of preeclampsia-associated biomarkers. Methods Mol. Biol. 2019, 1959, 39–50. [Google Scholar]
- Belov, L.; Matic, K.J.; Hallal, S.; Best, O.G.; Mulligan, S.P.; Christopherson, R.I. Extensive surface protein profiles of extracellular vesicles from cancer cells may provide diagnostic signatures from blood samples. J. Extracell. Vesicles 2016, 5, 25355. [Google Scholar] [CrossRef]
- Akagi, T.; Kato, K.; Kobayashi, M.; Kosaka, N.; Ochiya, T.; Ichiki, T. On-chip immunoelectrophoresis of extracellular vesicles released from human breast cancer cells. PLoS ONE 2015, 10, e0123603. [Google Scholar] [CrossRef]
- Kowal, J.; Arras, G.; Colombo, M.; Jouve, M.; Morath, J.P.; Primdal-Bengtson, B.; Dingli, F.; Loew, D.; Tkach, M.; Théry, C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. USA 2016, 113, E968–E977. [Google Scholar] [CrossRef] [Green Version]
- Matsumoto, A.; Takahashi, Y.; Chang, H.; Wu, Y.; Yamamoto, A.; Ishihama, Y.; Takakura, Y. Blood concentrations of small extracellular vesicles are determined by a balance between abundant secretion and rapid clearance. J. Extracell. Vesicles 2020, 9, 1696517. [Google Scholar] [CrossRef] [Green Version]
- Santucci, L.; Bruschi, M.; Del Zotto, G.; Antonini, F.; Ghiggeri, G.M.; Panfoli, I.; Candiano, G. Biological surface properties in extracellular vesicles and their effect on cargo proteins. Sci. Rep. 2019, 9, 13048. [Google Scholar] [CrossRef] [Green Version]
- Maroto, R.; Zhao, Y.; Jamaluddin, M.; Popov, V.L.; Wang, H.; Kalubowilage, M.; Zhang, Y.; Luisi, J.; Sun, H.; Culbertson, C.T.; et al. Effects of storage temperature on airway exosome integrity for diagnostic and functional analyses. J. Extracell. Vesicles 2017, 6, 1359478. [Google Scholar] [CrossRef] [Green Version]
- Sharma, S.; LeClaire, M.; Wohlschlegel, J.; Gimzewski, J. Impact of isolation methods on the biophysical heterogeneity of single extracellular vesicles. Sci. Rep. 2020, 10, 13327. [Google Scholar] [CrossRef]
- Chiang, C.-Y.; Chen, C. Toward characterizing extracellular vesicles at a single-particle level. J. Biomed. Sci. 2019, 26, 9. [Google Scholar] [CrossRef]
- Jung, A.L.; Møller Jørgensen, M.; Bæk, R.; Griss, K.; Han, M.; Auf Dem Brinke, K.; Timmesfeld, N.; Bertrams, W.; Greulich, T.; Koczulla, R.; et al. Surface proteome of plasma extracellular vesicles as biomarkers for pneumonia and acute exacerbation of chronic obstructive pulmonary disease. J. Infect. Dis. 2020, 221, 325–335. [Google Scholar] [CrossRef]
- Shao, S.; Fang, H.; Li, Q.; Wang, G. Extracellular vesicles in inflammatory skin disorders: From pathophysiology to treatment. Theranostics 2020, 10, 9937–9955. [Google Scholar] [CrossRef]
- Tsuno, H.; Arito, M.; Suematsu, N.; Sato, T.; Hashimoto, A.; Matsui, T.; Omoteyama, K.; Sato, M.; Okamoto, K.; Tohma, S.; et al. A proteomic analysis of serum-derived exosomes in rheumatoid arthritis. BMC Rheumatol. 2018, 2, 35. [Google Scholar] [CrossRef]
- Withrow, J.; Murphy, C.; Liu, Y.; Hunter, M.; Fulzele, S.; Hamrick, M.W. Extracellular vesicles in the pathogenesis of rheumatoid arthritis and osteoarthritis. Arthritis Res. Ther. 2016, 18, 286. [Google Scholar] [CrossRef] [Green Version]
- Andreu, Z.; Yáñez-Mó. Tetraspanins in extracellular vesicle formation and function. Front. Immunol. 2014, 5, 00442. [Google Scholar] [CrossRef] [Green Version]
- Brosseau, C.; Colas, L.; Magnan, A.; Brouard, S. CD9 Tetraspanin: A new pathway for the regulation of inflammation? Front. Immunol. 2018, 9, 2316. [Google Scholar] [CrossRef] [Green Version]
- Chettimada, S.; Lorenz, D.R.; Misra, V.; Dillon, S.T.; Reeves, R.K.; Manickam, C.; Morgello, S.; Kirk, G.D.; Mehta, S.H.; Gabuzda, D. Exosome markers associated with immune activation and oxidative stress in HIV patients on antiretroviral therapy. Sci. Rep. 2018, 8, 7227. [Google Scholar] [CrossRef]
- Di Meglio, P.; Villanova, F.; Nestle, F.O. Psoriasis. Cold Spring Harb. Perspect. Med. 2014, 4, a015354. [Google Scholar] [CrossRef] [Green Version]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the international society for extracellular vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [Green Version]
- Midekessa, G.; Godakumara, K.; Ord, J.; Viil, J.; Lättekivi, F.; Dissanayake, K.; Kopanchuk, S.; Rinken, A.; Andronowska, A.; Bhattacharjee, S.; et al. Zeta potential of extracellular vesicles: Toward understanding the attributes that determine colloidal stability. ACS Omega 2020, 5, 16701–16710. [Google Scholar] [CrossRef]
- Jørgensen, M.; Bæk, R.; Pedersen, S.; Søndergaard, E.K.L.; Kristensen, S.R.; Varming, K. Extracellular vesicle (EV) Array: Microarray capturing of exosomes and other extracellular vesicles for multiplexed phenotyping. J. Extracell. Vesicles 2013, 2, 20920. [Google Scholar] [CrossRef]
- Breakefield, X.O.; Das, S.; Gandhi, R.; Sood, A.K.; Balaj, L.; Filant, J.; Nejad, P.; Paul, A.; Simonson, B.; Srinivasan, S.; et al. Isolation of exosomal RNA from serum or plasma using the Qiagen miRNeasy Micro kit. Protocol. Exch. 2017. [Google Scholar] [CrossRef]
- Li, X.; Ben-Dov, I.Z.; Mauro, M.; Williams, Z. Lowering the quantification limit of the QubitTM RNA HS Assay using RNA spike-in. BMC Molec. Biol. 2015, 16, 9. [Google Scholar] [CrossRef] [Green Version]
- R Core Team. R: A language and environment for statistical computing. R Found. Stat. Comput. 2020. [Google Scholar]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 10 June 2021).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harrow, J.; Frankish, A.; Gonzalez, J.M.; Tapanari, E.; Diekhans, M.; Kokocinski, F.; Aken, B.L.; Barrell, D.; Zadissa, A.; Searle, S.; et al. GENCODE: The reference human genome annotation for The ENCODE Project. Genome Res. 2012, 22, 1760–1774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- The RNAcentral Consortium; Sweeney, B. A.; Petrov, A.I.; Burkov, B.; Finn, R.D.; Bateman, A.; Szymanski, M.; Karlowski, W.M.; Gorodkin, J.; Seemann, S.E.; et al. RNAcentral: A hub of information for non-coding RNA sequences. Nucleic Acids Res. 2019, 47, D221–D229. [Google Scholar]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Zhou, X.; Lindsay, H.; Robinson, M.D. Robustly detecting differential expression in RNA sequencing data using observation weights. Nucleic Acids Res. 2014, 42, e91. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, X. miRDB: An online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020, 48, D127–D131. [Google Scholar] [CrossRef] [Green Version]
- Yu, G.; He, Q.-Y. ReactomePA: An R/Bioconductor package for reactome pathway analysis and visualization. Mol. Biosyst. 2016, 12, 477–479. [Google Scholar] [CrossRef]
- Fabregat, A.; Jupe, S.; Matthews, L.; Sidiropoulos, K.; Gillespie, M.; Garapati, P.; Haw, R.; Jassal, B.; Korninger, F.; May, B.; et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2018, 46, D649–D655. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef] [Green Version]
- Meng, Y.; Asghari, M.; Aslan, M.K.; Yilmaz, A.; Mateescu, B.; Stavrakis, S.; deMello, A.J. Microfluidics for extracellular vesicle separation and mimetic synthesis: Recent advances and future perspectives. Chem. Eng. J. 2021, 404, 126110. [Google Scholar] [CrossRef]


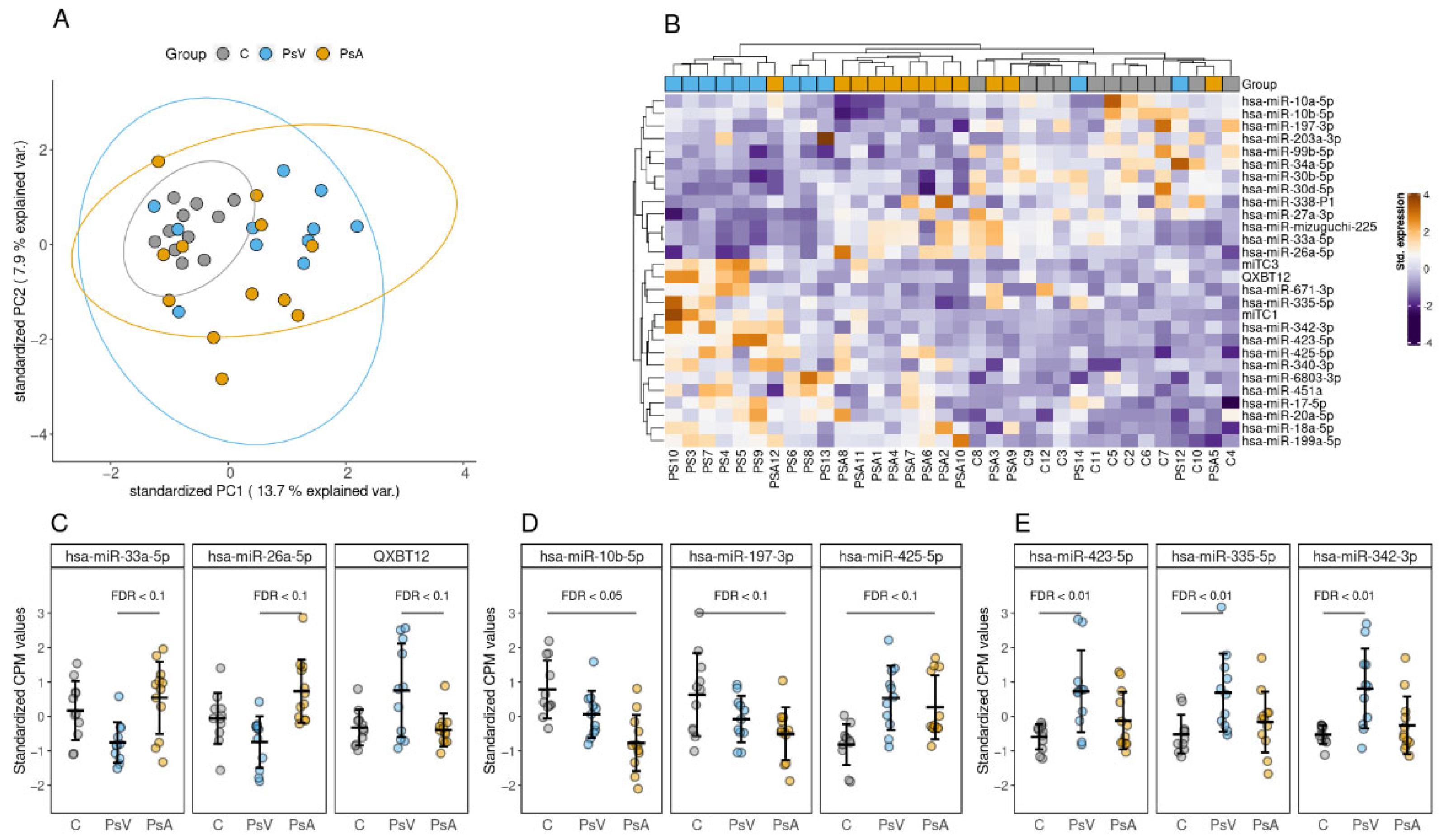
| Comparison | miRNA | FC | p-Value | FDR |
|---|---|---|---|---|
| PsA to PsV | QXBT12 | 0.46 | 0.00053 | 0.058 |
| hsa-miR-33a-5p | 1.82 | 0.000547 | 0.058 | |
| hsa-miR-26a-5p | 1.36 | 0.00101 | 0.0596 | |
| hsa-miR-mizuguchi-225 | 1.81 | 0.00113 | 0.0596 | |
| miTC3 | 0.43 | 0.00242 | 0.0879 | |
| hsa-miR-338-5p | 2.32 | 0.00273 | 0.0879 | |
| hsa-miR-671-3p | 0.43 | 0.0029 | 0.0879 | |
| hsa-miR-342-3p | 0.52 | 0.00375 | 0.0993 | |
| PsA to C | hsa-miR-10b-5p | 0.46 | 0.000204 | 0.0433 |
| hsa-miR-197-3p | 0.53 | 0.000873 | 0.0811 | |
| hsa-miR-425-5p | 1.51 | 0.00128 | 0.0811 | |
| hsa-miR-199a-5p | 1.57 | 0.00153 | 0.0811 | |
| hsa-miR-203a-3p | 0.35 | 0.002 | 0.0849 | |
| hsa-miR-10a-5p | 0.54 | 0.0025 | 0.0882 | |
| hsa-miR-34a-5p | 0.46 | 0.00292 | 0.0884 | |
| PsV to C | hsa-miR-423-5p | 2.09 | 3.58 × 10−5 | 0.00419 |
| hsa-miR-335-5p | 2.29 | 6.08 × 10−5 | 0.00419 | |
| hsa-miR-342-3p | 2.57 | 6.52 × 10−5 | 0.00419 | |
| miTC1 | 2.90 | 7.91 × 10−5 | 0.00419 | |
| has-miR-425-5p | 1.62 | 0.000183 | 0.00775 | |
| hsa-miR-99b-5p | 0.56 | 0.000898 | 0.0317 | |
| hsa-miR-17-5p | 1.44 | 0.00148 | 0.0384 | |
| hsa-miR-18a-5p | 1.55 | 0.00176 | 0.0384 | |
| hsa-miR-27a-3p | 0.72 | 0.0018 | 0.0384 | |
| hsa-miR-451a | 1.77 | 0.00181 | 0.0384 | |
| QXBT12 | 2.01 | 0.00204 | 0.0394 | |
| miTC3 | 2.48 | 0.00279 | 0.0493 | |
| hsa-miR-6803-3p | 1.94 | 0.00459 | 0.0697 | |
| hsa-miR-199a-5p | 1.50 | 0.0046 | 0.0697 | |
| hsa-miR-30b-5p | 0.67 | 0.00554 | 0.0783 | |
| hsa-miR-20a-5p | 1.37 | 0.00595 | 0.0789 | |
| hsa-miR-340-3p | 1.93 | 0.00641 | 0.0799 | |
| hsa-miR-30d-5p | 0.71 | 0.00728 | 0.0858 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lättekivi, F.; Guljavina, I.; Midekessa, G.; Viil, J.; Heath, P.R.; Bæk, R.; Jørgensen, M.M.; Andronowska, A.; Kingo, K.; Fazeli, A. Profiling Blood Serum Extracellular Vesicles in Plaque Psoriasis and Psoriatic Arthritis Patients Reveals Potential Disease Biomarkers. Int. J. Mol. Sci. 2022, 23, 4005. https://doi.org/10.3390/ijms23074005
Lättekivi F, Guljavina I, Midekessa G, Viil J, Heath PR, Bæk R, Jørgensen MM, Andronowska A, Kingo K, Fazeli A. Profiling Blood Serum Extracellular Vesicles in Plaque Psoriasis and Psoriatic Arthritis Patients Reveals Potential Disease Biomarkers. International Journal of Molecular Sciences. 2022; 23(7):4005. https://doi.org/10.3390/ijms23074005
Chicago/Turabian StyleLättekivi, Freddy, Irina Guljavina, Getnet Midekessa, Janeli Viil, Paul R. Heath, Rikke Bæk, Malene Møller Jørgensen, Aneta Andronowska, Kulli Kingo, and Alireza Fazeli. 2022. "Profiling Blood Serum Extracellular Vesicles in Plaque Psoriasis and Psoriatic Arthritis Patients Reveals Potential Disease Biomarkers" International Journal of Molecular Sciences 23, no. 7: 4005. https://doi.org/10.3390/ijms23074005







