Generation of a Library of Carbohydrate-Active Enzymes for Plant Biomass Deconstruction
Abstract
1. Introduction
2. Results and Discussion
2.1. Identification and Selection of CAZymes
2.2. Gene Production and Cloning
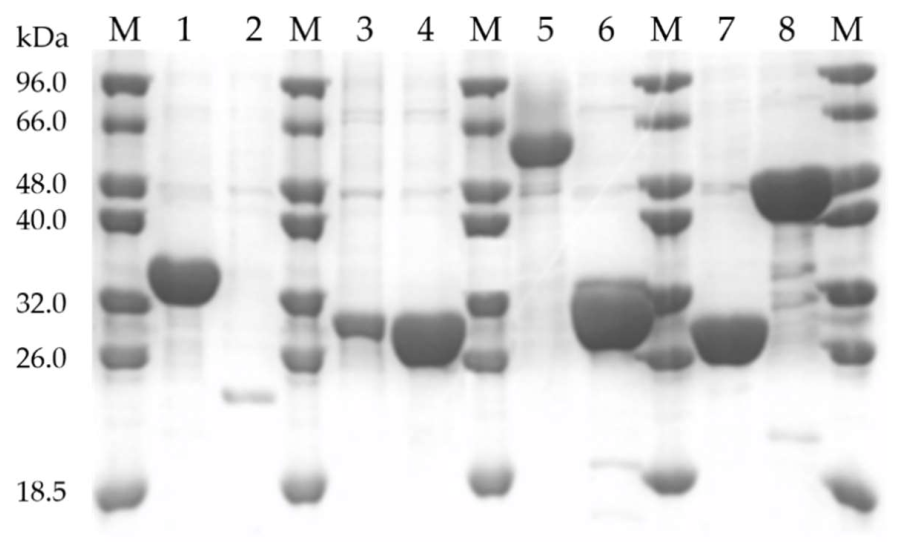
2.3. Protein Expression and Putative Factors Affecting Solubility
2.4. The Effect of Protein Origin on Recombinant Production
2.5. Influence of the Gene Production Strategy, Gene Synthesis, or PCR on Protein Production
2.6. Does the GFP Tag Promote Protein Production?
2.7. Influence of the Protein Molecular Weight on Protein Production
2.8. Influence of the Primary Sequence Composition on Protein Production
2.9. CAZymes Activity
2.10. Final Outputs
3. Materials and Methods
3.1. Identification and Selection of CAZymes
3.2. Polymerase Chain Reaction (PCR) and Gene Synthesis (GS)
3.3. High-Throughput Cloning, Transformation, and Sequencing
3.4. High-Throughput Protein Expression, Purification, and Quantification
3.5. Enzyme Activity
3.6. Statistical Analyses
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Caffall, K.H.; Mohnen, D. The structure, function, and biosynthesis of plant cell wall pectic polysaccharides. Carbohydr. Res. 2009, 344, 1879–1900. [Google Scholar] [CrossRef] [PubMed]
- Gray, K.A.; Zhao, L.; Emptage, M. Bioethanol. Curr. Opin. Chem. Biol. 2006, 10, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Lynd, L.R.; Laser, M.S.; Bransby, D.; Dale, B.E.; Davison, B.; Hamilton, R.; Himmel, M.; Keller, M.; McMillan, J.D.; Sheehan, J.; et al. How biotech can transform biofuels. Nat. Biotechnol. 2008, 26, 169–172. [Google Scholar] [CrossRef] [PubMed]
- Sticklen, M.B. Plant genetic engineering for biofuel production: Towards affordable cellulosic ethanol. Nat. Rev. Genet. 2008, 9, 433–443. [Google Scholar] [CrossRef] [PubMed]
- Himmel, M.E.; Bayer, E.A. Lignocellulose conversion to biofuels: Current challenges, global perspectives. Curr. Opin. Biotechnol. 2009, 20, 316–317. [Google Scholar] [CrossRef]
- Hervé, C.; Rogowski, A.; Blake, A.W.; Marcus, S.E.; Gilbert, H.J.; Knox, J.P. Carbohydrate-binding modules promote the enzymatic deconstruction of intact plant cell walls by targeting and proximity effects. Proc. Natl. Acad. Sci. USA 2010, 107, 15293–15298. [Google Scholar] [CrossRef]
- Chakraborty, S.; Rani, A.; Dhillon, A.; Goyal, A. Polysaccharide lyases. In Current Developments in Biotechnology and Bioengineering: Production, Isolation and Purification of Industrial Products; Elsevier B.V.: Gurgaon, India, 2017; pp. 527–539. [Google Scholar]
- Cantarel, B.L.; Coutinho, P.M.; Rancurel, C.; Bernard, T.; Lombard, V.; Henrissat, B. The carbohydrate-active enzymes database (CAZy): An expert resource for glycogenomics. Nucleic Acids Res. 2009, 37, 233–238. [Google Scholar] [CrossRef]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, 490–495. [Google Scholar] [CrossRef]
- Levasseur, A.; Drula, E.; Lombard, V.; Coutinho, P.M.; Henrissat, B. Expansion of the enzymatic repertoire of the CAZy database to integrate auxiliary redox enzymes. Biotechnol. Biofuels 2013, 6, 1–14. [Google Scholar] [CrossRef]
- Boraston, A.B.; Bolam, D.N.; Gilbert, H.J.; Davies, G.J. Carbohydrate-binding modules: Fine-tuning polysaccharide recognition. Biochem. J. 2004, 382, 769–781. [Google Scholar] [CrossRef]
- Hashimoto, H. Recent structural studies of carbohydrate-binding modules. Cell. Mol. Life Sci. 2006, 63, 2954–2967. [Google Scholar] [CrossRef] [PubMed]
- Guillén, D.; Sánchez, S.; Rodríguez-Sanoja, R. Carbohydrate-binding domains: Multiplicity of biological roles. Appl. Microbiol. Biotechnol. 2010, 85, 1241–1249. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, H.J. The biochemistry and structural biology of plant cell wall deconstruction. Plant Physiol. 2010, 153, 444–455. [Google Scholar] [CrossRef] [PubMed]
- Helbert, W.; Poulet, L.; Drouillard, S.; Mathieu, S.; Loiodice, M.; Couturier, M.; Lombard, V.; Terrapon, N.; Turchetto, J.; Vincentelli, R.; et al. Discovery of novel carbohydrate-active enzymes through the rational exploration of the protein sequences space. Proc. Natl. Acad. Sci. USA 2019, 116, 6063–6068. [Google Scholar] [CrossRef] [PubMed]
- Zeng, W.; Guo, L.; Xu, S.; Chen, J.; Zhou, J. High-throughput screening technology in industrial biotechnology. Trends Biotechnol. 2020, 38, 888–906. [Google Scholar] [CrossRef] [PubMed]
- Camilo, C.M.; Polikarpov, I. High-throughput cloning, expression and purification of glycoside hydrolases using ligation-independent cloning (LIC). Protein Expr. Purif. 2014, 99, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, V.D.; Hatahet, F.; Salo, K.E.H.; Enlund, E.; Zhang, C.; Ruddock, L.W. Pre-expression of a sulfhydryl oxidase significantly increases the yields of eukaryotic disulfide bond containing proteins expressed in the cytoplasm of E.coli. Microb. Cell Factories 2011, 10, 1. [Google Scholar] [CrossRef]
- Khow, O.; Suntrarachun, S. Strategies for production of active eukaryotic proteins in bacterial expression system. Asian Pac. J. Trop. Biomed. 2012, 2, 159–162. [Google Scholar] [CrossRef]
- Kim, S.; Lee, S.B. Rare codon clusters at 5′-end influence heterologous expression of archaeal gene in Escherichia coli. Protein Expr. Purif. 2006, 50, 49–57. [Google Scholar] [CrossRef]
- Lipinszki, Z.; Vernyik, V.; Farago, N.; Sari, T.; Puskas, L.G.; Blattner, F.R.; Posfai, G.; Gyorfy, Z. Enhancing the translational capacity of E. coli by resolving the codon bias. ACS Synth. Biol. 2018, 7, 2656–2664. [Google Scholar] [CrossRef]
- Kim, S.; Lee, S.B. Soluble expression of archaeal proteins in Escherichia coli by using fusion-partners. Protein Expr. Purif. 2008, 62, 116–119. [Google Scholar] [CrossRef]
- Piubelli, L.; Campa, M.; Temporini, C.; Binda, E.; Mangione, F.; Amicosante, M.; Terreni, M.; Marinelli, F.; Pollegioni, L. Optimizing Escherichia coli as a protein expression platform to produce Mycobacterium tuberculosis immunogenic proteins. Microb. Cell Factories 2013, 12, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Lai, G.H.; Lin, Y.C.; Tsai, Y.L.; Lien, Y.Y.; Lin, M.K.; Chen, H.J.; Chang, W.T.; Tzen, J.T.; Lee, M.S. High yield production of pigeon circovirus capsid protein in the E. coli by evaluating the key parameters needed for protein expression. BMC Vet. Res. 2014, 10, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Devi, N.; Adivitiya; Khasa, Y.P. A combinatorial approach of N-terminus blocking and codon optimization strategies to enhance the soluble expression of recombinant hIL-7 in E. coli fed-batch culture. Appl. Microbiol. Biotechnol. 2016, 100, 9979–9994. [Google Scholar] [CrossRef]
- Mirzaei, M.; Saffar, B.; Shareghi, B. Cloning, codon optimization, and expression of Yersinia intermedia phytase gene in E. coli. Iran. J. Biotechnol. 2016, 14, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Fahnert, B.; Lilie, H.; Neubauer, P. Inclusion bodies: Formation and utilisation. Adv. Biochem. Eng. Biotechnol. 2004, 89, 93–142. [Google Scholar] [PubMed]
- Zhao, M.; Wu, F.; Xu, P. Development of a rapid high-efficiency scalable process for acetylated Sus scrofa cationic trypsin production from Escherichia coli inclusion bodies. Protein Expr. Purif. 2015, 116, 120–126. [Google Scholar] [CrossRef] [PubMed]
- Mannall, G.J.; Titchener-Hooker, N.J.; Dalby, P.A. Factors affecting protein refolding yields in a fed-batch and batch-refolding system. Biotechnol. Bioeng. 2007, 97, 1523–1534. [Google Scholar] [CrossRef]
- de Groot, N.S.; Espargarö, A.; Morell, M.; Ventura, S. Studies on bacterial inclusion bodies. Future Microbiol. 2008, 3, 423–435. [Google Scholar] [CrossRef]
- Eiberle, M.K.; Jungbauer, A. Technical refolding of proteins: Do we have freedom to operate? Biotechnol. J. 2010, 5, 547–559. [Google Scholar] [CrossRef]
- Sahdev, S.; Khattar, S.K.; Saini, K.S. Production of active eukaryotic proteins through bacterial expression systems: A review of the existing biotechnology strategies. Mol. Cell. Biochem. 2008, 307, 249–264. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Upadhyay, V.; Upadhyay, A.K.; Singh, S.M.; Panda, A.K. Protein recovery from inclusion bodies of Escherichia coli using mild solubilization process. Microb. Cell Factories 2015, 14, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Voulgaridou, G.P.; Mantso, T.; Chlichlia, K.; Panayiotidis, M.I.; Pappa, A. Efficient E. coli expression strategies for production of soluble human crystallin ALDH3A1. PLoS ONE 2013, 8, 15–17. [Google Scholar] [CrossRef] [PubMed]
- Arya, R.; Sabir, J.S.M.; Bora, R.S.; Saini, K.S. Optimization of culture parameters and novel strategies to improve protein solubility. In Methods in Molecular Biology (Methods and Protocols); Humana Press: New York, NY, USA, 2015; pp. 45–63. [Google Scholar]
- Vera, A.; González-Montalbán, N.; Arís, A.; Villaverde, A. The conformational quality of insoluble recombinant proteins is enhanced at low growth temperatures. Biotechnol. Bioeng. 2007, 96, 1101–1106. [Google Scholar] [CrossRef]
- San-Miguel, T.; Pérez-Bermúdez, P.; Gavidia, I. Production of soluble eukaryotic recombinant proteins in E. coli is favoured in early log-phase cultures induced at low temperature. Springerplus 2013, 2, 89. [Google Scholar] [CrossRef]
- Huyen, D.T.; Giang, L.Q.; Hai, T.N. Expression of flagellin FLjB derived from Salmonella enterica serovar typhimurium in Escherichia coli BL21. Tap Chi Sinh Hoc 2014, 36, 506–514. [Google Scholar] [CrossRef][Green Version]
- Kaur, J.J.; Kumar, A.; Kaur, J.J. Strategies for optimization of heterologous protein expression in E. coli: Roadblocks and reinforcements. Int. J. Biol. Macromol. 2018, 106, 803–822. [Google Scholar] [CrossRef]
- Feilmeier, B.J.; Iseminger, G.; Schroeder, D.; Webber, H.; Phillips, G.J. Green fluorescent protein functions as a reporter for protein localization in Escherichia coli. J. Bacteriol. 2000, 182, 4068–4076. [Google Scholar] [CrossRef]
- Palacios, J.L.; Zaror, I.; Martίnez, P.; Uribe, F.; Opazo, P.; Socίas, T.; Gidekel, M.; Venegas, A. Subset of hybrid eukaryotic proteins is exported by the type I secretion system of Erwinia chrysanthemi. J. Bacteriol. 2001, 183, 1346–1358. [Google Scholar] [CrossRef]
- Bertone, P.; Kluger, Y.; Lan, N.; Zheng, D.; Christendat, D.; Yee, A.; Edwards, A.M.; Arrowsmith, C.H.; Montelione, G.T.; Gerstein, M. SPINE: An integrated tracking database and data mining approach for identifying feasible targets in high-throughput structural proteomics. Nucleic Acids Res. 2001, 29, 2884–2898. [Google Scholar] [CrossRef]
- Christendat, D.; Yee, A.; Dharamsi, A.; Kluger, Y.; Savchenko, A.; Cort, J.R.; Booth, V.; Mackereth, C.D.; Saridakis, V.; Ekiel, I.; et al. Structural proteomics of an archaeon. Nat. Struct. Biol. 2000, 7, 903–909. [Google Scholar]
- Niu, X.; Li, N.; Chen, D.; Wang, Z. Interconnection between the protein solubility and amino acid and dipeptide compositions. Protein Pept. Lett. 2013, 20, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Goh, C.S.; Lan, N.; Douglas, S.M.; Wu, B.; Echols, N.; Smith, A.; Milburn, D.; Montelione, G.T.; Zhao, H.; Gerstein, M. Mining the structural genomics pipeline: Identification of protein properties that affect high-throughput experimental analysis. J. Mol. Biol. 2004, 336, 115–130. [Google Scholar] [CrossRef] [PubMed]
- Petersen, T.N.; Brunak, S.; Heijne, G.V.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef]
- Yin, Y.; Mao, X.; Yang, J.; Chen, X.; Mao, F.; Xu, Y. dbCAN: A web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2012, 40, 445–451. [Google Scholar] [CrossRef] [PubMed]
- Sequeira, A.F.; Brás, J.L.A.; Fernandes, V.O.; Guerreiro, C.I.P.D.; Vincentelli, R.; Fontes, C.M.G.A. A novel platform for high-throughput gene synthesis to maximize recombinant expression in Escherichia coli. In Methods in Molecular Biology, 1620; Domingues, L., Ed.; Springer Science+Business Media: Berlin/Heidelberg, Germany, 2017; pp. 113–128. [Google Scholar]
- Sequeira, A.F.; Turchetto, J.; Saez, N.J.; Peysson, F.; Ramond, L.; Duhoo, Y.; Blémont, M.; Fernandes, V.O.; Gama, L.T.; Ferreira, L.; et al. Gene design, fusion technology and TEV cleavage conditions influence the purification of oxidized disulphide-rich venom peptides in Escherichia coli. Microb. Cell Factories 2017, 16, 4. [Google Scholar] [CrossRef]
- Turchetto, J.; Sequeira, A.F.; Ramond, L.; Peysson, F.; Brás, J.L.; Saez, N.J.; Duhoo, Y.; Blémont, M.; Guerreiro, C.I.; Quinton, L.; et al. High-throughput expression of animal venom toxins in Escherichia coli to generate a large library of oxidized disulphide-reticulated peptides for drug discovery. Microb. Cell Factories 2017, 16, 6. [Google Scholar] [CrossRef] [PubMed]
- Duhoo, Y.; GiraultJeremy, V.; Turchetto, J.; Ramond, L.; Durbesson, F.; Fourquet, P.; Nominé, Y.; Cardoso, V.; Sequeira, A.F.; Brás, J.L.A.; et al. High-throughput production of a new library of human single and tandem PDZ domains allows quantitative PDZ-peptide interaction screening through high-throughput holdup assay. In Methods in Molecular Biology; Springer: Berlin/Heidelberg, Germany, 2019. [Google Scholar]
- Correia, M.A.; Mazumder, K.; Brás, J.L.; Firbank, S.J.; Zhu, Y.; Lewis, R.J.; York, W.S.; Fontes, C.M.; Gilbert, H.J. Structure and function of an arabinoxylan-specific xylanase. J. Biol Chem. 2011, 286, 22510–22520. [Google Scholar] [CrossRef]
- Hüttner, S.; Klaubauf, S.; de Vries, R.P.; Olsson, L. Characterisation of three fungal glucuronoyl esterases on glucuronic acid ester model compounds. Appl. Microbiol. Biotechnol. 2017, 101, 5301–5311. [Google Scholar] [CrossRef]
- SAS. SAS User’s Guide: Statistics; SAS Institute: Cary, NC, USA, 2004. [Google Scholar]




| INS | SOL | SEM | p-Value | |
|---|---|---|---|---|
| (n) | 310 | 1166 | ||
| Amino acid groups (%) | ||||
| Non-polar amino acids 1 | 51.4 a | 50.2 b | 0.31 | 0.002 |
| Polar neutral amino acids 2 | 26.0 | 26.0 | 0.38 | 0.976 |
| Negatively charged amino acids 3 | 11.2 b | 12.2 a | 0.17 | 0.001 |
| Positively charged amino acids 4 | 11.4 | 11.5 | 0.17 | 0.420 |
| Amino acid contents (%) | ||||
| Isoleucine (I) | 5.0 | 5.2 | 0.11 | 0.051 |
| Leucine (L) | 7.6 | 7.4 | 0.15 | 0.213 |
| Lysine (K) | 4.4 b | 5.0 a | 0.15 | 0.001 |
| Methionine (M) | 1.9 | 2.0 | 0.06 | 0.138 |
| Phenylalanine (F) | 4.1 | 4.1 | 0.08 | 0.828 |
| Threonine (T) | 6.2 | 6.0 | 0.14 | 0.231 |
| Tryptophan (W) | 2.3 a | 2.2 b | 0.07 | 0.042 |
| Valine (V) | 6.5 | 6.5 | 0.10 | 0.610 |
| Arginine (R) | 4.7 a | 4.3 b | 0.12 | 0.005 |
| Histidine (H) | 2.3 | 2.3 | 0.07 | 0.659 |
| Alanine (A) | 9.0 a | 8.5 b | 0.20 | 0.014 |
| Asparagine (N) | 5.1 | 5.2 | 0.15 | 0.511 |
| Aspartic acid (D) | 6.0 b | 6.5 a | 0.10 | 0.001 |
| Cysteine (C) | 1.0 | 1.0 | 0.07 | 0.784 |
| Glutamic acid (E) | 5.2 b | 5.6 a | 0.12 | 0.002 |
| Glutamine (Q) | 3.6 | 3.4 | 0.09 | 0.287 |
| Glycine (G) | 8.9 a | 8.6 b | 0.13 | 0.016 |
| Proline (P) | 5.0 | 4.9 | 0.10 | 0.418 |
| Serine (S) | 6.6 | 6.5 | 0.15 | 0.246 |
| Tyrosine (Y) | 4.5 b | 4.9 a | 0.11 | 0.001 |
| CAZy Family | EC Number |
|---|---|
| AA1 | 1.10.3.2 |
| AA3 | 1.1.3.12 |
| AA7 | 1.1.3.- |
| AA10 | 1.-.-.-/1.14.99.54 |
| AA NC | 1.10.3.-/1.3.3.5 |
| CE1 | 3.1.1.73 |
| CE2 | 3.1.1.72 |
| CE3 | 3.1.1.72 |
| CE4 | 3.2.1.8/3.5.1.- |
| CE6 | 3.1.1.72 |
| CE7 | 3.1.1.41/3.1.1.72 |
| CE8 | 3.1.1.11 |
| CE9 | 3.5.1.25 |
| CE11 | 3.5.1.- |
| CE12 | 3.1.1.-/3.1.1.72 |
| CE14 | 3.5.1.-/3.5.1.89 |
| CE15 | 3.1.1.-/3.1.1.72 |
| GH1 | 3.2.1.-/3.2.1.21/3.2.1.23/3.2.1.25/3.2.1.37/3.2.1.74/3.2.1.85/3.2.1.86 |
| GH2 | 3.2.1.23/3.2.1.25/3.2.1.31/3.2.1.165 |
| GH3 | 3.2.1.21/3.2.1.37/3.2.1.45/3.2.1.52/3.2.1.74/3.2.1.120 |
| GH4 | 3.2.1.20/3.2.1.22/3.2.1.67/3.2.1.86/3.2.1.122/3.2.1.139 |
| GH5 | 3.2.1.4/3.2.1.8/3.2.1.73/3.2.1.74/3.2.1.78/3.2.1.91/3.2.1.123/3.2.1.132/3.2.1.151 |
| GH6 | 3.2.1.4 |
| GH8 | 3.2.1.4/3.2.1.73/3.2.1.132/3.2.1.156 |
| GH9 | 3.2.1.-/3.2.1.4/3.2.1.91/3.2.1.151/3.2.1.165 |
| GH10 | 3.2.1.4/3.2.1.8 |
| GH11 | 3.2.1.8 |
| GH12 | 3.2.1.4/3.2.1.151 |
| GH13 | 2.4.1.4/2.4.1.7/2.4.1.18/2.4.1.19/2.4.1.25/3.2.1.1/3.2.1.4/3.2.1.10/3.2.1.20/3.2.1.41/3.2.1.68/3.2.1.70/3.2.1.93/3.2.1.98/3.2.1.133/3.2.1.135/3.2.1.141/5.4.99.11/5.4.99.15/5.4.99.16 |
| GH14 | 3.2.1.2 |
| GH15 | 3.2.1.3/3.2.1.28/3.2.1.70 |
| GH16 | 3.2.1.-/3.2.1.4/3.2.1.6/3.2.1.39/3.2.1.73/3.2.1.81/3.2.1.83/3.2.1.103/3.2.1.178 |
| GH17 | 2.4.1.- |
| GH18 | 3.2.1.-/3.2.1.14/3.2.1.96 |
| GH19 | 3.2.1.14 |
| GH20 | 3.2.1.52 |
| GH23 | 3.2.1.17/4.2.2.n1 |
| GH24 | 3.2.1.17 |
| GH25 | 3.2.1.17 |
| GH26 | 3.2.1.78/3.2.1.100 |
| GH27 | 3.2.1.88/3.2.1.94 |
| GH28 | 3.2.1.15/3.2.1.67/3.2.1.82 |
| GH29 | 3.2.1.51/3.2.1.111 |
| GH30 | 3.2.1.8/3.2.1.31/3.2.1.38/3.2.1.136/3.2.1.164 |
| GH31 | 3.2.1.-/3.2.1.20/3.2.1.84/2.4.1.161/3.2.1.177 |
| GH32 | 3.2.1.26/3.2.1.64/3.2.1.65/3.2.1.80/3.2.1.153/4.2.2.16 |
| GH33 | 3.2.1.-/3.2.1.18/2.4.1.- |
| GH35 | 3.2.1.23/3.2.1.165 |
| GH36 | 3.2.1.22/3.2.1.49 |
| GH37 | 3.2.1.28 |
| GH38 | 3.2.1.-/3.2.1.24 |
| GH39 | 3.2.1.37 |
| GH42 | 3.2.1.-/3.2.1.23 |
| GH43 | 3.2.1.-/3.2.1.37/3.2.1.55/3.2.1.99 |
| GH44 | 3.2.1.4 |
| GH45 | 3.2.1.4 |
| GH46 | 3.2.1.132 |
| GH47 | 3.2.1.113 |
| GH48 | 3.2.1.4/3.2.1.176 |
| GH49 | 3.2.1.11/3.2.1.95 |
| GH50 | 3.2.1.23/3.2.1.81 |
| GH51 | 3.2.1.55 |
| GH52 | 3.2.1.37 |
| GH53 | 3.2.1.89 |
| GH55 | 3.2.1.39 |
| GH57 | 2.4.1.18/2.4.1.25/3.2.1.1/3.2.1.41/3.2.1.54 |
| GH62 | 3.2.1.55 |
| GH63 | 3.2.1.20/3.2.1.84/3.2.1.170 |
| GH64 | 3.2.1.39 |
| GH65 | 2.4.1.8/2.4.1.64/2.4.1.216/2.4.1.230/2.4.1.279/2.4.1.282 |
| GH66 | 3.2.1.11 |
| GH67 | 3.2.1.139 |
| GH68 | 2.4.1.9/2.4.1.10/3.2.1.26 |
| GH70 | 2.4.1.5/2.4.1.140/2.4.4.- |
| GH73 | 3.2.1.- / |
| GH74 | 3.2.1.-/3.2.1.4 |
| GH75 | 3.2.1.132 |
| GH76 | 3.2.1.101 |
| GH77 | 2.4.1.25 |
| GH78 | 3.2.1.40 |
| GH79 | 3.2.1.31 |
| GH80 | 3.2.1.132 |
| GH81 | 3.2.1.39 |
| GH82 | 3.2.1.157 |
| GH84 | 3.2.1.35/3.2.1.52/3.2.1.169 |
| GH85 | 3.2.1.96 |
| GH86 | 3.2.1.81/3.2.1.178 |
| GH87 | 3.2.1.59/3.2.1.61 |
| GH88 | 3.2.1.- |
| GH91 | 4.2.2.18 |
| GH92 | 3.2.1.-/3.2.1.24/3.2.1.113 |
| GH94 | 2.4.1.-/2.4.1.20/2.4.1.49 |
| GH95 | 3.2.1.51/3.2.1.63 |
| GH97 | 3.2.1.3/3.2.1.20/3.2.1.22 |
| GH98 | 3.2.1.102 |
| GH99 | 3.2.1.130 |
| GH100 | 3.2.1.26 |
| GH101 | 3.2.1.97 |
| GH102 | 4.2.2.n1 |
| GH103 | 4.2.2.n1 |
| GH104 | 4.2.2.n1 |
| GH105 | 3.2.1.-/3.2.1.172 |
| GH106 | 3.2.1.40 |
| GH107 | 3.2.1.- |
| GH108 | 3.2.1.17 |
| GH109 | 3.2.1.49 |
| GH110 | 3.2.1.-/3.2.1.22 |
| GH111 | 3.2.1.- |
| GH112 | 2.4.1.211/2.4.1.247 |
| GH113 | 3.2.1.78 |
| GH114 | 3.2.1.109 |
| GH115 | 3.2.1.- |
| GH116 | 3.2.1.21/3.2.1.37/3.2.1.52 |
| GH117 | 3.2.1.- |
| GH118 | 3.2.1.81 |
| GH119 | 3.2.1.1 |
| GH120 | 3.2.1.37 |
| GH121 | 3.2.1.- |
| GH122 | 3.2.1.20 |
| GH123 | 3.2.1.53 |
| GH125 | 3.2.1.- |
| GH126 | 3.2.1.- |
| GH127 | 3.2.1.185 |
| GH129 | 3.2.1.49 |
| GH130 | 2.4.1.281/2.4.1.319 |
| GH134 | 3.2.1.78 |
| GH137 | 3.2.1.31 |
| GH142 | 3.2.1.185 |
| GH143 | 3.2.1.185 |
| PL1 | 4.2.2.2/4.2.2.10 |
| PL2 | 4.2.2.2/4.2.2.9 |
| PL3 | 4.2.2.2 |
| PL4 | 4.2.2.- |
| PL5 | 4.2.2.3 |
| PL6 | 4.2.2.-/4.2.2.3 |
| PL7 | 4.2.2.-/4.2.2.3/4.2.2.11 |
| PL8 | 4.2.2.1/4.2.2.5/4.2.2.12/4.2.2.20 |
| PL9 | 4.2.2.2/4.2.2.9 |
| PL10 | 4.2.2.2 |
| PL11 | 4.2.2.23/4.2.2.24 |
| PL12 | 4.2.2.8 |
| PL13 | 4.2.2.7 |
| PL15 | 4.2.2.-/4.2.2.3 |
| PL17 | 4.2.2.- |
| PL18 | 4.2.2.3/4.2.2.11 |
| PL21 | 4.2.2.7/4.2.2.8 |
| PL22 | 4.2.2.6 |
| PL24 | 4.2.2.- |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cardoso, V.; Brás, J.L.A.; Costa, I.F.; Ferreira, L.M.A.; Gama, L.T.; Vincentelli, R.; Henrissat, B.; Fontes, C.M.G.A. Generation of a Library of Carbohydrate-Active Enzymes for Plant Biomass Deconstruction. Int. J. Mol. Sci. 2022, 23, 4024. https://doi.org/10.3390/ijms23074024
Cardoso V, Brás JLA, Costa IF, Ferreira LMA, Gama LT, Vincentelli R, Henrissat B, Fontes CMGA. Generation of a Library of Carbohydrate-Active Enzymes for Plant Biomass Deconstruction. International Journal of Molecular Sciences. 2022; 23(7):4024. https://doi.org/10.3390/ijms23074024
Chicago/Turabian StyleCardoso, Vânia, Joana L. A. Brás, Inês F. Costa, Luís M. A. Ferreira, Luís T. Gama, Renaud Vincentelli, Bernard Henrissat, and Carlos M. G. A. Fontes. 2022. "Generation of a Library of Carbohydrate-Active Enzymes for Plant Biomass Deconstruction" International Journal of Molecular Sciences 23, no. 7: 4024. https://doi.org/10.3390/ijms23074024
APA StyleCardoso, V., Brás, J. L. A., Costa, I. F., Ferreira, L. M. A., Gama, L. T., Vincentelli, R., Henrissat, B., & Fontes, C. M. G. A. (2022). Generation of a Library of Carbohydrate-Active Enzymes for Plant Biomass Deconstruction. International Journal of Molecular Sciences, 23(7), 4024. https://doi.org/10.3390/ijms23074024






