Carbon Nanotube-Mediated Plasmid DNA Delivery in Rice Leaves and Seeds
Abstract
:1. Introduction
2. Results
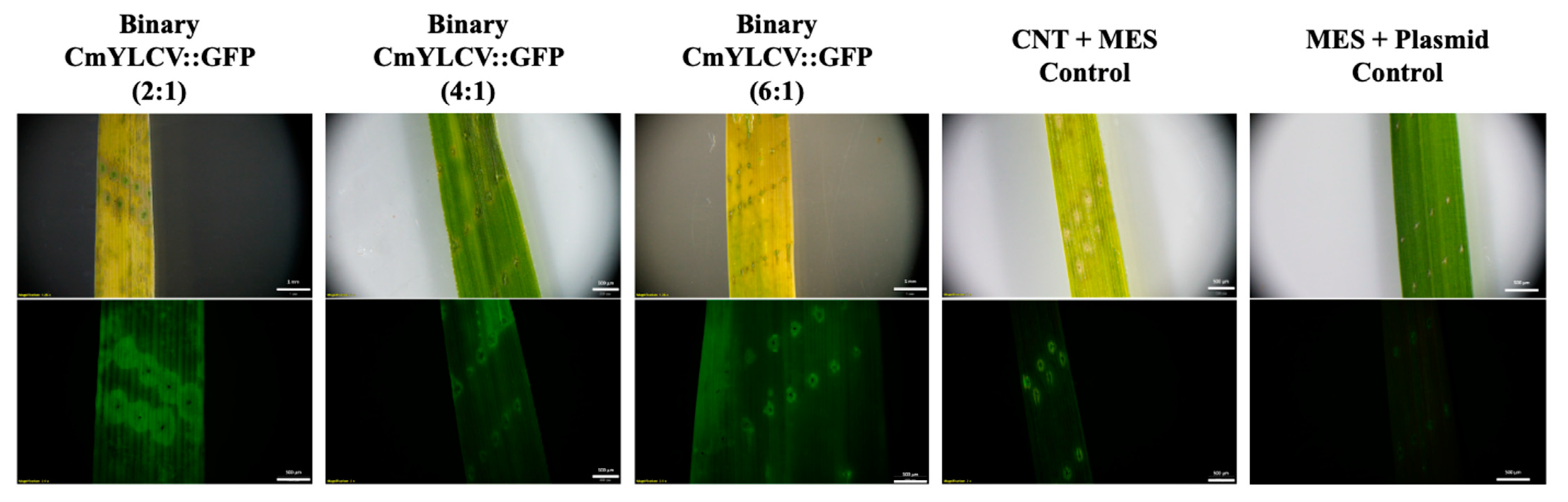
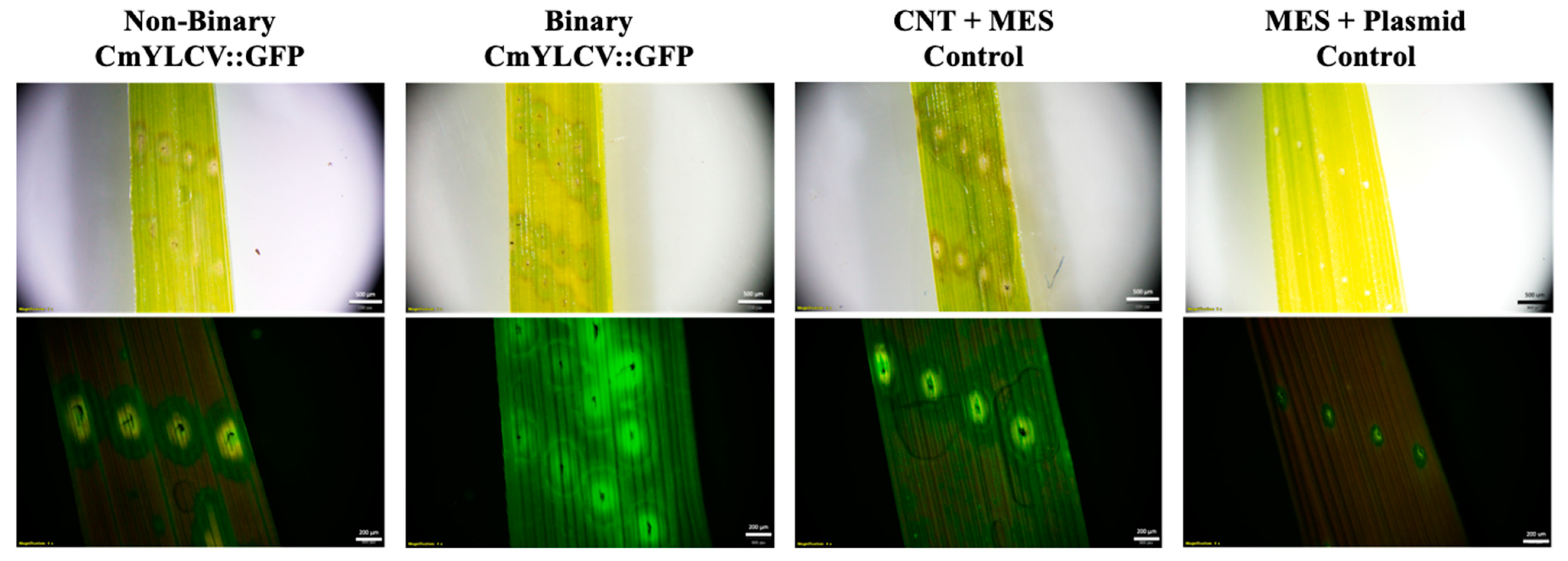
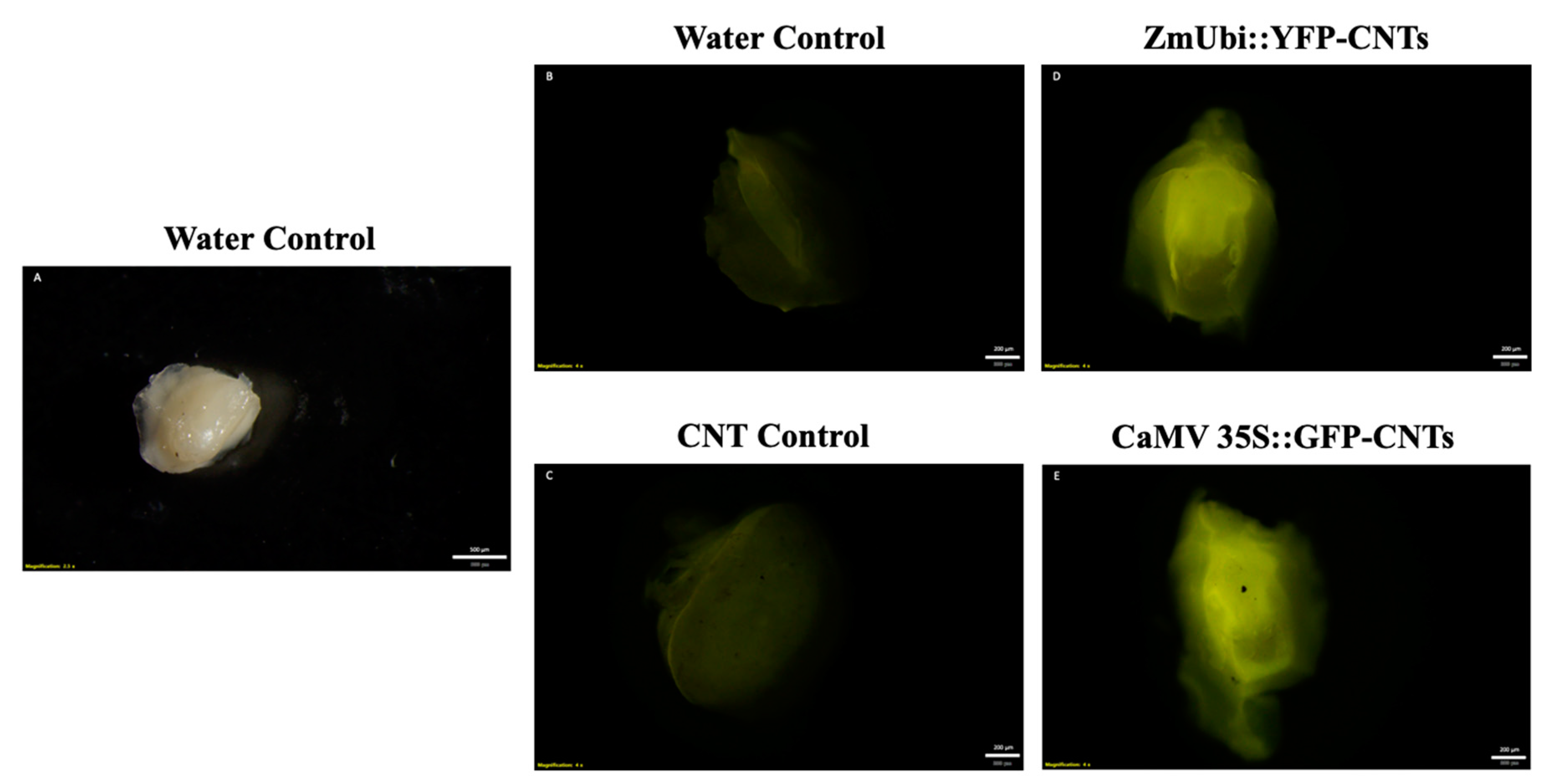
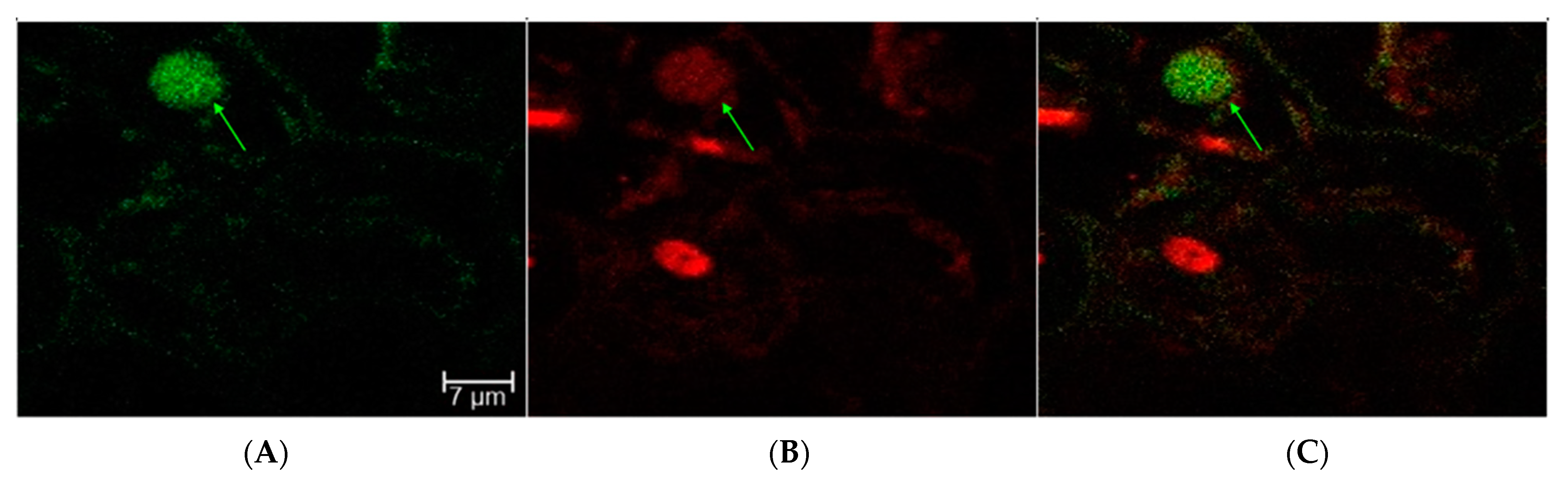
2.1. Transient Fluorescence in Rice Leaves and Embryos after CNT-Mediated Plasmid Delivery
2.2. Transient GUS Expression after CNT-Mediated Plasmid Delivery
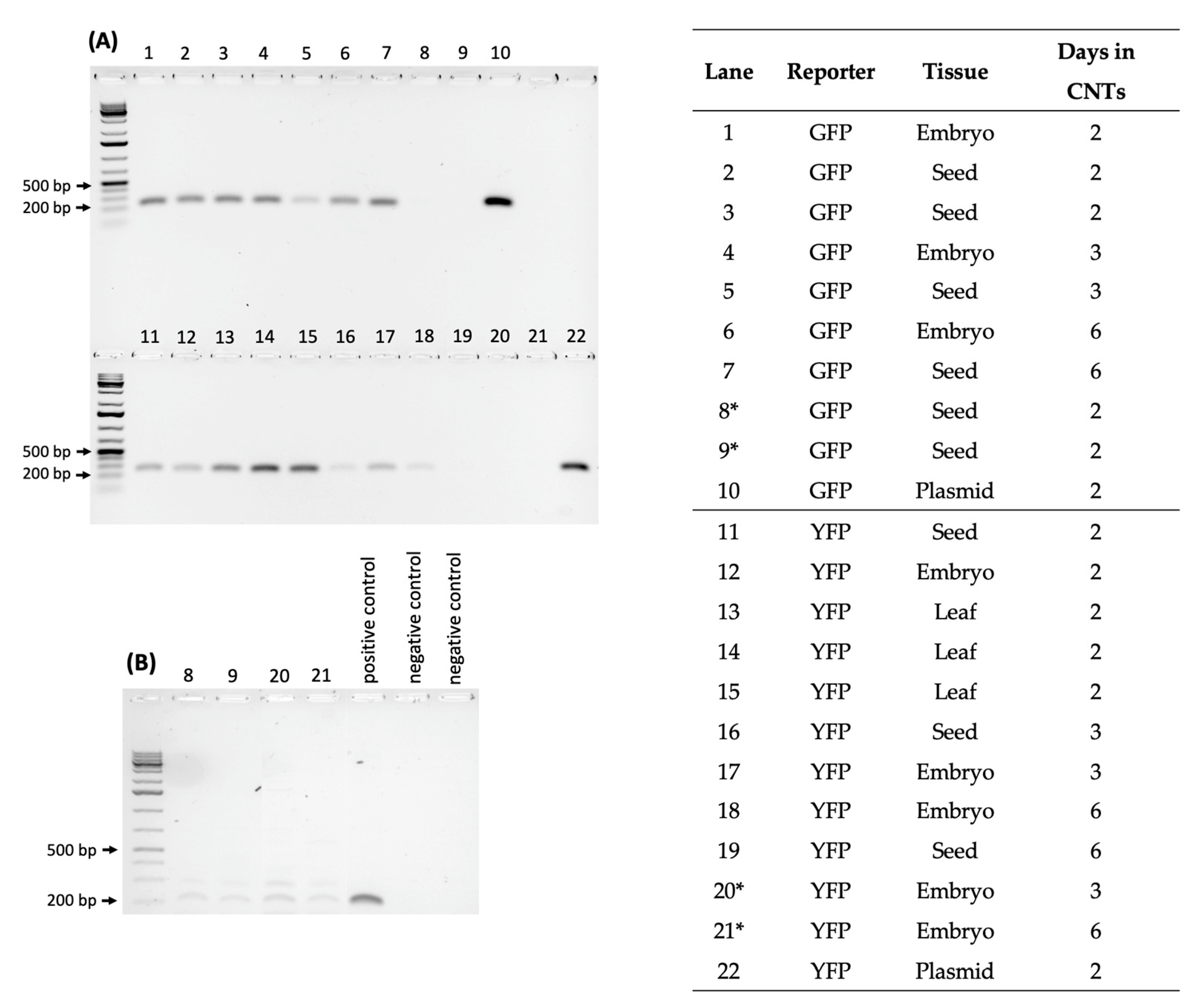
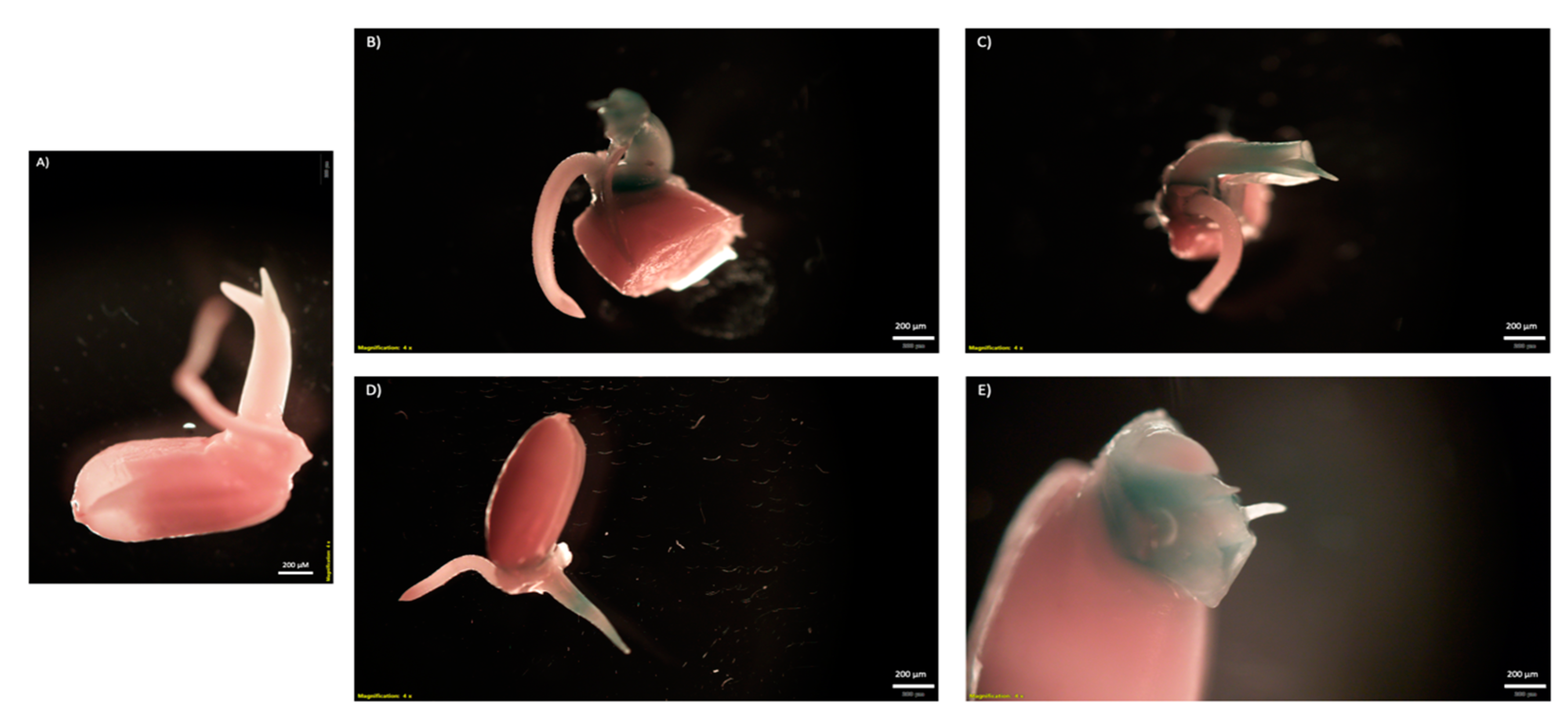
2.3. Testing CNT Delivery of CRISPR-Cas for Gene Editing of Rice Seeds and Embryos
2.4. DNA Sequencing of Phenotypically Aberrant Seedlings
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Plasmid-PEI-CNT Preparation
4.3. Plasmid Preparation
4.4. Leaf Infiltrations with CNTs
4.5. Rice Embryo Infiltrations with CNTs and Reporter Plasmids
4.6. Transcript Analysis
4.7. Confocal Imaging of NLS-GFP
4.8. GUSPlus Analysis
4.9. CRISPR/Cas9-CNT Seed and Embryo Infiltration
4.10. OsPDS Knockout Sequencing Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAO. The Future of Food and Agriculture: Trends and Challenges; FAO: Rome, Italy, 2017; Volume 296, pp. 1–180. [Google Scholar]
- United Nations. World Population Prospects: The 2015 Revision, Key Findings and Advance Tables. 2015. Available online: https://population.un.org/wpp/Publications/Files/Key_Findings_WPP_2015.pdf (accessed on 5 December 2020).
- Chen, K.; Wang, Y.; Zhang, R.; Zhang, H.; Gao, C. CRISPR/Cas Genome Editing and Precision Plant Breeding in Agriculture. Annu. Rev. Plant Biol. 2019, 70, 667–697. [Google Scholar] [CrossRef] [PubMed]
- Scheben, A.; Wolter, F.; Batley, J.; Puchta, H.; Edwards, D. Towards CRISPR/Cas crops–Bringing together genomics and genome editing. New Phytol. 2017, 216, 682–698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altpeter, F.; Springer, N.M.; Bartley, L.E.; Blechl, A.E.; Brutnell, T.P.; Citovsky, V.; Conrad, L.J.; Gelvin, S.B.; Jackson, D.P.; Kausch, A.P.; et al. Advancing Crop Transformation in the Era of Genome Editing. Plant Cell 2016, 28, 1510–1520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Atkins, P.A.; Voytas, D.F. Overcoming bottlenecks in plant gene editing. Curr. Opin. Plant Biol. 2020, 54, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Nasti, R.A.; Voytas, D.F. Attaining the promise of plant gene editing at scale. Proc. Natl. Acad. Sci. USA 2021, 118, e2004846117. [Google Scholar] [CrossRef]
- Demirer, G.S.; Zhang, H.; Matos, J.L.; Goh, N.S.; Cunningham, F.J.; Sung, Y.; Chang, R.; Aditham, A.J.; Chio, L.; Cho, M.-J.; et al. High aspect ratio nanomaterials enable delivery of functional genetic material without DNA integration in mature plants. Nat. Nanotechnol. 2019, 14, 456–464. [Google Scholar] [CrossRef]
- Demirer, G.S.; Silva, T.N.; Jackson, C.T.; Thomas, J.B.; Ehrhardt, D.W.; Rhee, S.Y.; Mortimer, J.C.; Landry, M.P. Nanotechnology to advance CRISPR–Cas genetic engineering of plants. Nat. Nanotechnol. 2021, 16, 243–250. [Google Scholar] [CrossRef]
- Kungulovski, G.; Jeltsch, A. Epigenome Editing: State of the Art, Concepts, and Perspectives. Trends Genet. 2015, 32, 101–113. [Google Scholar] [CrossRef]
- Marzec, M.; Brąszewska-Zalewska, A.; Hensel, G. Prime Editing: A New Way for Genome Editing. Trends Cell Biol. 2020, 30, 257–259. [Google Scholar] [CrossRef]
- Chen, Q.; Lai, H. Gene Delivery into Plant Cells for Recombinant Protein Production. BioMed Res. Int. 2015, 2015, 932161. [Google Scholar] [CrossRef] [Green Version]
- Ledford, H. Super-precise new CRISPR tool could tackle a plethora of genetic diseases. Nature 2019, 574, 464–465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, Y.; Biswas, S.; Kim, B.; Bailey-Serres, J.; Septiningsih, E. Improved Transformation and Regeneration of Indica Rice: Disruption of SUB1A as a Test Case via CRISPR-Cas9. Int. J. Mol. Sci. 2021, 22, 6989. [Google Scholar] [CrossRef] [PubMed]
- Biswas SWahl, N.J.; Thomson, M.J.; Cason, J.M.; McCutchen, B.F.; Septiningsih, E.M. Optimization of protoplast isolation and transformation for a pilot study of genome editing in peanut by targeting the allergen gene Ara h 2. Int. J. Mol. Sci. 2022. under review. [Google Scholar] [CrossRef]
- Himmel, M.E.; Ding, S.-Y.; Johnson, D.K.; Adney, W.S.; Nimlos, M.R.; Brady, J.W.; Foust, T.D. Biomass Recalcitrance: Engineering Plants and Enzymes for Biofuels Production. Science 2007, 315, 804–807. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Javed, M.R.; Noman, M.; Shahid, M.; Ahmed, T.; Khurshid, M.; Rashid, M.H.; Ismail, M.; Sadaf, M.; Khan, F. Current situation of biofuel production and its enhancement by CRISPR/Cas9-mediated genome engineering of microbial cells. Microbiol. Res. 2018, 219, 1–11. [Google Scholar] [CrossRef]
- Turnbull, C.; Lillemo, M.; Hvoslef-Eide, T.A.K. Global Regulation of Genetically Modified Crops Amid the Gene Edited Crop Boom–A Review. Front. Plant Sci. 2021, 12, 258. [Google Scholar] [CrossRef]
- Liu, J.; Nannas, N.J.; Fu, F.-F.; Shi, J.; Aspinwall, B.; Parrott, W.A.; Dawe, R.K. Genome-Scale Sequence Disruption Following Biolistic Transformation in Rice and Maize. Plant Cell 2019, 31, 368–383. [Google Scholar] [CrossRef] [Green Version]
- Banakar, R.; Wang, K. Biolistic Transformation of Japonica Rice Varieties. In Biolistic DNA Delivery in Plants: Methods and Protocols; Rustgi, S., Luo, H., Eds.; Springer: New York, NY, USA, 2020; pp. 163–176. [Google Scholar] [CrossRef]
- Hwang, H.-H.; Yu, M.; Lai, E.-M. Agrobacterium-Mediated Plant Transformation: Biology and Applications. Arab. Book 2017, 15, e0186. [Google Scholar] [CrossRef] [Green Version]
- Karthikeyan, A.; Pandian, S.T.K.; Ramesh, M. High frequency plant regeneration from embryogenic callus of a popular indica rice (Oryza sativa L.). Physiol. Mol. Biol. Plants Int. J. Funct. Plant Biol. 2009, 15, 371–375. [Google Scholar] [CrossRef] [Green Version]
- Wardrop, J.; Lowe, K.; Power, J.; Davey, M. Perfluorochemicals and plant biotechnology: An improved protocol for protoplast culture and plant regeneration in rice (Oryza sativa L.). J. Biotechnol. 1996, 50, 47–54. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. Cell Mol. Biol. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zlobin, N.E.; Lebedeva, M.V.; Taranov, V.V. CRISPR/Cas9 genome editing through in planta transformation. Crit. Rev. Biotechnol. 2020, 40, 153–168. [Google Scholar] [CrossRef] [PubMed]
- Qin, G.; Gu, H.; Ma, L.; Peng, Y.; Deng, X.W.; Chen, Z.; Qu, L.-J. Disruption of phytoene desaturase gene results in albino and dwarf phenotypes in Arabidopsis by impairing chlorophyll, carotenoid, and gibberellin biosynthesis. Cell Res. 2007, 17, 471–482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawahara, Y.; de la Bastide, M.; Hamilton, J.P.; Kanamori, H.; McCombie, W.R.; Ouyang, S.; Schwartz, D.C.; Tanaka, T.; Wu, J.; Zhou, S.; et al. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 2013, 6, 4. [Google Scholar] [CrossRef] [Green Version]
- UniProt Consortium. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef] [Green Version]
- Decaestecker, W.; Buono, R.A.; Pfeiffer, M.L.; Vangheluwe, N.; Jourquin, J.; Karimi, M.; Van Isterdael, G.; Beeckman, T.; Nowack, M.K.; Jacobs, T.B. CRISPR-TSKO: A Technique for Efficient Mutagenesis in Specific Cell Types, Tissues, or Organs in Arabidopsis. Plant Cell 2019, 31, 2868–2887. [Google Scholar] [CrossRef] [Green Version]
- Xu, R.; Qin, R.; Li, H.; Li, D.; Li, L.; Wei, P.; Yang, J. Generation of targeted mutant rice using a CRISPR-Cpf1 system. Plant Biotechnol. J. 2017, 15, 713–717. [Google Scholar] [CrossRef] [Green Version]
- Demirer, G.S.; Zhang, H.; Goh, N.S.; González-Grandío, E.; Landry, M.P. Carbon nanotube–mediated DNA delivery without transgene integration in intact plants. Nat. Protoc. 2019, 14, 2954–2971. [Google Scholar] [CrossRef]
- Cervera, M. Histochemical and Fluorometric Assays for uidA (GUS) Gene Detection. In Transgenic Plants: Methods and Protocols; Peña, L., Ed.; Humana Press: Totowa, NJ, USA, 2004; pp. 203–213. [Google Scholar] [CrossRef]
- Synthego Performance Analysis, ICE Analysis. v3.0. Synthego. 2019. Available online: https://www.synthego.com/products/bioinformatics/crispr-analysis (accessed on 27 September 2021).
- Čermák, T.; Curtin, S.J.; Gil-Humanes, J.; Čegan, R.; Kono, T.J.Y.; Konečná, E.; Belanto, J.J.; Starker, C.G.; Mathre, J.W.; Greenstein, R.L.; et al. A Multipurpose Toolkit to Enable Advanced Genome Engineering in Plants. Plant Cell 2017, 29, 1196–1217. [Google Scholar] [CrossRef] [Green Version]
- Molina-Risco, M.; Ibarra, O.; Faion-Molina, M.; Kim, B.; Septiningsih, E.M.; Thomson, M.J. Optimizing Agrobacterium-Mediated Transformation and CRISPR-Cas9 Gene Editing in the tropical japonica Rice Variety Presidio. Int. J. Mol. Sci. 2021, 22, 10909. [Google Scholar] [CrossRef]
- Andrieu, A.; Breitler, J.C.; Siré, C.; Meynard, D.; Gantet, P.; Guiderdoni, E. An in planta, Agrobacterium-mediated transient gene expression method for inducing gene silencing in rice (Oryza sativa L.) leaves. Rice 2012, 5, 23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamura, K.; Shimada, T.; Ono, E.; Tanaka, Y.; Nagatani, A.; Higashi, S.-I.; Watanabe, M.; Nishimura, M.; Hara-Nishimura, I. Why green fluorescent fusion proteins have not been observed in the vacuoles of higher plants. Plant J. 2003, 35, 545–555. [Google Scholar] [CrossRef] [PubMed]
- Valdez, M.; Cabrera-Ponce, J.L.; Sudhakar, D.; Herrera-Estrella, L.; Christou, P. Transgenic Central American, West African and Asian Elite Rice Varieties Resulting from Particle Bombardment of Foreign DNA into Mature Seed-derived Explants Utilizing Three Different Bombardment Devices. Ann. Bot. 1998, 82, 795–801. [Google Scholar] [CrossRef]
- Mazithulela, G.; Sudhakar, D.; Heckel, T.; Mehlo, L.; Christou, P.; Davies, J.; Boulton, M. The maize streak virus coat protein transcription unit exhibits tissue-specific expression in transgenic rice. Plant Sci. Int. J. Exp. Plant Biol. 2000, 155, 21–29. [Google Scholar] [CrossRef]
- Dutt, M.; Li, Z.T.; Dhekney, S.A.; Gray, D.J. Transgenic plants from shoot apical meristems of Vitis vinifera L. “Thompson Seedless” via Agrobacterium-mediated transformation. Plant Cell Rep. 2007, 26, 2101–2110. [Google Scholar] [CrossRef]
- Sato, S.; Newell, C.; Kolacz, K.; Tredo, L.; Finer, J.; Hinchee, M. Stable transformation via particle bombardment in two different soybean regeneration systems. Plant Cell Rep. 1993, 12, 408–413. [Google Scholar] [CrossRef] [PubMed]
- Caldana, C.; Scheible, W.-R.; Mueller-Roeber, B.; Ruzicic, S. A quantitative RT-PCR platform for high-throughput expression profiling of 2500 rice transcription factors. Plant Methods 2007, 3, 7. [Google Scholar] [CrossRef] [Green Version]
- Smith, P.J.; Wiltshire, M.; Errington, R.J. DRAQ 5 Labeling of Nuclear DNA in Live and Fixed Cells. Curr. Protoc. Cytom. 2004, 28, 7–25. [Google Scholar] [CrossRef]
- Slamet-Loedin, I.H.; Chadha-Mohanty, P.; Torrizo, L. Agrobacterium-Mediated Transformation: Rice Transformation. Methods Mol. Biol. 2013, 1099, 261–271. [Google Scholar] [CrossRef]
- Nekrasov, V.; Wang, C.; Win, J.; Lanz, C.; Weigel, D.; Kamoun, S. Rapid generation of a transgene-free powdery mildew resistant tomato by genome deletion. Sci. Rep. 2017, 7, 482. [Google Scholar] [CrossRef] [Green Version]
- Hsiau, T.; Maures, T.; Waite, K.; Yang, J.; Kelso, R.; Holden, K.; Stoner, R. Inference of CRISPR Edits from Sanger trace data. bioRxiv 2018, 251082. [Google Scholar] [CrossRef]
- Synthego’s ICE Tool Limitations. Synthego. 2020. Available online: https://www.synthego.com/help/ice-limitations (accessed on 17 September 2021).

| Vector Backbone | Reporter Gene | Promoter | Size (kb) |
|---|---|---|---|
| pPTN (binary) | YFP | ZmUbi | 12.0 |
| pCAMBIA1305.1 (binary) | GUSPlus | CaMV 35S | 11.8 |
| pTRANS210 (binary) | GFP | CmYLCV | 10.5 |
| pTRANS100 (nonbinary) | OsPDS sgRNA, Cas9 | OsU3, ZmUbi | 9.3 |
| pUC19 (nonbinary) | GUSPlus | CaMV 35S | 5.4 |
| pENTR (nonbinary) | Nuclear-Localized GFP | EL2 | 5.1 |
| pTRANS100 (nonbinary) | GFP | CmYLCV | 3.7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dunbar, T.; Tsakirpaloglou, N.; Septiningsih, E.M.; Thomson, M.J. Carbon Nanotube-Mediated Plasmid DNA Delivery in Rice Leaves and Seeds. Int. J. Mol. Sci. 2022, 23, 4081. https://doi.org/10.3390/ijms23084081
Dunbar T, Tsakirpaloglou N, Septiningsih EM, Thomson MJ. Carbon Nanotube-Mediated Plasmid DNA Delivery in Rice Leaves and Seeds. International Journal of Molecular Sciences. 2022; 23(8):4081. https://doi.org/10.3390/ijms23084081
Chicago/Turabian StyleDunbar, Tia, Nikolaos Tsakirpaloglou, Endang M. Septiningsih, and Michael J. Thomson. 2022. "Carbon Nanotube-Mediated Plasmid DNA Delivery in Rice Leaves and Seeds" International Journal of Molecular Sciences 23, no. 8: 4081. https://doi.org/10.3390/ijms23084081
APA StyleDunbar, T., Tsakirpaloglou, N., Septiningsih, E. M., & Thomson, M. J. (2022). Carbon Nanotube-Mediated Plasmid DNA Delivery in Rice Leaves and Seeds. International Journal of Molecular Sciences, 23(8), 4081. https://doi.org/10.3390/ijms23084081






