In Vitro Antimicrobial Potential of CAPE and Caffeamide Derivatives against Oral Microbes
Abstract
:1. Introduction
2. Results
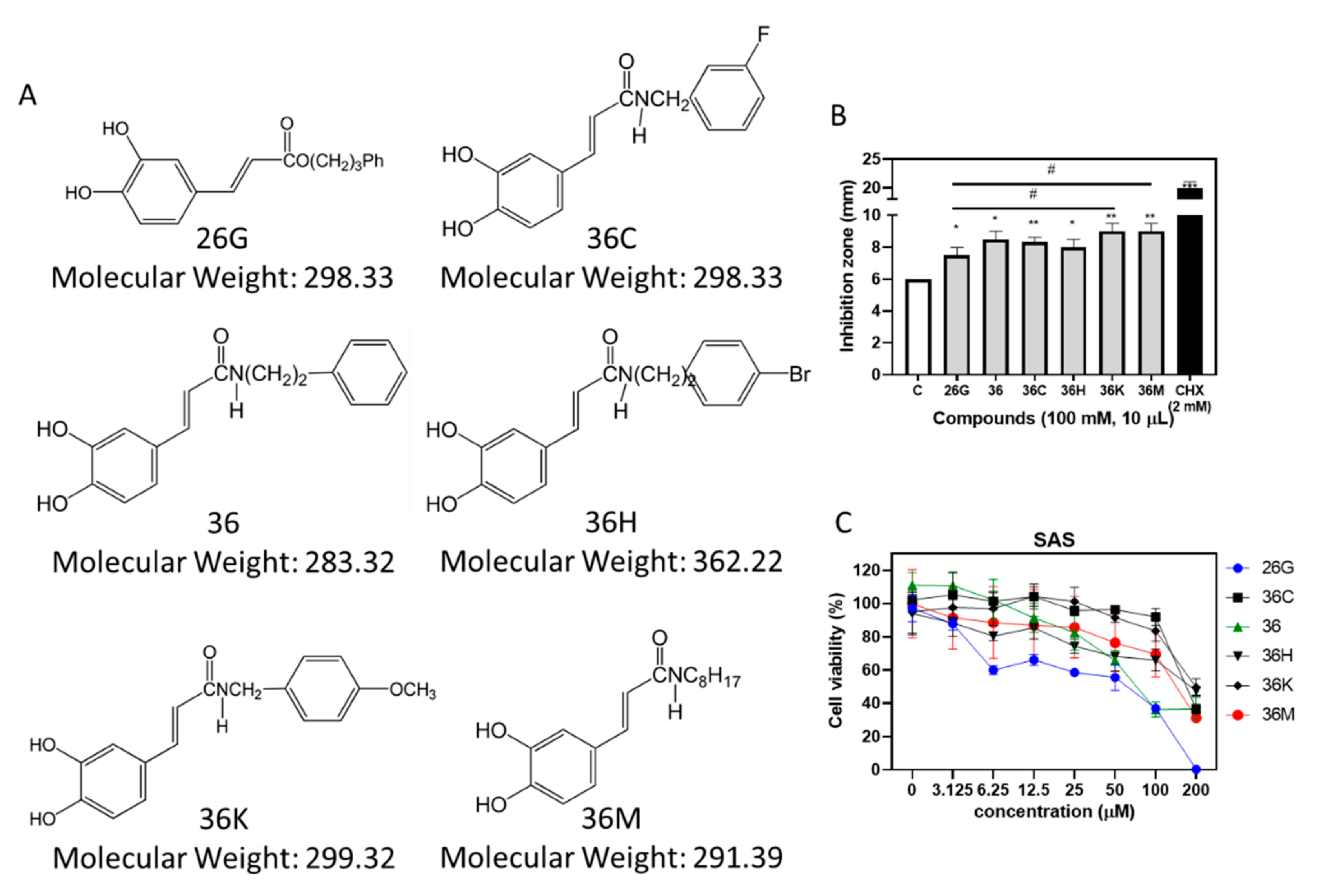
2.1. 26G and 36M Showed the Best Potential for Antibacterial and Anticancer Activities, Respectively, than other Caffeamide Derivatives
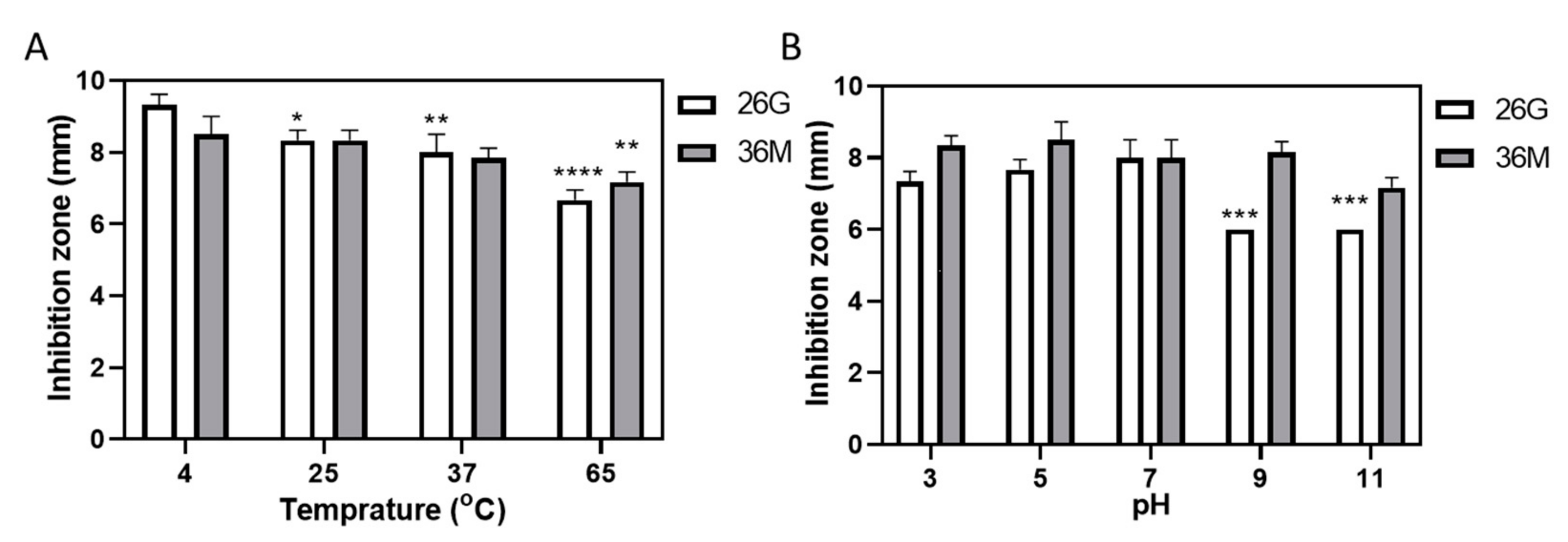
2.2. 36M Showed a Stable Antimicrobial Effect over a Wide pH Range but Weakened after Heating
2.3. Determining the Minimum Inhibitory Concentration (MIC) and Minimum Bactericidal Concentration (MBC) of 26G and 36M on Oral Microbes
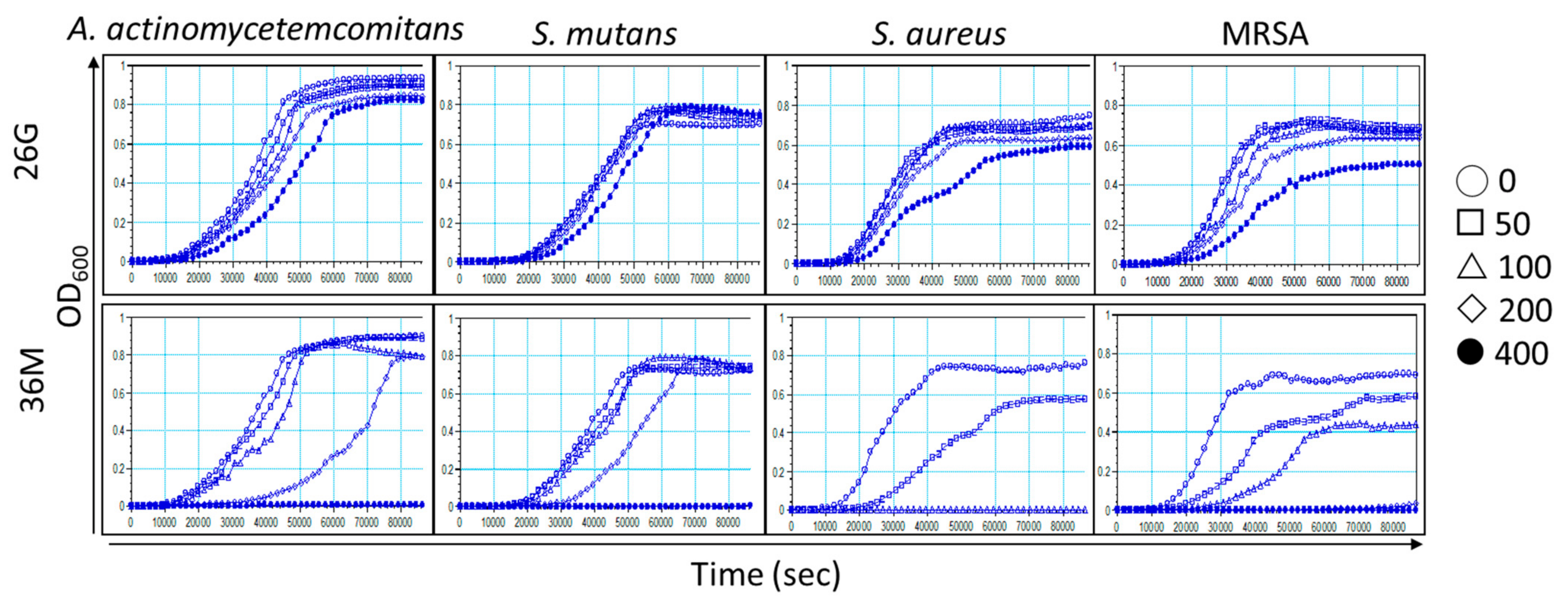
2.4. Microorganism Growth Is Delayed by 26G and 36M in a Concentration-Dependent Manner
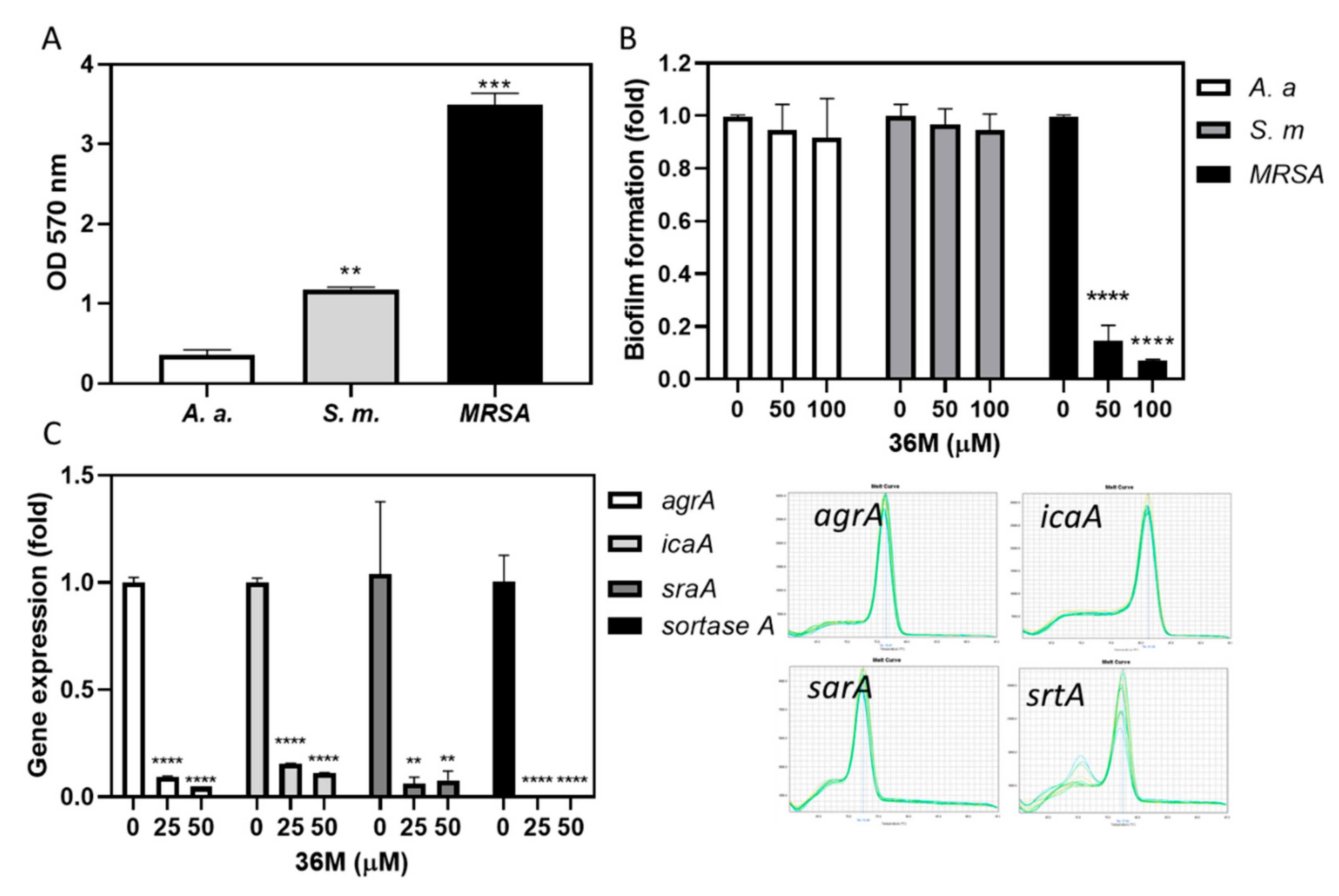
2.5. 36M Repressed the Drug-Resistance Gene Expression of MRSA
2.6. 36M Decreased MRSA Biofilm Formation
2.7. 36M Was More Cytotoxic Than 26G and Effectively Suppressed Pro-Inflammatory Gene Expression in RAW264.7 Cells
3. Discussion
4. Materials and Methods
4.1. Antimicrobial Agents and Other Chemicals
4.2. Microbial Cultures
4.3. Agar Diffusion Test
4.4. Cell Culture
4.5. Cell Viability Assay
4.6. Heat Stability Test
4.7. pH Value Stability Test
4.8. Determination of Minimum Inhibitory Concentration (MIC) and Minimum Bactericidal Concentration (MBC)
4.9. Growth Curve Assay
4.10. Drug Synergistic Test
4.11. Biofilm Formation Assay
4.12. Reverse Transcription-Quantitative Polymerase Chain Reaction
4.13. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hall-Stoodley, L.; Costerton, J.W.; Stoodley, P. Bacterial biofilms: From the natural environment to infectious diseases. Nat. Rev. Microbiol. 2004, 2, 95–108. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; Li, M.; Gregory, R.L. Bacterial interactions in dental biofilm. Virulence 2011, 2, 435–444. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez-Venegas, G.; Gomez-Mora, J.A.; Meraz-Rodriguez, M.A.; Flores-Sanchez, M.A.; Ortiz-Miranda, L.F. Effect of flavonoids on antimicrobial activity of microorganisms present in dental plaque. Heliyon 2019, 5, e03013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perez-Chaparro, P.J.; Goncalves, C.; Figueiredo, L.C.; Faveri, M.; Lobao, E.; Tamashiro, N.; Duarte, P.; Feres, M. Newly identified pathogens associated with periodontitis: A systematic review. J. Dent. Res. 2014, 93, 846–858. [Google Scholar] [CrossRef] [Green Version]
- Da Silva-Boghossian, C.M.; do Souto, R.M.; Luiz, R.R.; Colombo, A.P. Association of red complex, A. actinomycetemcomitans and non-oral bacteria with periodontal diseases. Arch. Oral Biol. 2011, 56, 899–906. [Google Scholar] [CrossRef] [Green Version]
- Srivastava, A.; Saha, S.; Sahu, C. Early and accurate detection of bacterial isolates from dental plaque in subjects with primary, mixed, and permanent dentition by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry technique. J. Indian Soc. Periodontol. 2020, 24, 104–108. [Google Scholar]
- Koukos, G.; Sakellari, D.; Arsenakis, M.; Tsalikis, L.; Slini, T.; Konstantinidis, A. Prevalence of Staphylococcus aureus and methicillin resistant Staphylococcus aureus (MRSA) in the oral cavity. Arch. Oral Biol. 2015, 60, 1410–1415. [Google Scholar] [CrossRef]
- Lu, W.; Wu, F.; Zhou, X.; Wu, L.; Li, M.; Ren, B.; Guo, Q.; Huang, R.; Li, J.; Xiao, L.; et al. Isolation and identification of aerobic and facultative anaerobic bacteria in the oral cavity. Nan Fang Yi Ke Da Xue Xue Bao 2015, 35, 1710–1714. [Google Scholar]
- Smith, K.; Robertson, D.P.; Lappin, D.F.; Ramage, G. Commercial mouthwashes are ineffective against oral MRSA biofilms. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2013, 115, 624–629. [Google Scholar] [CrossRef]
- Tada, A.; Senpuku, H.; Motozawa, Y.; Yoshihara, A.; Hanada, N.; Tanzawa, H. Association between commensal bacteria and opportunistic pathogens in the dental plaque of elderly individuals. Clin. Microbiol. Infect. 2006, 12, 776–781. [Google Scholar] [CrossRef] [Green Version]
- Valverde Villar, A.M.; Gutierrez Del Alamo Oms, J.; Neira Borrajo, I.; de Miguel Fernandez, S.; Flox Benitez, P.; Llopis Miro, R. Screening of MRSA colonization in patients undergoing total joint arthroplasty. J. Infect. Prev. 2021, 22, 283–288. [Google Scholar] [CrossRef]
- Miyake, M.; Obbayashi, Y.; Iwasaki, A.; Ogawa, T.; Nagahata, S. Risk factors for methicillin-resistant Staphylococcus aureus (MRSA) and use of a nasal mupirocin ointment in oral cancer inpatients. J. Oral Maxil. Surg. 2007, 65, 2159–2163. [Google Scholar] [CrossRef]
- Levy, S.B. The challenge of antibiotic resistance. Sci. Am. 1998, 278, 46–53. [Google Scholar] [CrossRef]
- Burdock, G.A. Review of the biological properties and toxicity of bee propolis (propolis). Food Chem. Toxicol. 1998, 36, 347–363. [Google Scholar] [CrossRef]
- Khurshid, Z.; Naseem, M.; Zafar, M.S.; Najeeb, S.; Zohaib, S. Propolis: A natural biomaterial for dental and oral healthcare. J. Dent. Res. Dent. Clin. Dent. Prospect. 2017, 11, 265–274. [Google Scholar]
- Liu, X.; Du, Q.; Tian, C.; Tang, M.; Jiang, Y.; Wang, Y.; Cao, Y.; Wang, Z.; Wang, Z.; Yang, J.; et al. Discovery of CAPE derivatives as dual EGFR and CSK inhibitors with anticancer activity in a murine model of hepatocellular carcinoma. Bioorg. Chem. 2021, 107, 104536. [Google Scholar] [CrossRef]
- Niu, Y.; Wang, K.; Zheng, S.; Wang, Y.; Ren, Q.; Li, H.; Ding, L.; Li, W.; Zhang, L. Antibacterial Effect of Caffeic Acid Phenethyl Ester on Cariogenic Bacteria and Streptococcus mutans Biofilms. Antimicrob. Agents Chemother. 2020, 64, e00251-20. [Google Scholar] [CrossRef]
- De Carvalho Furtado, J.H., Jr.; Rocha Valadas, L.A.; Mendonca, K.S.; de Oliveira Filho, R.D.; Gadelha, L.M.U.; de Mello Fiallos, N.; Neto, E.M.R.; de Mello Fiallos, A.C.; de Franca Fonteles, M.M. Propolis and its Dental Applications: A Technological Prospection. Recent. Pat. Biotechnol. 2018, 12, 288–296. [Google Scholar] [CrossRef]
- Celli, N.; Dragani, L.K.; Murzilli, S.; Pagliani, T.; Poggi, A. In vitro and in vivo stability of caffeic acid phenethyl ester, a bioactive compound of propolis. J. Agric. Food Chem. 2007, 55, 3398–3407. [Google Scholar] [CrossRef]
- Celli, N.; Mariani, B.; Dragani, L.K.; Murzilli, S.; Rossi, C.; Rotilio, D. Development and validation of a liquid chromatographic-tandem mass spectrometric method for the determination of caffeic acid phenethyl ester in rat plasma and urine. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2004, 810, 129–136. [Google Scholar] [CrossRef]
- Arasoglu, T.; Derman, S.; Mansuroglu, B. Comparative evaluation of antibacterial activity of caffeic acid phenethyl ester and PLGA nanoparticle formulation by different methods. Nanotechnology 2016, 27, 025103. [Google Scholar] [CrossRef]
- Baliga, S.; Muglikar, S.; Kale, R. Salivary pH: A diagnostic biomarker. J. Indian Soc. Periodontol. 2013, 17, 461–465. [Google Scholar] [CrossRef]
- Lingstrom, P.; van Ruyven, F.O.; van Houte, J.; Kent, R. The pH of dental plaque in its relation to early enamel caries and dental plaque flora in humans. J. Dent. Res. 2000, 79, 770–777. [Google Scholar] [CrossRef]
- Hayacibara, M.F.; Koo, H.; Rosalen, P.L.; Duarte, S.; Franco, E.M.; Bowen, W.H.; Ikegaki, M.; Cury, J.A. In vitro and in vivo effects of isolated fractions of Brazilian propolis on caries development. J. Ethnopharmacol. 2005, 101, 110–115. [Google Scholar] [CrossRef]
- Tuon, F.F.; Gavrilko, O.; Almeida, S.; Sumi, E.R.; Alberto, T.; Rocha, J.L.; Rosa, E.A. Prospective, randomised, controlled study evaluating early modification of oral microbiota following admission to the intensive care unit and oral hygiene with chlorhexidine. J. Glob. Antimicrob. Resist. 2017, 8, 159–163. [Google Scholar] [CrossRef]
- Stepanovic, S.; Antic, N.; Dakic, I.; Svabic-Vlahovic, M. In vitro antimicrobial activity of propolis and synergism between propolis and antimicrobial drugs. Microbiol. Res. 2003, 158, 353–357. [Google Scholar] [CrossRef]
- Oliveira, D.C.; de Lencastre, H. Methicillin-resistance in Staphylococcus aureus is not affected by the overexpression in trans of the mecA gene repressor: A surprising observation. PLoS ONE 2011, 6, e23287. [Google Scholar] [CrossRef]
- Stapleton, P.D.; Taylor, P.W. Methicillin resistance in Staphylococcus aureus: Mechanisms and modulation. Sci. Prog. 2002, 85 Pt 1, 57–72. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, N.; Taniguchi, K.; Urasawa, S. Analysis of diversity of mutations in the mecI gene and mecA promoter/operator region of methicillin-resistant Staphylococcus aureus and Staphylococcus epidermidis. Antimicrob. Agents Chemother. 1998, 42, 717–720. [Google Scholar] [CrossRef] [Green Version]
- Kalogeropoulos, N.; Konteles, S.J.; Troullidou, E.; Mourtzinos, I.; Karathanos, V.T. Chemical composition, antioxidant activity and antimicrobial properties of propolis extracts from Greece and Cyprus. Food Chem. 2009, 116, 452–461. [Google Scholar] [CrossRef]
- Spirig, T.; Weiner, E.M.; Clubb, R.T. Sortase enzymes in Gram-positive bacteria. Mol. Microbiol. 2011, 82, 1044–1059. [Google Scholar] [CrossRef] [PubMed]
- Coelho, L.R.; Souza, R.R.; Ferreira, F.A.; Guimaraes, M.A.; Ferreira-Carvalho, B.T.; Figueiredo, A.M.S. agr RNAIII divergently regulates glucose-induced biofilm formation in clinical isolates of Staphylococcus aureus. Microbiology 2008, 154 Pt 11, 3480–3490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vuong, C.; Otto, M. Staphylococcus epidermidis infections. Microbes Infect. 2002, 4, 481–489. [Google Scholar] [CrossRef]
- Selvaraj, C.; Priya, R.B.; Singh, S.K. Exploring the Biology and Structural Architecture of Sortase Role on Biofilm Formation in Gram Positive Pathogens. Curr. Top. Med. Chem. 2018, 18, 2462–2480. [Google Scholar] [CrossRef]
- Levesque, C.M.; Voronejskaia, E.; Huang, Y.C.; Mair, R.W.; Ellen, R.P.; Cvitkovitch, D.G. Involvement of sortase anchoring of cell wall proteins in biofilm formation by Streptococcus mutans. Infect. Immun. 2005, 73, 3773–3777. [Google Scholar] [CrossRef] [Green Version]
- Waykole, Y.P.; Doiphode, S.S.; Rakhewar, P.S.; Mhaske, M. Anticytokine therapy for periodontal diseases: Where are we now? J. Indian Soc. Periodontol. 2009, 13, 64–68. [Google Scholar] [CrossRef]
- Tsai, T.H.; Yu, C.H.; Chang, Y.P.; Lin, Y.T.; Huang, C.J.; Kuo, Y.H.; Tsai, P.J. Protective Effect of Caffeic Acid Derivatives on tert-Butyl Hydroperoxide-Induced Oxidative Hepato-Toxicity and Mitochondrial Dysfunction in HepG2 Cells. Molecules 2017, 22, 702. [Google Scholar] [CrossRef]
- Choi, W.; Villegas, V.; Istre, H.; Heppler, B.; Gonzalez, N.; Brusman, N.; Snider, L.; Hogle, E.; Tucker, J.; Onate, A.; et al. Synthesis and characterization of CAPE derivatives as xanthine oxidase inhibitors with radical scavenging properties. Bioorg. Chem. 2019, 86, 686–695. [Google Scholar] [CrossRef]
- Weng, Y.C.; Chiu, H.L.; Lin, Y.C.; Chi, T.C.; Kuo, Y.H.; Su, M.J. Antihyperglycemic Effect of a Caffeamide Derivative, KS370G, in Normal and Diabetic Mice. J. Agric. Food Chem. 2010, 58, 10033–10038. [Google Scholar] [CrossRef]
- Moosavi, F.; Hosseini, R.; Rajaian, H.; Silva, T.; Silva, D.M.E.; Saso, L.; Edraki, N.; Miri, R.; Borges, F.; Firuzi, O. Derivatives of caffeic acid, a natural antioxidant, as the basis for the discovery of novel nonpeptidic neurotrophic agents. Bioorgan. Med. Chem. 2017, 25, 3233. [Google Scholar] [CrossRef]
- Liu, W.H.; Liu, T.C.; Mong, M.C. Antibacterial effects and action modes of asiatic acid. Biomedicine 2015, 5, 16. [Google Scholar] [CrossRef]
- Shih, Y.H.; Chang, K.W.; Hsia, S.M.; Yu, C.C.; Fuh, L.J.; Chi, T.Y.; Shieh, T.M. In vitro antimicrobial and anticancer potential of hinokitiol against oral pathogens and oral cancer cell lines. Microbiol. Res. 2013, 168, 254–262. [Google Scholar] [CrossRef]
- Tu, M.G.; Sun, K.T.; Wang, T.H.; He, Y.Z.; Hsia, S.M.; Tsai, B.H.; Shih, Y.H.; Shieh, T.M. Effects of mineral trioxide aggregate and bioceramics on macrophage differentiation and polarization in vitro. J. Formos Med. Assoc. 2019, 118, 1458–1465. [Google Scholar] [CrossRef]
- Tu, H.F.; Chen, M.Y.; Lai, J.C.; Chen, Y.L.; Wong, Y.W.; Yang, C.C.; Chen, H.Y.; Hsia, S.M.; Shih, Y.H.; Shieh, T.M. Arecoline-regulated ataxia telangiectasia mutated expression level in oral cancer progression. Head Neck 2019, 41, 2525–2537. [Google Scholar] [CrossRef]
- Shih, Y.H.; Lin, D.J.; Chang, K.W.; Hsia, S.M.; Ko, S.Y.; Lee, S.Y.; Hsue, S.S.; Wang, T.H.; Chen, Y.L.; Shieh, T.M. Evaluation physical characteristics and comparison antimicrobial and anti-inflammation potentials of dental root canal sealers containing hinokitiol in vitro. PLoS ONE 2014, 9, e94941. [Google Scholar] [CrossRef] [Green Version]
- Chiu, K.C.; Shih, Y.H.; Wang, T.H.; Lan, W.C.; Li, P.J.; Jhuang, H.S.; Hsia, S.M.; Shen, Y.W.; Yuan-Chien Chen, M.; Shieh, T.M. In vitro antimicrobial and antipro-inflammation potential of honokiol and magnolol against oral pathogens and macrophages. J. Formos Med. Assoc. 2021, 120, 827–837. [Google Scholar] [CrossRef]
- O’Toole, G.A. Microtiter dish biofilm formation assay. J. Vis. Exp. 2011, 47, e2437. [Google Scholar] [CrossRef]
- Wu, Z.; Hong, H.; Zhao, X.; Wang, X. Efficient expression of sortase A from Staphylococcus aureus in Escherichia coli and its enzymatic characterizations. Bioresour. Bioprocess. 2017, 4, 13. [Google Scholar] [CrossRef] [Green Version]
- Yan, X.; Gu, S.; Cui, X.; Shi, Y.; Wen, S.; Chen, H.; Ge, J. Antimicrobial, anti-adhesive and anti-biofilm potential of biosurfactants isolated from Pediococcus acidilactici and Lactobacillus plantarum against Staphylococcus aureus CMCC26003. Microb. Pathog. 2019, 127, 12–20. [Google Scholar] [CrossRef]
- Li, C.; Wang, M.H. Anti-inflammatory effect of the water fraction from hawthorn fruit on LPS-stimulated RAW 264.7 cells. Nutr. Res. Pract. 2011, 5, 101–106. [Google Scholar] [CrossRef] [Green Version]


| MIC/MBC | |||
|---|---|---|---|
| 26G (μM) | 36M (μM) | CHX (μg/mL) | |
| A. actinomycetemcomitans (G−) | >400/>400 | 400/400 | <1/<1 |
| S. mutans (G+) | >400/>400 | 400/400 | <1/<1 |
| S. aureus (G+) | >400/400 | 100/200 | 1</<1 |
| MRSA (G+) | >400/>400 | 200/200 | 1</2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shih, Y.-H.; Hsia, S.-M.; Chiu, K.-C.; Wang, T.-H.; Chien, C.-Y.; Li, P.-J.; Kuo, Y.-H.; Shieh, T.-M. In Vitro Antimicrobial Potential of CAPE and Caffeamide Derivatives against Oral Microbes. Int. J. Mol. Sci. 2022, 23, 4099. https://doi.org/10.3390/ijms23084099
Shih Y-H, Hsia S-M, Chiu K-C, Wang T-H, Chien C-Y, Li P-J, Kuo Y-H, Shieh T-M. In Vitro Antimicrobial Potential of CAPE and Caffeamide Derivatives against Oral Microbes. International Journal of Molecular Sciences. 2022; 23(8):4099. https://doi.org/10.3390/ijms23084099
Chicago/Turabian StyleShih, Yin-Hwa, Shih-Min Hsia, Kuo-Chou Chiu, Tong-Hong Wang, Chi-Ying Chien, Po-Jung Li, Yueh-Hsiung Kuo, and Tzong-Ming Shieh. 2022. "In Vitro Antimicrobial Potential of CAPE and Caffeamide Derivatives against Oral Microbes" International Journal of Molecular Sciences 23, no. 8: 4099. https://doi.org/10.3390/ijms23084099
APA StyleShih, Y.-H., Hsia, S.-M., Chiu, K.-C., Wang, T.-H., Chien, C.-Y., Li, P.-J., Kuo, Y.-H., & Shieh, T.-M. (2022). In Vitro Antimicrobial Potential of CAPE and Caffeamide Derivatives against Oral Microbes. International Journal of Molecular Sciences, 23(8), 4099. https://doi.org/10.3390/ijms23084099









