Cell-in-Cell Phenomena in Wall-Less Bacteria: Is It Possible?
Abstract
:1. Introduction
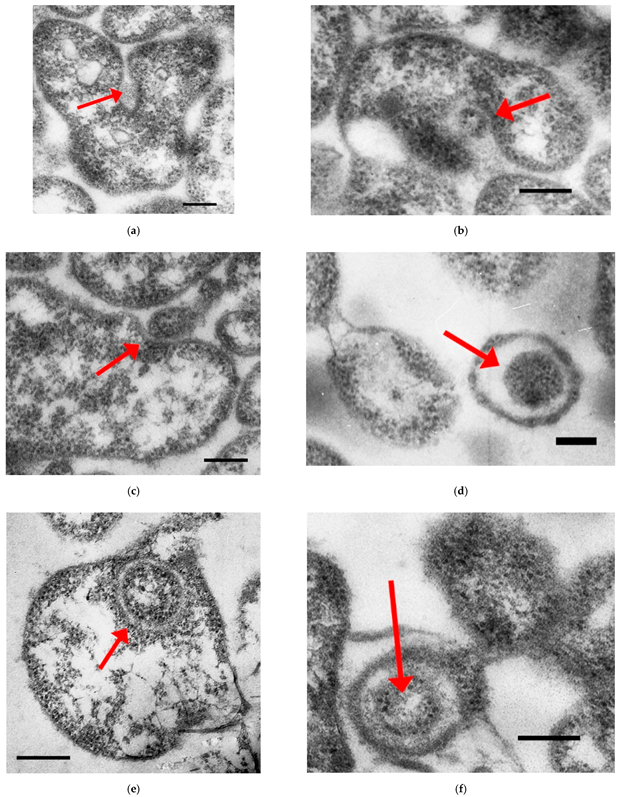
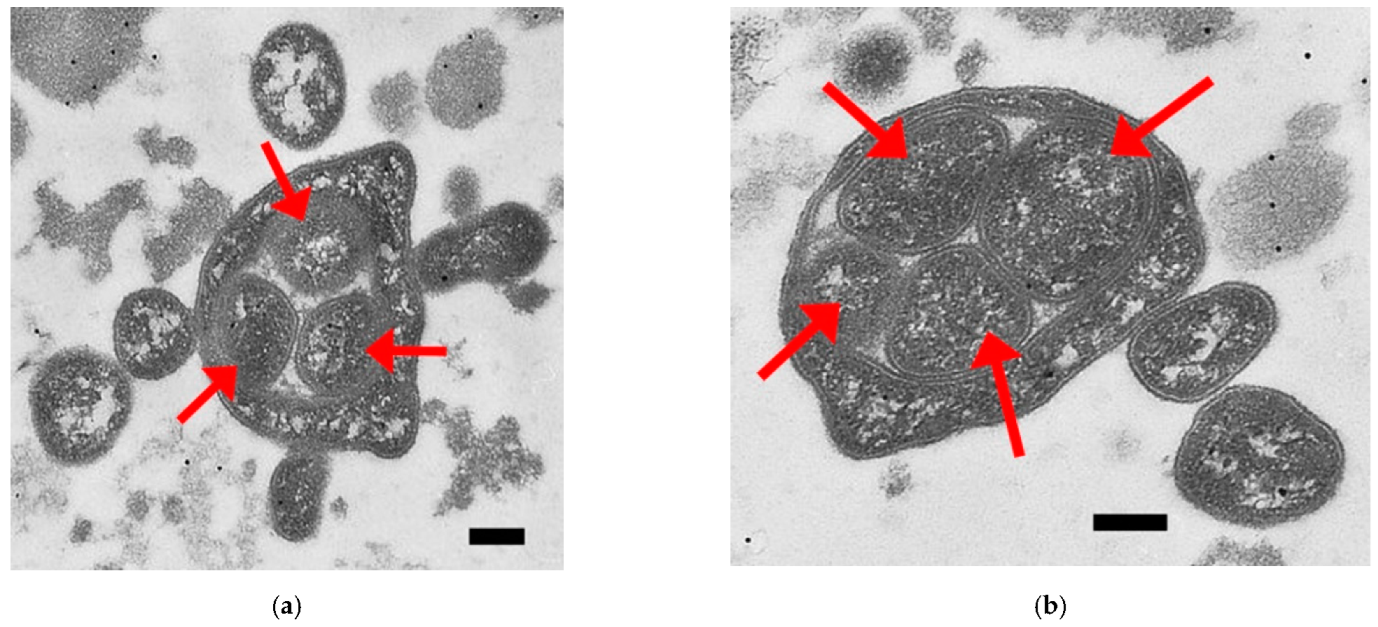
2. Results
3. Discussion
4. Materials and Methods
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Demin, S.; Berdieva, M.; Goodkov, A. Cell-cell fusions and cell-in-cell phenomena in healthy cells and cancer: Lessons from protists and invertebrates. Semin. Cancer Biol. 2022, 81, 96–105. [Google Scholar] [CrossRef] [PubMed]
- Davies, S.P.; Terry, L.V.; Wilkinson, A.L.; Stamataki, Z. Cell-in-Cell Structures in the Liver: A Tale of Four E’s. Front. Immunol. 2020, 11, 650. [Google Scholar] [CrossRef] [PubMed]
- Fais, S.; Overholtzer, M. Cell-in-cell phenomena in cancer. Nat. Rev. Cancer 2018, 18, 758–766. [Google Scholar] [CrossRef] [PubMed]
- Duelli, D.; Lazebnik, Y. Cell-to-cell fusion as a link between viruses and cancer. Nat. Rev. Cancer 2007, 7, 968–976. [Google Scholar] [CrossRef]
- Brukman, N.G.; Uygur, B.; Podbilewicz, B.; Chernomordik, L.V. How cells fuse. J. Cell Biol. 2019, 218, 1436–1451. [Google Scholar] [CrossRef] [Green Version]
- Cabezón, E.; Ripoll-Rozada, J.; Peña, A.; de la Cruz, F.; Arechaga, I. Towards an integrated model of bacterial conjugation. FEMS Microbiol. Rev. 2015, 39, 81–95. [Google Scholar] [CrossRef] [Green Version]
- Graf, F.E.; Palm, M.; Warringer, J.; Farewell, A. Inhibiting conjugation as a tool in the fight against antibiotic resistance. Drug Dev. Res. 2019, 80, 19–23. [Google Scholar] [CrossRef] [Green Version]
- Borchsenius, S.N.; Chernova, O.A.; Chernov, V.M.; Vishnyakov, I.E. Mikoplazmy v Biologii i Meditsine Nachala 21 Veka (Mycoplasmas in Biology and Medicine of the Early 21st Century, in Russian); Nauka: St. Petersburg, Russia, 2016; pp. 1–334. [Google Scholar]
- García-Galán, A.; Baranowski, E.; Hygonenq, M.C.; Walch, M.; Croville, G.; Citti, C.; De la Fe, C.; Nouvel, L.X. Genome mosaicism in field strains of Mycoplasma bovis as footprints of in-host horizontal chromosomal transfer. Appl. Environ. Microbiol. 2021, 20, e01661-21. [Google Scholar] [CrossRef]
- Franzoso, G.; Dimitrov, D.S.; Blumenthal, R.; Barile, M.F.; Rottem, S. Fusion of Mycoplasma fermentans strain incognitus with T-lymphocytes. FEBS Lett. 1992, 303, 251–254. [Google Scholar] [CrossRef] [Green Version]
- Rottem, S.; Naot, Y. Subversion and exploitation of host cells by mycoplasmas. Trends Microbiol. 1998, 6, 436–440. [Google Scholar] [CrossRef]
- Rottem, S.; Yogev, D. Mycoplasma interaction with eukaryotic cells. Sub-Cell. Biochem. 2000, 33, 199–227. [Google Scholar] [CrossRef]
- Rottem, S. Choline-containing lipids in mycoplasmas. Microbes. Infect. 2002, 4, 963–968. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, H.; Xie, X.; Chen, R.; Li, J.; Ni, B.; Yu, P.; Liu, Z.; Shao, G.; Xiong, Q.; et al. Long-Residence Pneumonia Vaccine Developed Using PEG-Grafted Hybrid Nanovesicles from Cell Membrane Fusion of Mycoplasma and IFN-γ-Primed Macrophages. Small 2021, 17, e2101183. [Google Scholar] [CrossRef] [PubMed]
- Tarshis, M.A.; Ladygina, V.G.; Migoushina, V.L.; Klebanov, G.I.; Rakovskaya, I.V. Interactions of Acholeplasma laidlawii cells with mouse spleen lymphocytes. Zentralbl. Bakteriol. Mikrobiol. Hyg. A 1981, 250, 153–156. [Google Scholar] [CrossRef]
- Rechnitzer, H.; Rottem, S. Reconstituted proteolipid vesicles prepared from Mycoplasma fermentans membranes are able to bind and fuse with Molt-3 cells. Curr. Microbiol. 2006, 53, 293–297. [Google Scholar] [CrossRef] [PubMed]
- Tarshis, M.; Salman, M.; Rottem, S. Fusion of mycoplasmas: The formation of cell hybrids. FEMS Microbiol. Lett. 1991, 6, 67–71. [Google Scholar] [CrossRef]
- Brunner, H.; Schaar, H.; Krauss, H. Giant cell formation in Acholeplasma laidlawii, strain JA1. Zentralbl. Bakteriol. A 1980, 248, 120–128. [Google Scholar] [CrossRef]
- Vishnyakov, I.E.; Levitskii, S.A.; Borchsenius, S.N. The effect of heat shock on phytopathogenic mycoplasma Acholeplasma laidlawii PG-8A. Cell Tissue Biol. 2015, 9, 149–157. [Google Scholar] [CrossRef]
- Chernov, V.M.; Mouzykantov, A.A.; Baranova, N.B.; Medvedeva, E.S.; Grygorieva, T.Y.; Trushin, M.V.; Vishnyakov, I.E.; Sabantsev, A.V.; Borchsenius, S.N.; Chernova, O.A. Extracellular membrane vesicles secreted by mycoplasma Acholeplasma laidlawii PG8 are enriched in virulence proteins. J. Proteom. 2014, 110, 117–128. [Google Scholar] [CrossRef]
- Sweeney, E.L.; Dando, S.J.; Kallapur, S.G.; Knox, C.L. The Human Ureaplasma Species as Causative Agents of Chorioamnionitis. Clin. Microbiol. Rev. 2016, 30, 349–379. [Google Scholar] [CrossRef] [Green Version]
- Kukekova, A.V.; Malinin, A.Y.; Ayala, J.A.; Borchsenius, S.N. Characterization of Acholeplasma laidlawii ftsZ gene and its gene product. Biochem. Biophys. Res. Commun. 1999, 262, 44–49. [Google Scholar] [CrossRef] [PubMed]
- Chernova, L.S.; Bogachev, M.I.; Chasov, V.V.; Vishnyakov, I.E.; Kayumov, A.R. N-and C-terminal regions of the small heat shock protein IbpA from Acholeplasma laidlawii competitively govern its oligomerization pattern and chaperone-like activity. RSC Adv. 2020, 10, 8364–8376. [Google Scholar] [CrossRef]
- Glass, J.I.; Lefkowitz, E.J.; Glass, J.S.; Heiner, C.R.; Chen, E.Y.; Cassell, G.H. The complete sequence of the mucosal pathogen Ureaplasma urealyticum. Nature 2000, 407, 757–762. [Google Scholar] [CrossRef] [PubMed]
- Vedyaykin, A.D.; Ponomareva, E.V.; Khodorkovskii, M.A.; Borchsenius, S.N.; Vishnyakov, I.E. Mechanisms of Bacterial Cell Division. Microbiology 2019, 88, 245–260. [Google Scholar] [CrossRef]
- Salman, M.; Tarshis, M.; Rottem, S. Small unilamellar vesicles are able to fuse with Mycoplasma capricolum cells. Biochim. Biophys. Acta 1991, 1063, 209–216. [Google Scholar] [CrossRef]
- Mlynarczuk-Bialy, I.; Dziuba, I.; Sarnecka, A.; Platos, E.; Kowalczyk, M.; Pels, K.K.; Wilczynski, G.M.; Wojcik, C.; Bialy, L.P. Entosis: From Cell Biology to Clinical Cancer Pathology. Cancers 2020, 12, 2481. [Google Scholar] [CrossRef] [PubMed]
- Rastogi, V.; Sharma, R.; Misra, S.R.; Yadav, L.; Sharma, V. Emperipolesis—A review. J. Clin. Diagn. Res. 2014, 8, ZM01. [Google Scholar] [CrossRef]
- Durgan, J.; Florey, O. Cancer cell cannibalism: Multiple triggers emerge for entosis. Biochim. Biophys. Acta Mol. Cell Res. 2018, 1865, 831–841. [Google Scholar] [CrossRef]
- Citti, C.; Baranowski, E.; Dordet-Frisoni, E.; Faucher, M.; Nouvel, L.X. Genomic Islands in Mycoplasmas. Genes 2020, 11, 836. [Google Scholar] [CrossRef]
- Marenda, M.; Barbe, V.; Gourgues, G.; Mangenot, S.; Sagne, E.; Citti, C. A new integrative conjugative element occurs in Mycoplasma agalactiae as chromosomal and free circular forms. J. Bacteriol. 2006, 188, 4137–4141. [Google Scholar] [CrossRef] [Green Version]
- Meygret, A.; Peuchant, O.; Dordet-Frisoni, E.; Sirand-Pugnet, P.; Citti, C.; Bébéar, C.; Béven, L.; Pereyre, S. High Prevalence of Integrative and Conjugative Elements Encoding Transcription Activator-Like Effector Repeats in Mycoplasma hominis. Front. Microbiol. 2019, 10, 2385. [Google Scholar] [CrossRef] [PubMed]
- Tardy, F.; Mick, V.; Dordet-Frisoni, E.; Marenda, M.S.; Sirand-Pugnet, P.; Blanchard, A.; Citti, C. Integrative conjugative elements are widespread in field isolates of Mycoplasma species pathogenic for ruminants. Appl. Environ. Microbiol. 2015, 81, 1634–1643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Citti, C.; Dordet-Frisoni, E.; Nouvel, L.X.; Kuo, C.H.; Baranowski, E. Horizontal Gene Transfers in Mycoplasmas (Mollicutes). Curr. Issues Mol. Biol. 2018, 29, 3–22. [Google Scholar] [CrossRef] [Green Version]
- Faucher, M.; Nouvel, L.X.; Dordet-Frisoni, E.; Sagné, E.; Baranowski, E.; Hygonenq, M.C.; Marenda, M.S.; Tardy, F.; Citti, C. Mycoplasmas under experimental antimicrobial selection: The unpredicted contribution of horizontal chromosomal transfer. PLoS Genet. 2019, 15, e1007910. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dordet-Frisoni, E.; Faucher, M.; Sagné, E.; Baranowski, E.; Tardy, F.; Nouvel, L.X.; Citti, C. Mycoplasma Chromosomal Transfer: A Distributive, Conjugative Process Creating an Infinite Variety of Mosaic Genomes. Front. Microbiol. 2019, 10, 2441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sirand-Pugnet, P.; Lartigue, C.; Marenda, M.; Jacob, D.; Barré, A.; Barbe, V.; Schenowitz, C.; Mangenot, S.; Couloux, A.; Segurens, B.; et al. Being pathogenic, plastic, and sexual while living with a nearly minimal bacterial genome. PLoS Genet. 2007, 3, e75. [Google Scholar] [CrossRef]
- Naito, M.; Pawlowska, T.E. The role of mobile genetic elements in evolutionary longevity of heritable endobacteria. Mob. Genet. Elem. 2015, 6, e1136375. [Google Scholar] [CrossRef] [Green Version]
- Freundt, E. Culture media for classical mycoplasmas. In Methods in Mycoplasmology; Razin, S., Ed.; Academic Press: New York, NY, USA, 1983; Volume 1, pp. 127–136. ISBN 9780323147132. [Google Scholar]
- Vishnyakov, I.E.; Borchsenius, S.N.; Basovskii, Y.I.; Levitskii, S.A.; Lazarev, V.N.; Snigirevskaya, E.S.; Komissarchik, Y.Y. Localization of division protein FtsZ in Mycoplasma hominis. Cell Tissue Biol. 2009, 3, 254–262. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vishnyakov, I.E. Cell-in-Cell Phenomena in Wall-Less Bacteria: Is It Possible? Int. J. Mol. Sci. 2022, 23, 4345. https://doi.org/10.3390/ijms23084345
Vishnyakov IE. Cell-in-Cell Phenomena in Wall-Less Bacteria: Is It Possible? International Journal of Molecular Sciences. 2022; 23(8):4345. https://doi.org/10.3390/ijms23084345
Chicago/Turabian StyleVishnyakov, Innokentii E. 2022. "Cell-in-Cell Phenomena in Wall-Less Bacteria: Is It Possible?" International Journal of Molecular Sciences 23, no. 8: 4345. https://doi.org/10.3390/ijms23084345
APA StyleVishnyakov, I. E. (2022). Cell-in-Cell Phenomena in Wall-Less Bacteria: Is It Possible? International Journal of Molecular Sciences, 23(8), 4345. https://doi.org/10.3390/ijms23084345





