A Novel FGFR1 Missense Mutation in a Portuguese Family with Congenital Hypogonadotropic Hypogonadism
Abstract
1. Introduction
2. Results
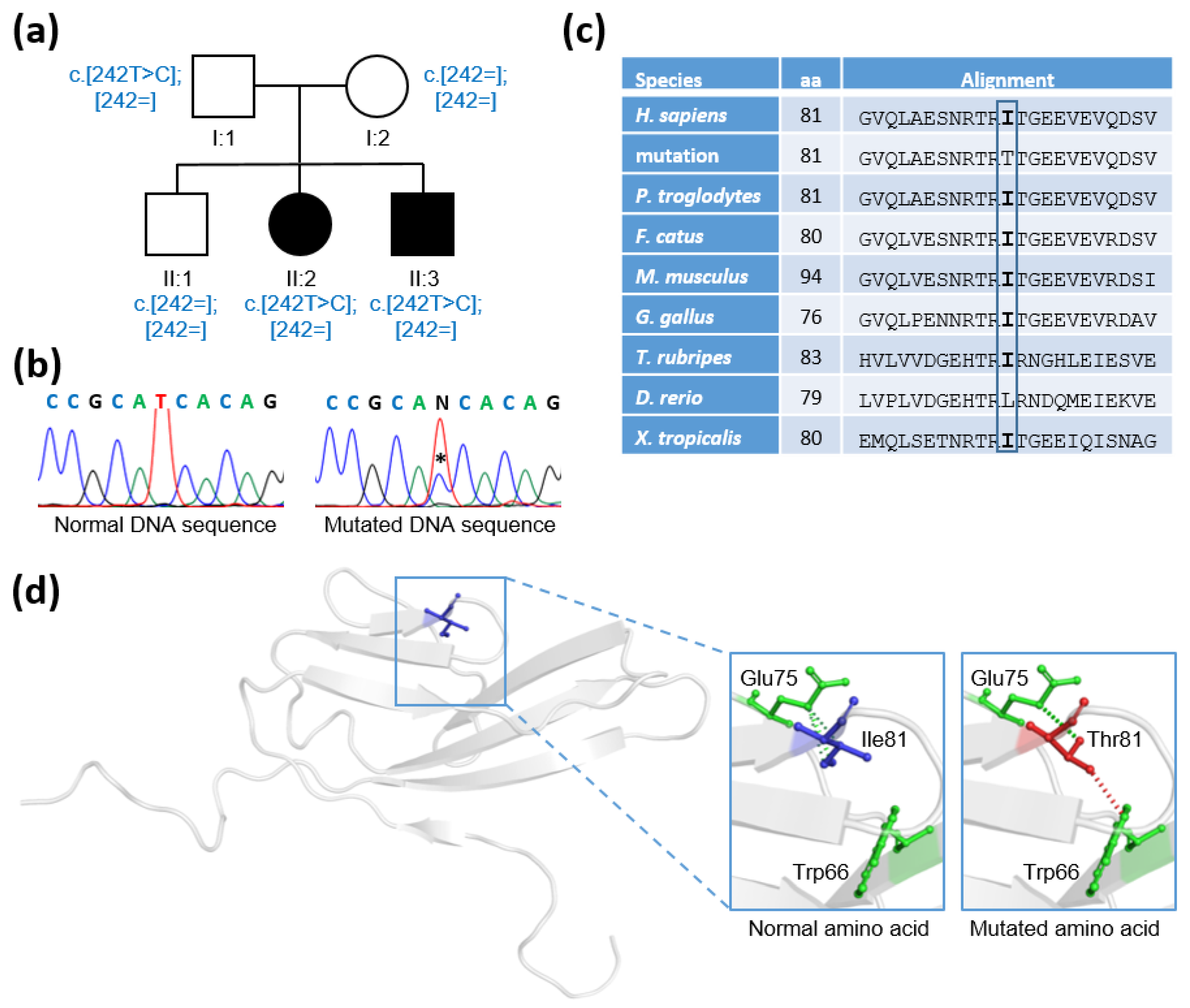
2.1. Clinical Studies
2.2. Genetic Studies
3. Discussion
4. Materials and Methods
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Young, J.; Xu, C.; Papadakis, G.E.; Acierno, J.S.; Maione, L.; Hietamaki, J.; Raivio, T.; Pitteloud, N. Clinical Management of Congenital Hypogonadotropic Hypogonadism. Endocr. Rev. 2019, 40, 669–710. [Google Scholar] [CrossRef] [PubMed]
- Boehm, U.; Bouloux, P.M.; Dattani, M.T.; de Roux, N.; Dode, C.; Dunkel, L.; Dwyer, A.A.; Giacobini, P.; Hardelin, J.P.; Juul, A.; et al. Expert consensus document: European Consensus Statement on congenital hypogonadotropic hypogonadism—Pathogenesis, diagnosis and treatment. Nat. Rev. Endocrinol. 2015, 11, 547–564. [Google Scholar] [CrossRef] [PubMed]
- Raivio, T.; Miettinen, P.J. Constitutional delay of puberty versus congenital hypogonadotropic hypogonadism: Genetics, management and updates. Best Pract. Res. Clin. Endocrinol. Metab. 2019, 33, 101316. [Google Scholar] [CrossRef] [PubMed]
- Grinspon, R.P. Genetics of congenital central hypogonadism. Best Pract. Res. Clin. Endocrinol. Metab. 2021, 36, 101599. [Google Scholar] [CrossRef]
- Louden, E.D.; Poch, A.; Kim, H.G.; Ben-Mahmoud, A.; Kim, S.H.; Layman, L.C. Genetics of hypogonadotropic Hypogonadism-Human and mouse genes, inheritance, oligogenicity, and genetic counseling. Mol. Cell Endocrinol. 2021, 534, 111334. [Google Scholar] [CrossRef]
- Butz, H.; Nyiro, G.; Kurucz, P.A.; Liko, I.; Patocs, A. Molecular genetic diagnostics of hypogonadotropic hypogonadism: From panel design towards result interpretation in clinical practice. Hum. Genet. 2021, 140, 113–134. [Google Scholar] [CrossRef]
- Karczewski, K.J.; Francioli, L.C.; Tiao, G.; Cummings, B.B.; Alfoldi, J.; Wang, Q.; Collins, R.L.; Laricchia, K.M.; Ganna, A.; Birnbaum, D.P.; et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020, 581, 434–443. [Google Scholar] [CrossRef]
- Adzhubei, I.A.; Schmidt, S.; Peshkin, L.; Ramensky, V.E.; Gerasimova, A.; Bork, P.; Kondrashov, A.S.; Sunyaev, S.R. A method and server for predicting damaging missense mutations. Nat. Methods 2010, 7, 248–249. [Google Scholar] [CrossRef]
- Kircher, M.; Witten, D.M.; Jain, P.; O’Roak, B.J.; Cooper, G.M.; Shendure, J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014, 46, 310–315. [Google Scholar] [CrossRef]
- Cooper, G.M.; Stone, E.A.; Asimenos, G.; Program, N.C.S.; Green, E.D.; Batzoglou, S.; Sidow, A. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005, 15, 901–913. [Google Scholar] [CrossRef]
- Schwarz, J.M.; Cooper, D.N.; Schuelke, M.; Seelow, D. MutationTaster2: Mutation prediction for the deep-sequencing age. Nat. Methods 2014, 11, 361–362. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, C.H.; Pires, D.E.; Ascher, D.B. DynaMut: Predicting the impact of mutations on protein conformation, flexibility and stability. Nucleic Acids Res. 2018, 46, W350–W355. [Google Scholar] [CrossRef] [PubMed]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef] [PubMed]
- Good, D.J. New gene targets in the study of hypogonadotropic hypogonadism. Mol. Cell Endocrinol. 2021, 520, 111077. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi, M.; Olsen, S.K.; Ibrahimi, O.A. Structural basis for fibroblast growth factor receptor activation. Cytokine Growth Factor Rev. 2005, 16, 107–137. [Google Scholar] [CrossRef]
- Oleari, R.; Massa, V.; Cariboni, A.; Lettieri, A. The Differential Roles for Neurodevelopmental and Neuroendocrine Genes in Shaping GnRH Neuron Physiology and Deficiency. Int. J. Mol. Sci. 2021, 22, 9425. [Google Scholar] [CrossRef]
- Dode, C.; Levilliers, J.; Dupont, J.M.; De Paepe, A.; Le Du, N.; Soussi-Yanicostas, N.; Coimbra, R.S.; Delmaghani, S.; Compain-Nouaille, S.; Baverel, F.; et al. Loss-of-function mutations in FGFR1 cause autosomal dominant Kallmann syndrome. Nat. Genet. 2003, 33, 463–465. [Google Scholar] [CrossRef]
- Stenson, P.D.; Mort, M.; Ball, E.V.; Evans, K.; Hayden, M.; Heywood, S.; Hussain, M.; Phillips, A.D.; Cooper, D.N. The Human Gene Mutation Database: Towards a comprehensive repository of inherited mutation data for medical research, genetic diagnosis and next-generation sequencing studies. Hum. Genet. 2017, 136, 665–677. [Google Scholar] [CrossRef]
- Goncalves, C.; Bastos, M.; Pignatelli, D.; Borges, T.; Aragues, J.M.; Fonseca, F.; Pereira, B.D.; Socorro, S.; Lemos, M.C. Novel FGFR1 mutations in Kallmann syndrome and normosmic idiopathic hypogonadotropic hypogonadism: Evidence for the involvement of an alternatively spliced isoform. Fertil. Steril. 2015, 104, 1261–1267. [Google Scholar] [CrossRef]
- Goncalves, C.I.; Aragues, J.M.; Bastos, M.; Barros, L.; Vicente, N.; Carvalho, D.; Lemos, M.C. GNRHR biallelic and digenic mutations in patients with normosmic congenital hypogonadotropic hypogonadism. Endocr. Connect. 2017, 6, 360–366. [Google Scholar] [CrossRef][Green Version]
- Goncalves, C.I.; Fonseca, F.; Borges, T.; Cunha, F.; Lemos, M.C. Expanding the genetic spectrum of ANOS1 mutations in patients with congenital hypogonadotropic hypogonadism. Hum. Reprod. 2017, 32, 704–711. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Goncalves, C.I.; Patriarca, F.M.; Aragues, J.M.; Carvalho, D.; Fonseca, F.; Martins, S.; Marques, O.; Pereira, B.D.; Martinez-de-Oliveira, J.; Lemos, M.C. High frequency of CHD7 mutations in congenital hypogonadotropic hypogonadism. Sci. Rep. 2019, 9, 1597. [Google Scholar] [CrossRef] [PubMed]
- Pitteloud, N.; Meysing, A.; Quinton, R.; Acierno, J.S., Jr.; Dwyer, A.A.; Plummer, L.; Fliers, E.; Boepple, P.; Hayes, F.; Seminara, S.; et al. Mutations in fibroblast growth factor receptor 1 cause Kallmann syndrome with a wide spectrum of reproductive phenotypes. Mol. Cell Endocrinol. 2006, 254–255, 60–69. [Google Scholar] [CrossRef] [PubMed]
- Raivio, T.; Sidis, Y.; Plummer, L.; Chen, H.; Ma, J.; Mukherjee, A.; Jacobson-Dickman, E.; Quinton, R.; Van Vliet, G.; Lavoie, H.; et al. Impaired fibroblast growth factor receptor 1 signaling as a cause of normosmic idiopathic hypogonadotropic hypogonadism. J. Clin. Endocrinol. Metab. 2009, 94, 4380–4390. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009, 15, 1754–1760. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef]
- Liu, X.; Wu, C.; Li, C.; Boerwinkle, E. dbNSFP v3.0: A One-Stop Database of Functional Predictions and Annotations for Human Nonsynonymous and Splice-Site SNVs. Hum Mutat. 2016, 37, 235–241. [Google Scholar] [CrossRef]
- Van der Auwera, G.A.; Carneiro, M.O.; Hartl, C.; Poplin, R.; Del Angel, G.; Levy-Moonshine, A.; Jordan, T.; Shakir, K.; Roazen, D.; Thibault, J.; et al. From FastQ data to high confidence variant calls: The Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinform. 2013, 43, 11.10.1–11.10.33. [Google Scholar] [CrossRef]
- Kopanos, C.; Tsiolkas, V.; Kouris, A.; Chapple, C.E.; Albarca Aguilera, M.; Meyer, R.; Massouras, A. VarSome: The human genomic variant search engine. Bioinformatics 2019, 35, 1978–1980. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fadiga, L.; Lavrador, M.; Vicente, N.; Barros, L.; Gonçalves, C.I.; Al-Naama, A.; Saraiva, L.R.; Lemos, M.C. A Novel FGFR1 Missense Mutation in a Portuguese Family with Congenital Hypogonadotropic Hypogonadism. Int. J. Mol. Sci. 2022, 23, 4423. https://doi.org/10.3390/ijms23084423
Fadiga L, Lavrador M, Vicente N, Barros L, Gonçalves CI, Al-Naama A, Saraiva LR, Lemos MC. A Novel FGFR1 Missense Mutation in a Portuguese Family with Congenital Hypogonadotropic Hypogonadism. International Journal of Molecular Sciences. 2022; 23(8):4423. https://doi.org/10.3390/ijms23084423
Chicago/Turabian StyleFadiga, Lúcia, Mariana Lavrador, Nuno Vicente, Luísa Barros, Catarina I. Gonçalves, Asma Al-Naama, Luis R. Saraiva, and Manuel C. Lemos. 2022. "A Novel FGFR1 Missense Mutation in a Portuguese Family with Congenital Hypogonadotropic Hypogonadism" International Journal of Molecular Sciences 23, no. 8: 4423. https://doi.org/10.3390/ijms23084423
APA StyleFadiga, L., Lavrador, M., Vicente, N., Barros, L., Gonçalves, C. I., Al-Naama, A., Saraiva, L. R., & Lemos, M. C. (2022). A Novel FGFR1 Missense Mutation in a Portuguese Family with Congenital Hypogonadotropic Hypogonadism. International Journal of Molecular Sciences, 23(8), 4423. https://doi.org/10.3390/ijms23084423







