Chaperone Hsp70 (HSPA1) Is Involved in the Molecular Mechanisms of Sleep Cycle Integration
Abstract
:1. Introduction
2. Results
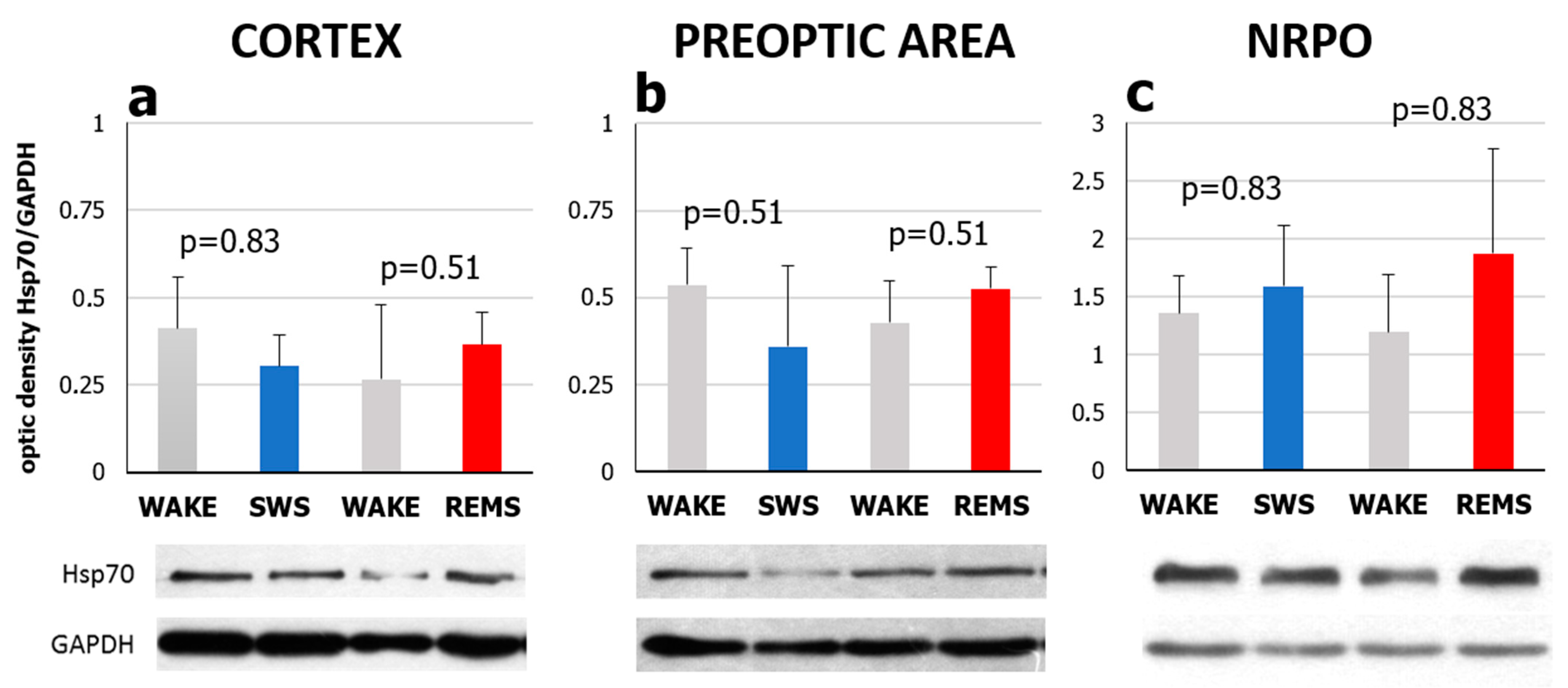
2.1. Natural REMS Is Accompanied by Hspa1 Upregulation
2.2. Short-Term Selective REMS Deprivation Suppresses Deep SWS and Induces REMS Rebound but Does Not Affect Hspa1 Expression
3. Discussion
3.1. Hspa1 Expression Is Upregulated in the NRPO during the Natural Daily Maximum of REM Sleep in Rats
3.2. Natural SWS-REMS Relation Should Be Preserved for the Hspa1 Upregulation to Occur
4. Materials and Methods
4.1. Animals
4.2. Surgical Procedure
4.3. Sleep Recording and Selective REM Sleep Deprivation (REMSD) Procedure
4.4. Sleep–Wake Cycle Scoring and Analysis
4.5. Experimental Procedure
4.6. Real-Time PCR and Western Blotting
4.7. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zepelin, H.; Siegel, J.M.; Tobler, I. Mammalian Sleep. In Principles and Practice of Sleep Medicine; Kryger, M.H., Roth, T., Dement, W.C., Eds.; Elsevier Saunders: St. Louis, MO, USA, 2005; pp. 91–100. ISBN 07970081/6/0. [Google Scholar]
- Pastukhov, Y.F. Slow-Wave Sleep and Molecular Chaperones. J. Evol. Biochem. Physiol. 2016, 52, 87–101. [Google Scholar] [CrossRef]
- Aserinsky, E.; Kleitman, N. Regularly Occurring Periods of Eye Motility, and Concomitant Phenomena, during Sleep. Science 1953, 118, 273–274. [Google Scholar] [CrossRef] [Green Version]
- Aserinsky, E. The Discovery of REM Sleep. J. Hist. Neurosci. 1996, 5, 213–227. [Google Scholar] [CrossRef]
- Jouvet, M. Paradoxical Sleep—A Study of Its Nature and Mechanisms. Prog. Brain Res. 1965, 18, 20–62. [Google Scholar] [CrossRef]
- Jouvet, M. Correlations Electromyographiques Du Sommeil Chez Le Chat Decortique et Mesencephalique Chronique. Comptes Rendus Soc. Biol. 1959, 153, 422–425. [Google Scholar]
- Brown, R.E.; Basheer, R.; McKenna, J.T.; Strecker, R.E.; McCarley, R.W. Control of Sleep and Wakefulness. Physiol. Rev. 2012, 92, 1087–1187. [Google Scholar] [CrossRef] [Green Version]
- Dement, W.; Kleitman, N. Cyclic Variations in EEG during Sleep and Their Relation to Eye Movements, Body Motility, and Dreaming. Electroencephalogr. Clin. Neurophysiol. 1957, 9, 673–690. [Google Scholar] [CrossRef]
- Jouvet, M. Recherches Sur Les Structures Nerveuses et Les Mecanismes Responsables Des Differentes Phases Du Sommeil Physiologique. Arch. Ital. Biol. 1962, 100, 125–206. [Google Scholar] [PubMed]
- Frank, M.G.; Heller, H.C. The Function(s) of Sleep. In Sleep-Wake Neurobiology and Pharmacology. Handbook of Experimental Pharmacology; Landolt, H.P., Dijk, D.J., Eds.; Springer: Berlin/Heidelberg, Germany, 2018; pp. 3–34. [Google Scholar]
- Blumberg, M.S.; Lesku, J.A.; Libourel, P.-A.; Schmidt, M.H.; Rattenborg, N.C. What Is REM Sleep? Curr. Biol. Rev. 2020, 30, R38–R49. [Google Scholar] [CrossRef] [PubMed]
- Thakkar, M.M.; Datta, S. The Evolution of REM Sleep. In Evolution of Sleep: Phylogenetic and Functional Perspectives; McNamara, P., Barton, R.A., Nunn, C.L., Eds.; Cambridge University Press: New York, NY, USA, 2010; pp. 197–215. [Google Scholar]
- Siegel, J.M. Sleep in Animals: A State of Adaptive Inactivity. In Principles and Practice of Sleep Medicine, 5th ed.; Elsevier Saunders: St. Louis, MO, USA, 2010; pp. 126–138. ISBN 9781416066453. [Google Scholar]
- Vyazovskiy, V.V.; Delogu, A. NREM and REM Sleep: Complementary Roles in Recovery after Wakefulness. Neuroscientist 2014, 20, 203–219. [Google Scholar] [CrossRef]
- Dworak, M.; McCarley, R.W.; Kim, T.; Kalinchuk, A.V.; Basheer, R. Sleep and Brain Energy Levels: ATP Changes during Sleep. J. Neurosci. 2010, 30, 9007–9016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mackiewicz, M.; Shockley, K.R.; Romer, M.A.; Galante, R.J.; Zimmerman, J.E.; Naidoo, N.N.; Baldwin, D.A.; Jensen, S.T.; Churchill, G.A.; Pack, A.I. Macromolecule Biosynthesis: A Key Function of Sleep. Physiol. Genom. 2007, 31, 441–457. [Google Scholar] [CrossRef] [PubMed]
- Maquet, P. Sleep Function(s) and Cerebral Metabolism. Behav. Brain Res. 1995, 69, 75–83. [Google Scholar] [CrossRef]
- Nakanishi, H.; Sun, Y.; Nakamura, R.K.; Mori, K.; Ito, M.; Suda, S.; Namba, H.; Storch, F.I.; Dang, T.P.; Mendelson, W. Positive Correlations between Cerebral Protein Synthesis Rates and Deep Sleep in Macaca Mulatta. Eur. J. Neurosci. 1997, 9, 271–279. [Google Scholar] [CrossRef]
- Ramm, P.; Smith, C.T. Rates of Cerebral Protein Synthesis Are Linked to Slow Wave Sleep in the Rat. Physiol. Behav. 1990, 48, 749–753. [Google Scholar] [CrossRef]
- Schubert, U.; Anton, L.C.; Gibbs, J.; Norbury, C.C.; Yewdell, J.W.; Bennink, J.R. Rapid Degradation of a Large Fraction of Newly Synthesized Proteins by Proteasomes. Nature 2000, 404, 770–774. [Google Scholar] [CrossRef]
- Hartl, F.U.; Bracher, A.; Hayer-Hartl, M. Molecular Chaperones in Protein Folding and Proteostasis. Nature 2011, 475, 324–332. [Google Scholar] [CrossRef] [PubMed]
- Cirelli, C.; Tononi, G. Gene Expression in the Brain across the Sleep–Waking Cycle. Brain Res. 2000, 885, 303–321. [Google Scholar] [CrossRef]
- Terao, A.; Steininger, T.L.; Hyder, K.; Apte-Deshpande, A.; Ding, J.; Rishipathak, D.; Davis, R.W.; Heller, H.C.; Kilduff, T.S. Differential Increase in the Expression of Heat Shock Protein Family Members during Sleep Deprivation and during Sleep. Neuroscience 2003, 116, 187–200. [Google Scholar] [CrossRef]
- Terao, A.; Wisor, J.P.; Peyron, C.; Apte-Deshpande, A.; Wurts, S.W.; Edgar, D.M.; Kilduff, T.S. Gene Expression in the Rat Brain during Sleep Deprivation and Recovery Sleep: An Affymetrix GeneChip Study. Neuroscience 2006, 137, 593–605. [Google Scholar] [CrossRef] [Green Version]
- Naidoo, N.N. Cellular Stress/the Unfolded Protein Response: Relevance to Sleep and Sleep Disorders. Sleep Med. Rev. 2009, 13, 195–204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hasan, S.; Dauvilliers, Y.; Mongrain, V.; Franken, P.; Tafti, M. Age-Related Changes in Sleep in Inbred Mice Are Genotype Dependent. Neurobiol. Aging 2012, 33, 195.e13–195.e26. [Google Scholar] [CrossRef] [PubMed]
- Cirelli, C.; Gutierrez, C.M.; Tononi, G. Extensive and Divergent Effects of Sleep and Wakefulness on Brain Gene Expression. Neuron 2004, 41, 35–43. [Google Scholar] [CrossRef] [Green Version]
- Calasso, M.; Parmeggiani, P.L. Carotid Blood Flow during REM Sleep. Sleep 2008, 31, 701–707. [Google Scholar] [CrossRef] [Green Version]
- Pastukhov, Y.F.; Ekimova, I.V. The Thermophysiology of Paradoxical Sleep. Neurosci. Behav. Physiol. 2012, 42, 933–947. [Google Scholar] [CrossRef]
- Ellis, R.J. The Molecular Chaperone Concept. Semin. Cell Biol. 1990, 1, 1–9. [Google Scholar]
- Balchin, D.; Hayer-Hartl, M.; Hartl, F.U. In Vivo Aspects of Protein Folding and Quality Control. Science 2016, 353, aac4354. [Google Scholar] [CrossRef]
- Morozov, A.V.; Astakhova, T.M.; Garbuz, D.G.; Krasnov, G.S.; Bobkova, N.V.; Zatsepina, O.G.; Karpov, V.L.; Evgen’ev, M.B. Interplay between Recombinant Hsp70 and Proteasomes: Proteasome Activity Modulation and Ubiquitin-Independent Cleavage of Hsp70. Cell Stress Chaperones 2017, 22, 687–697. [Google Scholar] [CrossRef] [Green Version]
- Mogk, A.; Kummer, E.; Bukau, B. Cooperation of Hsp70 and Hsp100 Chaperone Machines in Protein Disaggregation. Front. Mol. Biosci. 2015, 2, 22. [Google Scholar] [CrossRef] [Green Version]
- Pastukhov, Y.F.; Ekimova, I.V.; Khudik, K.A.; Guzhova, I.V. Protein 70 KDa in the Control of Sleep and Thermoregulation. J. Evol. Biochem. Physiol. 2008, 44, 74–81. [Google Scholar] [CrossRef]
- Ekimova, I.V. Somnogenic Effect of Exogenous Heat Shock Protein 70 KDa Is Mediated by GABA(A) Receptors in the Preoptic Area of the Hypothalamus. Dokl. Biol. Sci. 2013, 449, 89–92. [Google Scholar] [CrossRef] [PubMed]
- Ekimova, I.V.; Pastukhov, Y.F. The Role of Adenosine A-2A Receptors of the Preoptic Area in Somnogenic Activity of 70 KDa Protein in Pigeons. J. Evol. Biochem. Physiol. 2014, 50, 492–499. [Google Scholar] [CrossRef]
- Pastukhov, Y.F.; Simonova, V.V.; Guzeev, M.A.; Meshalkina, D.A.; Guzhova, I.V.; Ekimova, I.V. Chaperone Hsp70 Is Involved in the Molecular Mechanisms of Slow Wave Sleep Regulation. Dokl. Biochem. Biophys. 2015, 461, 76. [Google Scholar] [CrossRef] [PubMed]
- Naidoo, N.N.; Giang, W.; Galante, R.J.; Pack, A.I. Sleep Deprivation Induces the Unfolded Protein Response in Mouse Cerebral Cortex. J. Neurochem. 2005, 92, 1150–1157. [Google Scholar] [CrossRef]
- Naidoo, N.N.; Zhu, J.; Galante, R.J.; Lian, J.; Strus, E.; Lee, A.; Keenan, B.T.; Pack, A.I. Reduction of the Molecular Chaperone BiP Accentuates the Effect of Aging on Sleep-Wake Behavior. Neurobiol. Aging 2018, 69, 10–25. [Google Scholar] [CrossRef] [PubMed]
- Benington, J.H.; Woudenberg, M.C.; Heller, H.C. REM-Sleep Propensity Accumulates during 2-h REM-Sleep Deprivation in the Rest Period in Rats. Neurosci. Lett. 1994, 180, 76–80. [Google Scholar] [CrossRef]
- Ocampo-Garcé, A.N.; Molina, E.; Guez, A.R.; Vivaldi, E.A. Homeostasis of REM Sleep after Total and Selective Sleep Deprivation in the Rat. J. Neurophysiol. 2000, 84, 2699–2702. [Google Scholar] [CrossRef]
- Benington, J.H.; Heller, H.C. REM-Sleep Timing Is Controlled Homeostatically by Accumulation of REM-Sleep Propensity in Non-REM Sleep. Am. J. Physiol. Regul. Integr. Comp. Physiol. 1994, 266, R1992–R2000. [Google Scholar] [CrossRef]
- Tobler, I.; Borbely, A.A. The Effect of 3-h and 6-h Sleep Deprivation on Sleep and EEG Spectra of the Rat. Behav. Brain Res. 1990, 36, 73–78. [Google Scholar] [CrossRef]
- Lasunskaia, E.B.; Fridlianskaia, I.I.; Guzhova, I.V.; Bozhkov, V.M.; Margulis, B.A. Accumulation of Major Stress Protein 70kDa Protects Myeloid and Lymphoid Cells from Death by Apoptosis. Apoptosis 1997, 2, 156–163. [Google Scholar] [CrossRef]
- Tabuchi, Y.; Takasaki, I.; Wada, S.; Zhao, Q.-L.; Hori, T.; Nomura, T.; Ohtsuka, K.; Kondo, T. Genes and Genetic Networks Responsive to Mild Hyperthermia in Human Lymphoma U937 Cells. Int. J. Hyperth. 2008, 24, 613–622. [Google Scholar] [CrossRef] [PubMed]
- McLean, P.J.; Klucken, J.; Shin, Y.; Hyman, B.T. Geldanamycin Induces Hsp70 and Prevents α-Synuclein Aggregation and Toxicity In Vitro. Biochem. Biophys. Res. Commun. 2004, 321, 665–669. [Google Scholar] [CrossRef]
- Ekimova, I.V.; Plaksina, D.V.; Pastukhov, Y.F.; Lapshina, K.V.; Lazarev, V.F.; Mikhaylova, E.R.; Polonik, S.G.; Pani, B.; Margulis, B.A.; Guzhova, I.V.; et al. New HSF1 Inducer as a Therapeutic Agent in a Rodent Model of Parkinson’s Disease. Exp. Neurol. 2018, 306, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Sistonen, L.; Sarge, K.D.; Morimoto, R.I. Human Heat Shock Factors 1 and 2 Are Differentially Activated and Can Synergistically Induce Hsp70 Gene Transcription. Mol. Cell. Biol. 1994, 14, 2087–2099. [Google Scholar] [CrossRef] [Green Version]
- Jones, B.E. Paradoxical Sleep and Its Chemical/Structural Substrates in the Brain. Neuroscience 1991, 40, 637–656. [Google Scholar] [CrossRef]
- Reinoso-Suarez, F.; de Andrés, I.; Rodrigo-Angulo, M.L.; Garzón, M. Brain Structures and Mechanisms Involved in the Generation of REM Sleep. Sleep Med. Rev. 2001, 5, 63–77. [Google Scholar] [CrossRef] [PubMed]
- Deurveilher, S.; Hars, B.; Hennevin, E. Pontine Microinjection of Carbachol Does Not Reliably Enhance Paradoxical Sleep in Rats. Sleep 1997, 20, 593–607. [Google Scholar] [CrossRef] [Green Version]
- Luppi, P.-H.; Fort, P. Neuroanatomical and Neurochemical Bases of Vigilance States. In Sleep-Wake Neurobiology and Pharmacology. Handbook of Experimental Pharmacology; Landolt, H.P., Dijk, D.J., Eds.; Springer: Berlin/Heidelberg, Germany, 2018; pp. 35–58. [Google Scholar]
- Cissé, Y.; Toossi, H.; Ishibashi, M.; Mainville, L.; Leonard, C.S.; Adamantidis, A.R.; Jones, B.E. Discharge and Role of Acetylcholine Pontomesencephalic Neurons in Cortical Activity and Sleep-Wake States Examined by Optogenetics and Juxtacellular Recording in Mice. Eneuro 2018, 5, ENEURO.0270-18.2018. [Google Scholar] [CrossRef] [Green Version]
- Rashotte, M.E.; Pastukhov, Y.F.; Poliakov, E.L.; Henderson, R.P. Vigilance States and Body Temperature during the Circadian Cycle in Fed and Fasted Pigeons (Columba livia). Am. J. Physiol. Integr. Comp. Physiol. 1998, 275, R1690. [Google Scholar] [CrossRef]
- Endo, T.; Schwierin, B.; Borbely, A.A.; Tobler, I. Selective and Total Sleep Deprivation: Effect on the Sleep EEG in the Rat. Psychiatry Res. 1997, 66, 97–110. [Google Scholar] [CrossRef]
- Ellis, J. Proteins as Molecular Chaperones. Nature 1987, 328, 378–379. [Google Scholar] [CrossRef] [PubMed]
- Dunker, A.K.; Silman, I.; Uversky, V.N.; Sussman, J.L. Function and Structure of Inherently Disordered Proteins. Curr. Opin. Struct. Biol. 2008, 18, 756–764. [Google Scholar] [CrossRef]
- Xie, L.; Kang, H.; Xu, Q.; Chen, M.J.; Liao, Y.; Thiyagarajan, M.; O’Donnell, J.; Christensen, D.J.; Nicholson, C.; Iliff, J.J.; et al. Sleep Drives Metabolite Clearance from the Adult Brain. Science 2013, 342, 373–377. [Google Scholar] [CrossRef] [Green Version]
- Shapiro, C.; Girdwood, P. Protein Synthesis in Rat Brain during Sleep. Neuropharmacology 1981, 20, 457–460. [Google Scholar] [CrossRef]
- Bobillier, P.; Sakai, F.; Seguin, S.; Jouvet, M. The Effect of Sleep Deprivation upon the in Vivo and in Vitro Incorporation of Tritiated Amino Acids into Brain Proteins in the Rat at Three Different Age Levels. J. Neurochem. 1974, 22, 23–31. [Google Scholar] [CrossRef]
- Uchida, S.; Hara, K.; Kobayashi, A.; Fujimoto, M.; Otsuki, K.; Yamagata, H.; Hobara, T.; Abe, N.; Higuchi, F.; Shibata, T. Impaired Hippocampal Spinogenesis and Neurogenesis and Altered Affective Behavior in Mice Lacking Heat Shock Factor 1. Proc. Natl. Acad. Sci. USA 2011, 108, 1681–1686. [Google Scholar] [CrossRef] [Green Version]
- Hooper, P.L.; Durham, H.D.; Török, Z.; Hooper, P.L.; Crul, T.; Vígh, L. The Central Role of Heat Shock Factor 1 in Synaptic Fidelity and Memory Consolidation. Cell Stress Chaperones 2016, 21, 745–753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pastukhov, Y.F.; Simonova, V.V.; Shemyakova, T.S.; Guzeev, M.A.; Polonik, S.G.; Ekimova, I.V. U-133, a Chaperone Inducer, Eliminates Sleep Disturbances in a Model of the Preclinical Stage of Parkinson’s Disease in Aged Rats. Adv. Gerontol. 2020, 10, 254–259. [Google Scholar] [CrossRef]
- Pastukhov, Y.F.; Simonova, V.V.; Chernyshev, M.V.; Guzeev, M.A.; Shemyakova, T.S.; Ekimova, I.V. Signs of Sleep and Behavior Disorders Indicating the Initial Stage of Neurodegeneration in a Rat Model of Parkinson’s Disease. J. Evol. Biochem. Physiol. 2017, 53, 431–434. [Google Scholar] [CrossRef]
- Hafycz, J.M.; Naidoo, N.N. Sleep, Aging, and Cellular Health: Aged-Related Changes in Sleep and Protein Homeostasis Converge in Neurodegenerative Diseases. Front. Aging Neurosci. 2019, 11, 140. [Google Scholar] [CrossRef]
- Arthaud, S.; Libourel, P.-A.; Luppi, P.-H.; Peyron, C. Insights into Paradoxical (REM) Sleep Homeostatic Regulation in Mice Using an Innovative Automated Sleep Deprivation Method. Sleep 2020, 43, aa003. [Google Scholar] [CrossRef] [PubMed]
- Revel, F.G.; Gottowik, J.; Gatti, S.; Wettstein, J.G.; Moreau, J.L. Rodent Models of Insomnia: A Review of Experimental Procedures That Induce Sleep Disturbances. Neurosci. Biobehav. Rev. 2009, 33, 874–899. [Google Scholar] [CrossRef] [PubMed]
- Fujihara, H.; Serino, R.; Ueta, Y.; Sei, H.; Morita, Y. Six-Hour Selective REM Sleep Deprivation Increases the Expression of the Galanin Gene in the Hypothalamus of Rats. Mol. Brain Res. 2003, 119, 152–159. [Google Scholar] [CrossRef] [PubMed]
- Paxinos, G.; Watson, C. The Rat Brain in Stereotaxic Coordinates (EBook), 6th ed.; Elsevier: Amsterdam, The Netherlands; Academic Press: Amsterdam, The Netherland, 2007; ISBN 9780123741219. [Google Scholar]
- Lee, K.S.; Alvarenga, T.A.; Guindalini, C.; Andersen, M.L.; Castro, R.M.R.P.S.; Tufik, S. Validation of Commonly Used Reference Genes for Sleep-Related Gene Expression Studies. BMC Mol. Biol. 2009, 10, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ravassard, P.; Hamieh, A.M.; Malleret, G.; Salin, P.-A.A. Paradoxical Sleep: A Vigilance State to Gate Long-Term Brain Plasticity? Neurobiol. Learn. Mem. 2015, 122, 4–10. [Google Scholar] [CrossRef] [PubMed]









| State and Time, ZT | Total Time 1, % of Registration Time | Episode Duration 1, s | Number of Episodes 1 | ||||
|---|---|---|---|---|---|---|---|
| Control 2 | REMSD 2 | Control | REMSD | Control | REMSD | ||
| Wakefulness | 00-02 | 35.3 ± 4.92 | 34.5 ± 4.73 | 244 ± 34.7 | 124 ± 27.2 * | 11 ± 0.9 | 22 ± 2.1 * |
| 02-04 | 19.4 ± 4.08 | 25.8 ± 5.05 | 115 ± 21.6 | 78 ± 18.6 | 14 ± 2.2 | 27 ± 4.6 * | |
| 04-06 | 29.5 ± 3.54 | 30.7 ± 4.33 | 174 ± 21.8 | 91 ± 17.1 * | 14 ± 2.0 | 27 ± 4.0 * | |
| 06-08 | 24.3 ± 4.25 | 19.9 ± 3.39 | 159 ± 40.8 | 131 ± 28.9 | 12 ± 1.4 | 11 ± 0.9 | |
| 08-10 | 22.7 ± 4.05 | 21.5 ± 3.86 | 150 ± 24.5 | 147 ± 41.6 | 10 ± 0.9 | 12 ± 1.8 | |
| 10-12 | 37.3 ± 5.46 | 32.8 ± 5.62 | 273 ± 49.7 | 249 ± 60.0 | 10 ± 1.5 | 11 ± 1.1 | |
| Drowsiness | 00-02 | 16.1 ± 2.44 | 23.3 ± 2.89 | 41 ± 3.8 | 41 ± 3.3 | 28 ± 3.0 | 41 ± 4.7 * |
| 02-04 | 17.4 ± 2.10 | 28.2.2 ± 2.66 * | 39 ± 2.2 | 35 ± 2.8 | 36 ± 5.2 | 61 ± 6.3 * | |
| 04-06 | 18.6 ± 2.60 | 27.6 ± 3.21 * | 44 ± 5.0 | 34 ± 2.7 | 32 ± 3.2 | 59 ± 4.4 * | |
| 06-08 | 19.4 ± 1.82 | 17.3 ± 2.52 | 43 ± 2.9 | 40 ± 3.7 | 33 ± 2.8 | 30 ± 2.3 | |
| 08-10 | 20.1 ± 3.52 | 19.5 ± 3.30 | 42 ± 3.7 | 42 ± 4.7 | 32 ± 3.7 | 34 ± 3.9 | |
| 10-12 | 21.5 ± 4.35 | 20.0 ± 2.48 | 50 ± 4.8 | 44 ± 5.3 | 29 ± 3.1 | 34 ± 2.9 | |
| Slow-wave sleep | 00-02 | 40.0 ± 3.19 | 39.7 ± 2.39 | 138 ± 14.8 | 101 ± 7.8 * | 23 ± 2.9 | 30 ± 3.3 |
| 02-04 | 50.4 ± 3.95 | 38.0 ± 1.85 * | 155 ± 25.0 | 73 ± 7.0 * | 26 ± 2.6 | 40 ± 4.3 * | |
| 04-06 | 41.3 ± 2.72 | 33.4 ± 2.58 * | 136 ± 15.1 | 64 ± 8.2 * | 24 ± 2.0 | 40 ± 4.3 * | |
| 06-08 | 41.7 ± 2.28 | 43.7 ± 2.67 | 125 ± 11.9 | 132 ± 11.5 | 26 ± 2.5 | 24 ± 1.8 | |
| 08-10 | 42.6 ± 2.84 | 41.6 ± 2.15 | 129 ± 13.9 | 126 ± 24.4 | 25 ± 2.8 | 27 ± 2.8 | |
| 10-12 | 30.3 ± 3.29 | 34.2 ± 3.80 | 107 ± 21.0 | 97 ± 8.4 | 22 ± 2.5 | 26 ± 2.8 | |
| REM sleep | 00-02 | 8.5 ± 1.27 | 2.4 ± 0.79 * | 128 ± 12.3 | 17 ± 2.0 * | 5 ± 0.9 | 10 ± 3.0 |
| 02-04 | 12.9 ± 1.21 | 8.0 ± 1.62 * | 166 ± 14.5 | 18 ± 1.9 * | 9 ± 4.1 | 32 ± 5.5 * | |
| 04-06 | 10.6 ± 1.35 | 8.4 ± 1.58 | 145 ± 11.1 | 21 ± 5.3 * | 8 ± 2.5 | 35 ± 6.7 * | |
| 06-08 | 14.5 ± 1.51 | 19.1 ± 1.42 * | 149 ± 13.3 | 169 ± 13.5 | 7 ± 0.7 | 8 ± 1.0 | |
| 08-10 | 14.6 ± 1.62 | 17.4 ± 1.33 | 157 ± 10.3 | 151 ± 14.7 | 7 ± 0.9 | 9 ± 0.8 | |
| 10-12 | 10.9 ± 1.80 | 13.0 ± 1.82 | 161 ± 15.2 | 172 ± 12.1 | 5 ± 1.8 | 6 ± 1.3 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Simonova, V.V.; Guzeev, M.A.; Ekimova, I.V.; Pastukhov, Y.F. Chaperone Hsp70 (HSPA1) Is Involved in the Molecular Mechanisms of Sleep Cycle Integration. Int. J. Mol. Sci. 2022, 23, 4464. https://doi.org/10.3390/ijms23084464
Simonova VV, Guzeev MA, Ekimova IV, Pastukhov YF. Chaperone Hsp70 (HSPA1) Is Involved in the Molecular Mechanisms of Sleep Cycle Integration. International Journal of Molecular Sciences. 2022; 23(8):4464. https://doi.org/10.3390/ijms23084464
Chicago/Turabian StyleSimonova, Valentina V., Mikhail A. Guzeev, Irina V. Ekimova, and Yuri F. Pastukhov. 2022. "Chaperone Hsp70 (HSPA1) Is Involved in the Molecular Mechanisms of Sleep Cycle Integration" International Journal of Molecular Sciences 23, no. 8: 4464. https://doi.org/10.3390/ijms23084464
APA StyleSimonova, V. V., Guzeev, M. A., Ekimova, I. V., & Pastukhov, Y. F. (2022). Chaperone Hsp70 (HSPA1) Is Involved in the Molecular Mechanisms of Sleep Cycle Integration. International Journal of Molecular Sciences, 23(8), 4464. https://doi.org/10.3390/ijms23084464






