Functional Characterization of Cardiac Actin Mutants Causing Hypertrophic (p.A295S) and Dilated Cardiomyopathy (p.R312H and p.E361G)
Abstract
:1. Introduction
2. Results
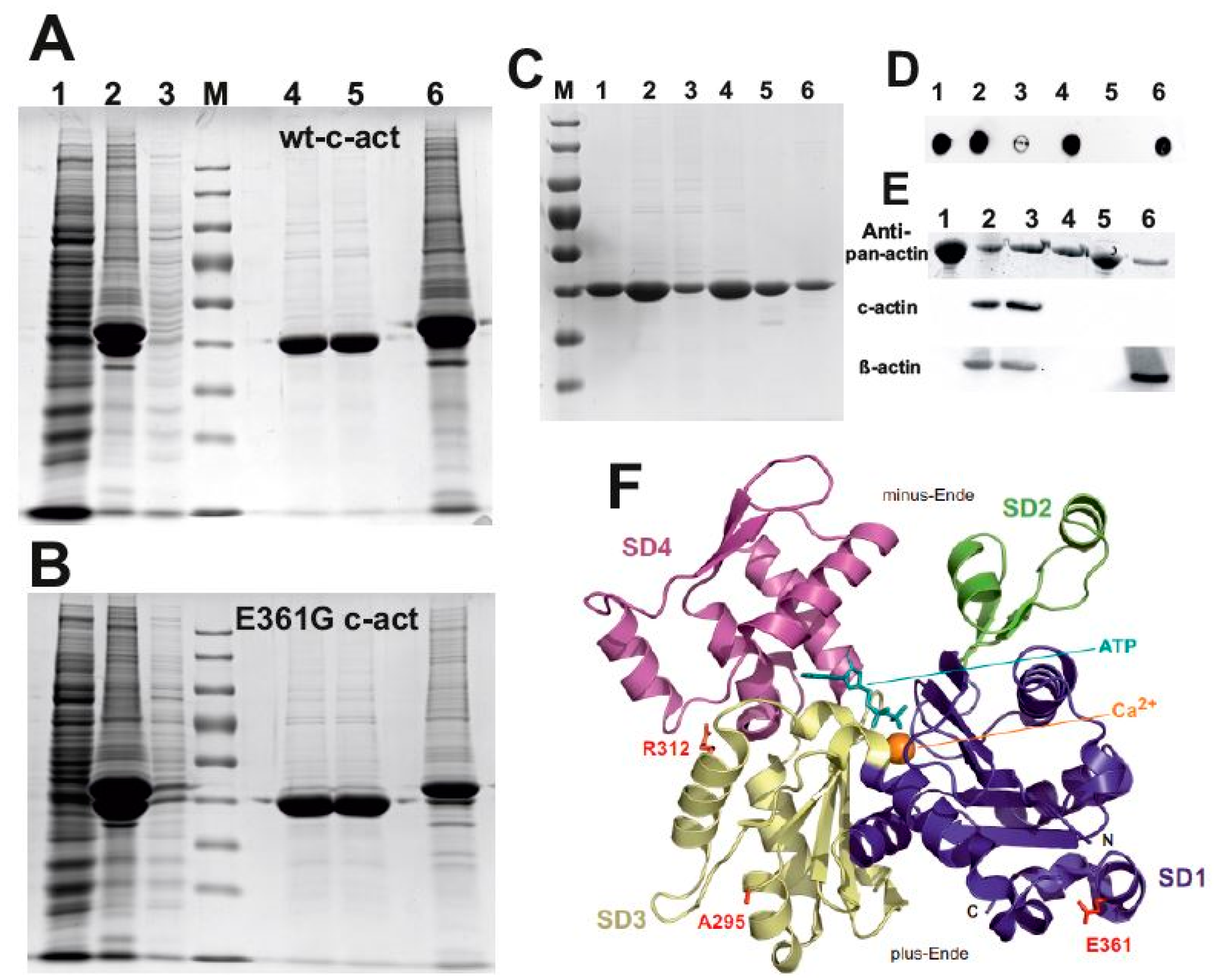
2.1. Expression and Purification of the Cardiac α-Actin Variants
2.2. Tests for Native Configuration of the Cardiac Actin Variants
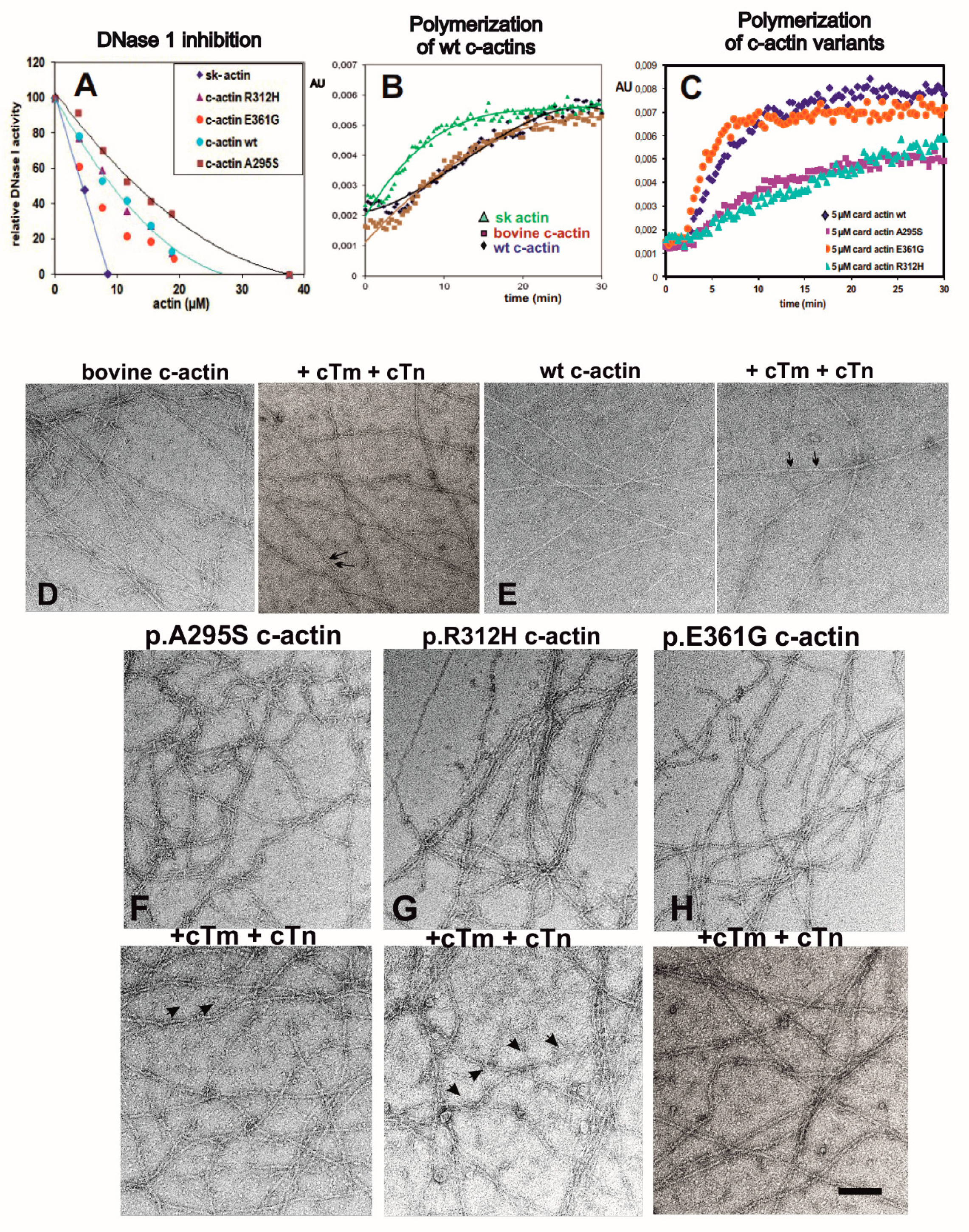
2.3. Polymerization Behavior of Recombinant c-α-Actins
2.4. Electron Microscopy of Filaments Formed by the c-Actin Variants
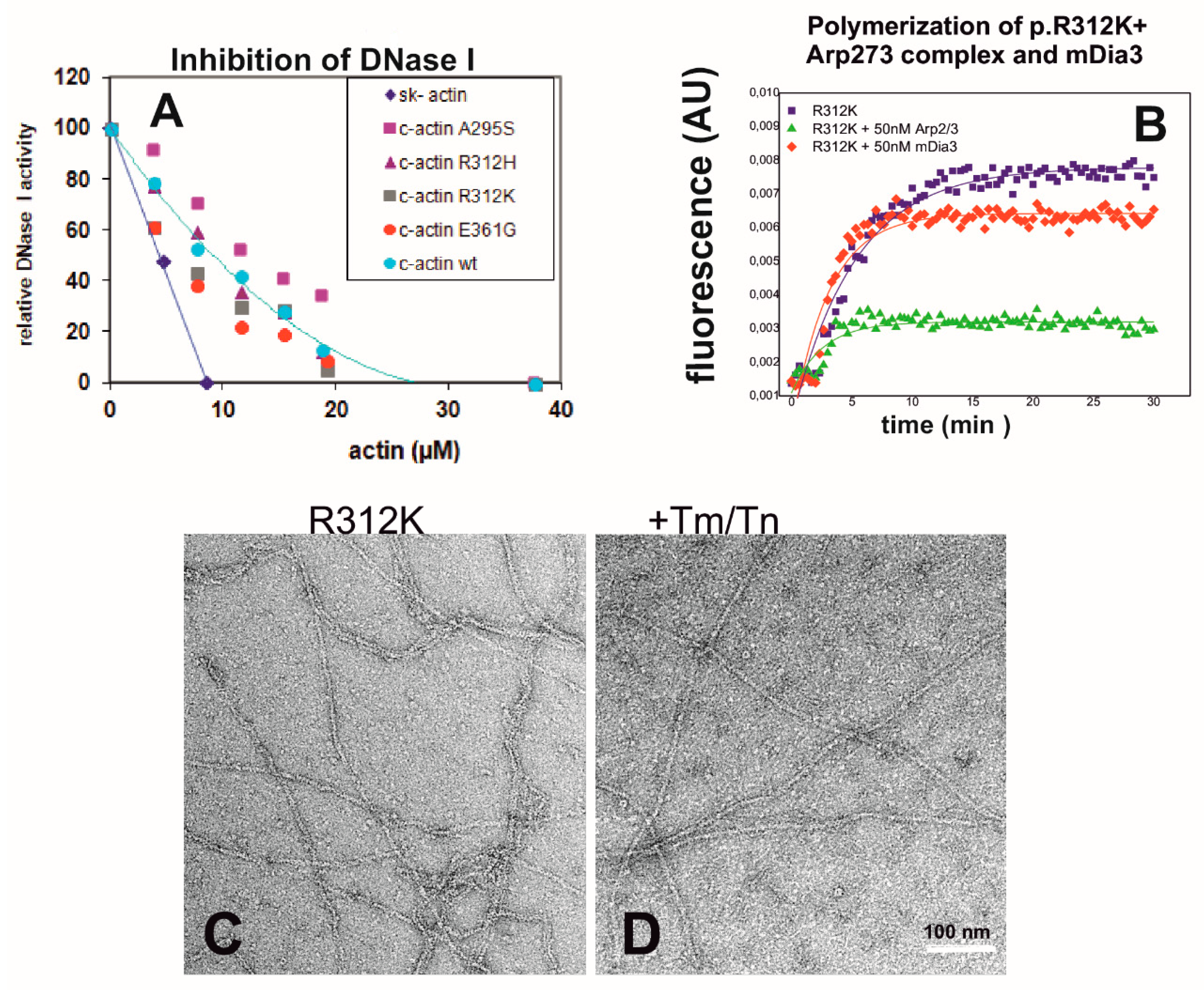
2.5. Properties of p.R312K c-Actin—Probably a Polymorphic Variant
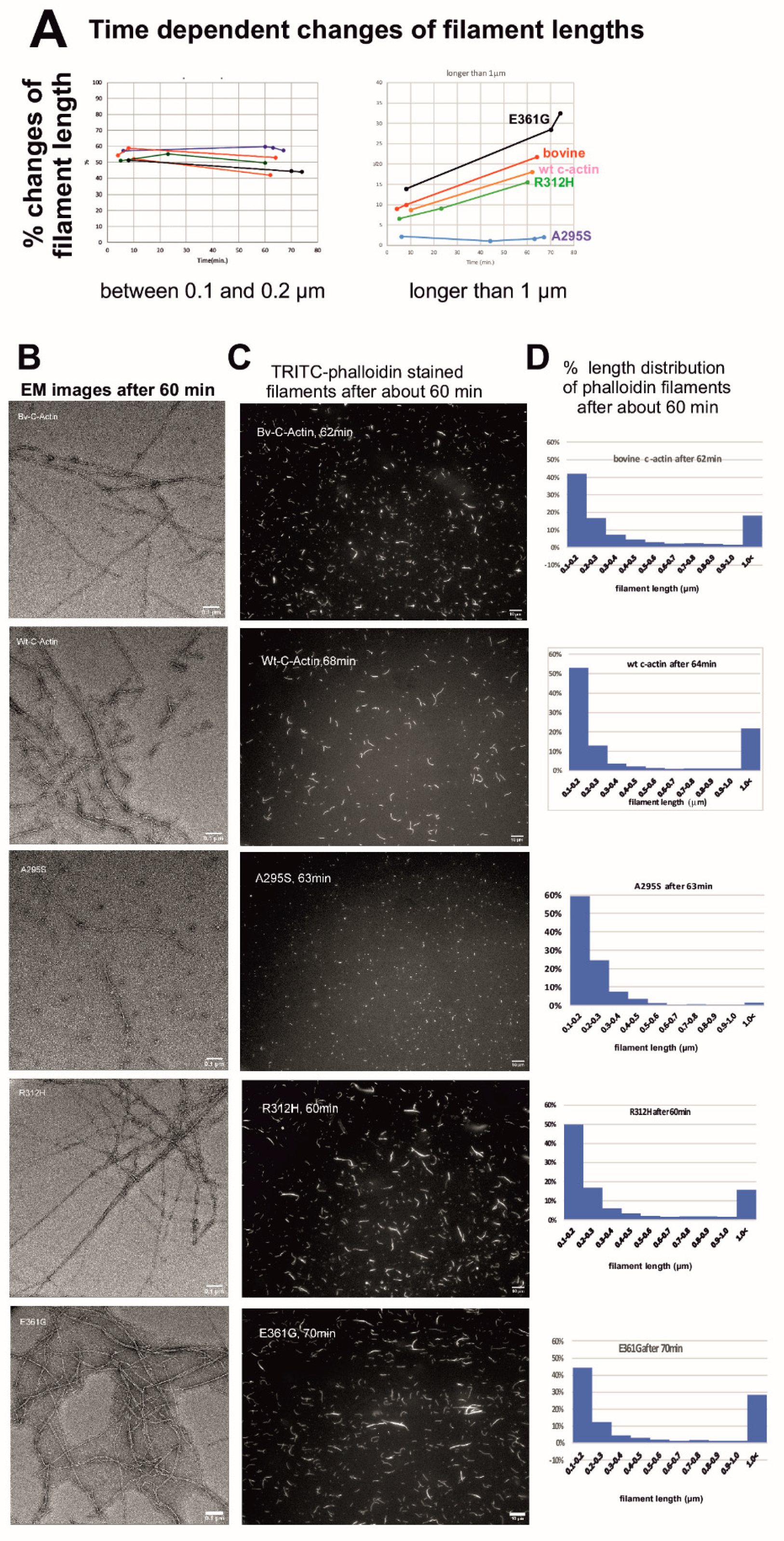
2.6. Determination of Filament Lengths by Fluorescence Microscopy
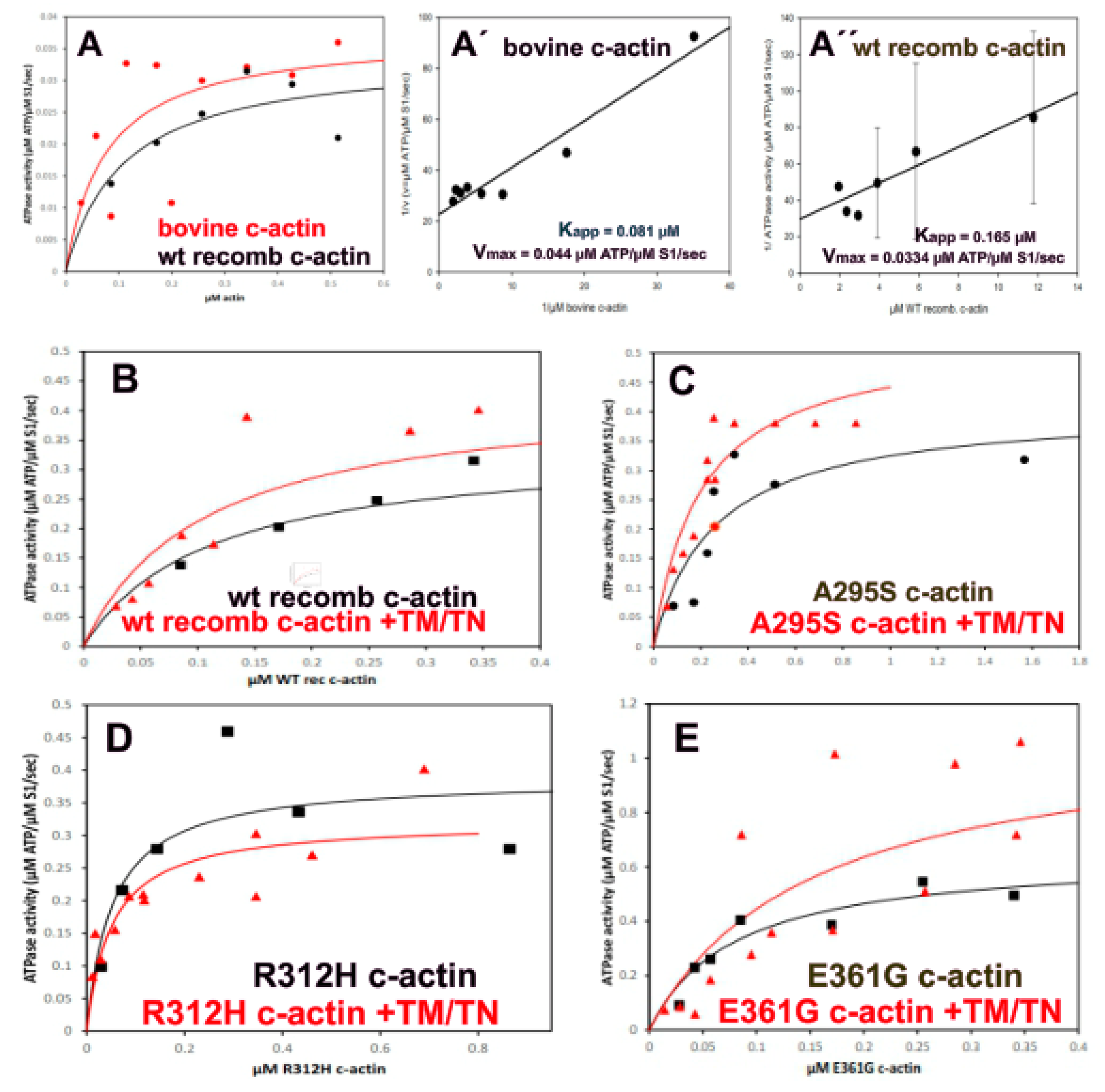
2.7. Stimulation of the Myosin-S1 ATPase Activity by Filamentous c-Actins
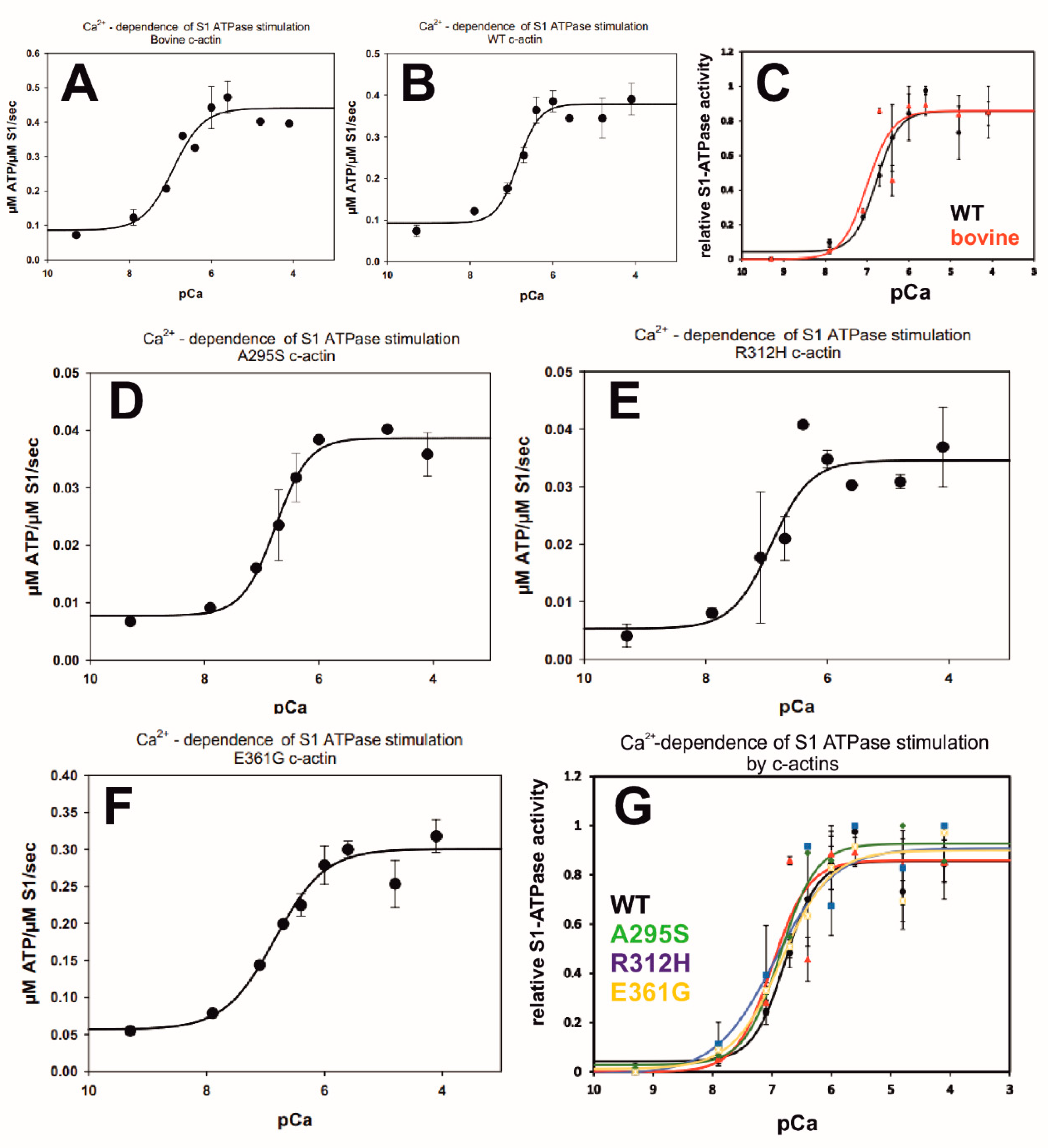
2.8. Ca2+-Dependence of the Myosin-S1 ATPase Stimulation by c-α-Actin Variants
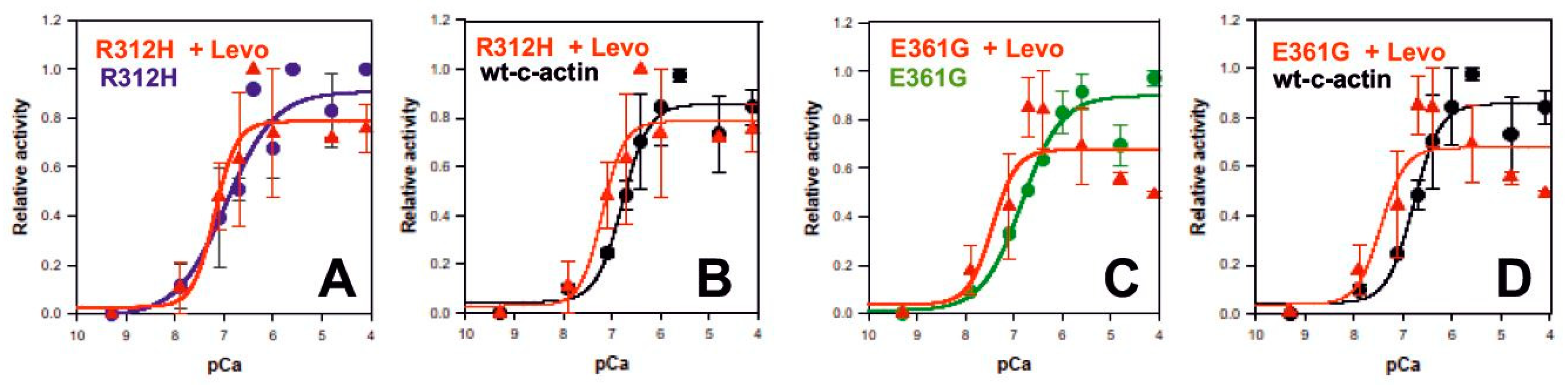
2.9. Levosimendan Affects the Ca2+-Dependence of Myosin-S1 ATPase Stimulation by the DCM c-Actin Mutants
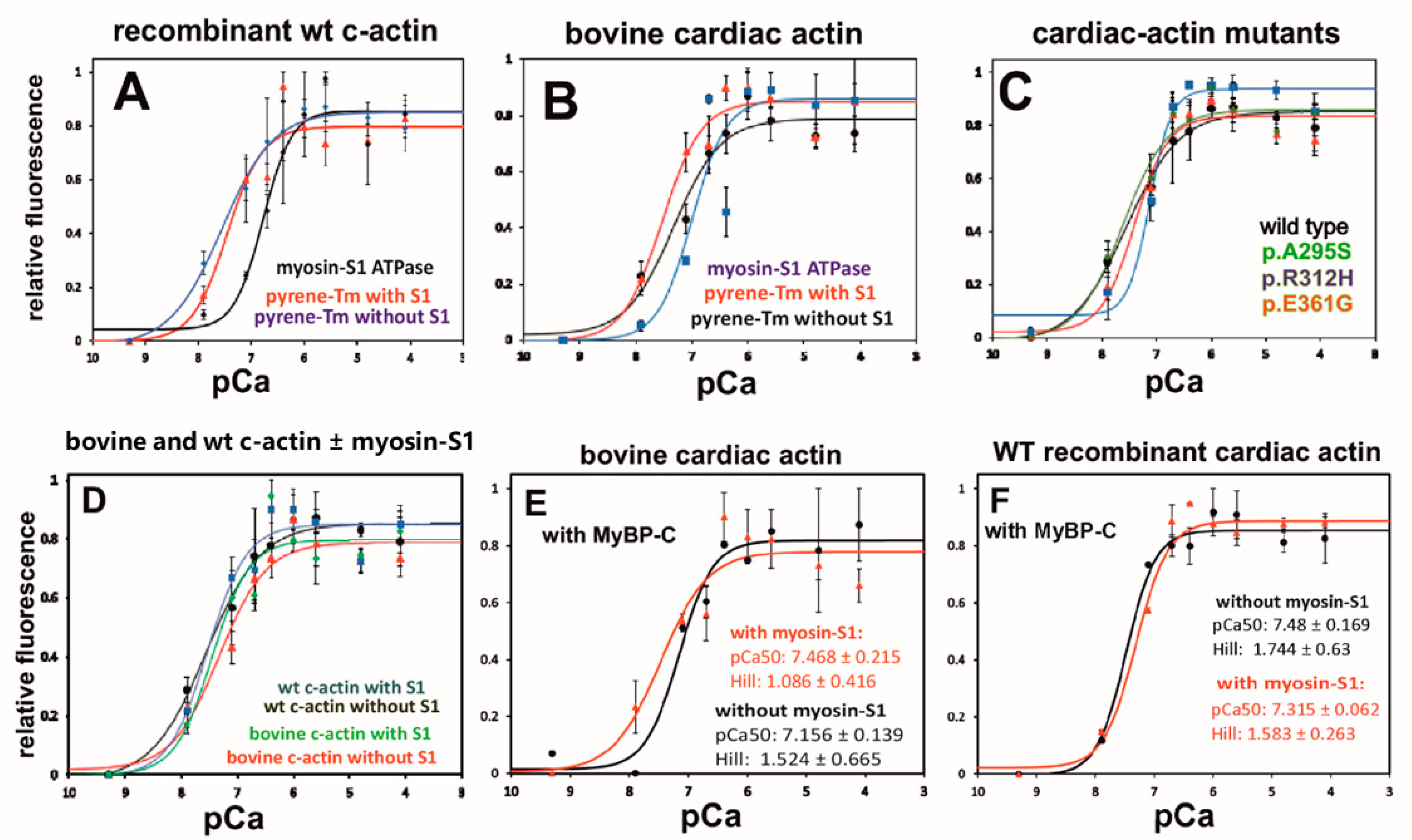
2.10. Ca2+-Dependence of Tropomyosin Movement on c-α-Actin Variants
2.11. Effect of the N-Terminal C0-C2 Segment of Myosin-Binding Protein-C on the Ca2+-Dependency of Pyrene-cTm Movement
3. Discussion
4. Materials and Methods
4.1. Antibodies and Reagents
4.2. Clones
4.3. Protein Expression and Purification
4.4. ATPase Assay
4.5. DNase I Inhibition Assay
4.6. Gel Electrophoresis and Immunoblotting
4.7. Actin Polymerization Assays
4.8. Electron Microscopy
4.9. Determination of the Ca2+-Dependence of Tm Movement on Cardiac F-Actins
4.10. Determination of Filaments Length by Fluorescence Microscopy
4.11. Imaging Techniques
4.12. Analytical Tools
4.13. Data Evaluation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sabater-Molina, M.; Pérez-Sánchez, I.; Hernández Del Rincón, J.P.; Gimeno, J.R. Genetics of Hypertrophic Cardiomyopathy: A Review of Current State. Clin. Genet. 2018, 93, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Barry, S.P.; Davidson, S.M.; Townsend, P.A. Molecular Regulation of Cardiac Hypertrophy. Int. J. Biochem. Cell Biol. 2008, 40, 2023–2039. [Google Scholar] [CrossRef] [PubMed]
- Weintraub, R.G.; Semsarian, C.; Macdonald, P. Dilated Cardiomyopathy. Lancet 2017, 390, 400–414. [Google Scholar] [CrossRef]
- Lynn, M.L.; Lehman, S.J.; Tardiff, J.C. Biophysical Derangements in Genetic Cardiomyopathies. Heart Fail. Clin. 2018, 14, 147–159. [Google Scholar] [CrossRef] [PubMed]
- Olson, T.M.; Doan, T.P.; Kishimoto, N.Y.; Whitby, F.G.; Ackerman, M.J.; Fananapazir, L. Inherited and de Novo Mutations in the Cardiac Actin Gene Cause Hypertrophic Cardiomyopathy. J. Mol. Cell. Cardiol. 2000, 32, 1687–1694. [Google Scholar] [CrossRef] [PubMed]
- Marston, S.B. How Do Mutations in Contractile Proteins Cause the Primary Familial Cardiomyopathies? J. Cardiovasc. Transl. Res. 2011, 4, 245–255. [Google Scholar] [CrossRef]
- Yamada, Y.; Namba, K.; Fujii, T. Cardiac Muscle Thin Filament Structures Reveal Calcium Regulatory Mechanism. Nat. Commun. 2020, 11, 153. [Google Scholar] [CrossRef]
- Ehler, E. Actin-Associated Proteins and Cardiomyopathy-the “unknown” beyond Troponin and Tropomyosin. Biophys. Rev. 2018, 10, 1121–1128. [Google Scholar] [CrossRef] [Green Version]
- Chow, M.L.; Shaffer, J.F.; Harris, S.P.; Dawson, J.F. Altered Interactions between Cardiac Myosin Binding Protein-C and α-Cardiac Actin Variants Associated with Cardiomyopathies. Arch. Biochem. Biophys. 2014, 550–551, 28–32. [Google Scholar] [CrossRef] [Green Version]
- Perriard, J.-C.; Hirschy, A.; Ehler, E. Dilated Cardiomyopathy: A Disease of the Intercalated Disc? Trends Cardiovasc. Med. 2003, 13, 30–38. [Google Scholar] [CrossRef]
- Herman, D.S.; Lam, L.; Taylor, M.R.G.; Wang, L.; Teekakirikul, P.; Christodoulou, D.; Conner, L.; DePalma, S.R.; McDonough, B.; Sparks, E.; et al. Truncations of Titin Causing Dilated Cardiomyopathy. N. Engl. J. Med. 2012, 366, 619–628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Müller, M.; Mazur, A.J.; Behrmann, E.; Diensthuber, R.P.; Radke, M.B.; Qu, Z.; Littwitz, C.; Raunser, S.; Schoenenberger, C.-A.; Manstein, D.J.; et al. Functional Characterization of the Human α-Cardiac Actin Mutations Y166C and M305L Involved in Hypertrophic Cardiomyopathy. Cell. Mol. Life Sci. 2012, 69, 3457–3479. [Google Scholar] [CrossRef] [PubMed]
- Debold, E.P.; Saber, W.; Cheema, Y.; Bookwalter, C.S.; Trybus, K.M.; Warshaw, D.M.; Vanburen, P. Human Actin Mutations Associated with Hypertrophic and Dilated Cardiomyopathies Demonstrate Distinct Thin Filament Regulatory Properties in Vitro. J. Mol. Cell. Cardiol. 2010, 48, 286–292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, J.-J.; Marston, S. Genotype–Phenotype Correlations in ACTA1 Mutations That Cause Congenital Myopathies. Neuromuscul. Disord. 2009, 19, 6–16. [Google Scholar] [CrossRef] [PubMed]
- Olson, T.M.; Michels, V.V.; Thibodeau, S.N.; Tai, Y.S.; Keating, M.T. Actin Mutations in Dilated Cardiomyopathy, a Heritable Form of Heart Failure. Science 1998, 280, 750–752. [Google Scholar] [CrossRef] [Green Version]
- Mogensen, J.; Klausen, I.C.; Pedersen, A.K.; Egeblad, H.; Bross, P.; Kruse, T.A.; Gregersen, N.; Hansen, P.S.; Baandrup, U.; Borglum, A.D. Alpha-Cardiac Actin Is a Novel Disease Gene in Familial Hypertrophic Cardiomyopathy. J. Clin. Invest. 1999, 103, R39–R43. [Google Scholar] [CrossRef] [Green Version]
- Parker, F.; Baboolal, T.G.; Peckham, M. Actin Mutations and Their Role in Disease. Int. J. Mol. Sci. 2020, 21, 3371. [Google Scholar] [CrossRef]
- Bookwalter, C.S.; Trybus, K.M. Functional Consequences of a Mutation in an Expressed Human α-Cardiac Actin at a Site Implicated in Familial Hypertrophic Cardiomyopathy. J. Biol. Chem. 2006, 281, 16777–16784. [Google Scholar] [CrossRef] [Green Version]
- Spudich, J.A. The Myosin Mesa and a Possible Unifying Hypothesis for the Molecular Basis of Human Hypertrophic Cardiomyopathy. Biochem. Soc. Trans. 2015, 43, 64–72. [Google Scholar] [CrossRef] [Green Version]
- Viswanathan, M.C.; Schmidt, W.; Rynkiewicz, M.J.; Agarwal, K.; Gao, J.; Katz, J.; Lehman, W.; Cammarato, A. Distortion of the Actin A-Triad Results in Contractile Disinhibition and Cardiomyopathy. Cell Rep. 2017, 20, 2612–2625. [Google Scholar] [CrossRef] [Green Version]
- Bai, F.; Caster, H.M.; Rubenstein, P.A.; Dawson, J.F.; Kawai, M. Using Baculovirus/Insect Cell Expressed Recombinant Actin to Study the Molecular Pathogenesis of HCM Caused by Actin Mutation A331P. J. Mol. Cell. Cardiol. 2014, 74, 64–75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mazur, A.J. Expression of Constructs of WT-Alpha-Cardiac Actin and Its Mutants in Different Cell Lines and Primary Rat Cardiac Myocytes. Ph.D Thesis, Ruhr-University Bochum, Bochum, Germany, 2008. [Google Scholar]
- Al Haj, A.; Mazur, A.J.; Radaszkiewicz, K.; Radaszkiewicz, T.; Makowiecka, A.; Stopschinski, B.E.; Schönichen, A.; Geyer, M.; Mannherz, H.G. Distribution of Formins in Cardiac Muscle: FHOD1 Is a Component of Intercalated Discs and Costameres. Eur. J. Cell Biol. 2015, 94, 101–113. [Google Scholar] [CrossRef]
- Bennett, P.M. Riding the Waves of the Intercalated Disc of the Heart. Biophys. Rev. 2018, 10, 955–959. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kabsch, W.; Mannherz, H.G.; Suck, D.; Pai, E.F.; Holmes, K.C. Atomic Structure of the Actin:DNase I Complex. Nature 1990, 347, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Mannherz, H.G.; Goody, R.S.; Konrad, M.; Nowak, E. The Interaction of Bovine Pancreatic Deoxyribonuclease I and Skeletal Muscle Actin. Eur. J. Biochem. 1980, 104, 367–379. [Google Scholar] [CrossRef] [PubMed]
- Kouyama, T.; Mihashi, K. Fluorimetry Study of N-(1-Pyrenyl)Iodoacetamide-Labelled F-Actin. Local Structural Change of Actin Protomer Both on Polymerization and on Binding of Heavy Meromyosin. Eur. J. Biochem. 1981, 114, 33–38. [Google Scholar] [CrossRef]
- Behrmann, E.; Müller, M.; Penczek, P.A.; Mannherz, H.G.; Manstein, D.J.; Raunser, S. Structure of the Rigor Actin-Tropomyosin-Myosin Complex. Cell 2012, 150, 327–338. [Google Scholar] [CrossRef] [Green Version]
- Erdmann, C.; Hassoun, R.; Schmitt, S.; Kikuti, C.; Houdusse, A.; Mazur, A.J.; Mügge, A.; Hamdani, N.; Geyer, M.; Jaquet, K.; et al. Integration of Cardiac Actin Mutants Causing Hypertrophic (p.A295S) and Dilated Cardiomyopathy (p.R312H and p.E361G) into Cellular Structures. Antioxidants 2021, 10, 1082. [Google Scholar] [CrossRef]
- Ebashi, S.; Endo, M.; Otsuki, I. Control of Muscle Contraction. Q. Rev. Biophys. 1969, 2, 351–384. [Google Scholar] [CrossRef]
- Mannherz, H.G.; Brehme, H.; Lamp, U. Depolymerisation of F-Actin to G-Actin and Its Repolymerisation in the Presence of Analogs of Adenosine Triphosphate. Eur. J. Biochem. 1975, 60, 109–116. [Google Scholar] [CrossRef]
- Geeves, M.A.; Lehrer, S.S. Dynamics of the Muscle Thin Filament Regulatory Switch: The Size of the Cooperative Unit. Biophys. J. 1994, 67, 273–282. [Google Scholar] [CrossRef] [Green Version]
- Maytum, R.; Lehrer, S.S.; Geeves, M.A. Cooperativity and Switching within the Three-State Model of Muscle Regulation. Biochemistry 1999, 38, 1102–1110. [Google Scholar] [CrossRef] [PubMed]
- Geeves, M.A.; Lehrer, S.S.; Lehman, W. The Mechanism of Thin Filament Regulation: Models in Conflict? J. Gen. Physiol. 2019, 151, 1265–1271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lehman, W. Thin Filament Structure and the Steric Blocking Model. Compr. Physiol. 2016, 6, 1043–1069. [Google Scholar] [PubMed]
- Lehman, W.; Rynkiewicz, M.J.; Moore, J.R. A New Twist on Tropomyosin Binding to Actin Filaments: Perspectives on Thin Filament Function, Assembly and Biomechanics. J. Muscle Res. Cell Motil. 2020, 41, 23–38. [Google Scholar] [CrossRef]
- Graceffa, P.; Lehrer, S.S. The Excimer Fluorescence of Pyrene-Labeled Tropomyosin. A Probe of Conformational Dynamics. J. Biol. Chem. 1980, 255, 11296–11300. [Google Scholar] [CrossRef]
- Ishii, Y.; Lehrer, S.S. Fluorescence Studies of the Conformation of Pyrene-Labeled Tropomyosin: Effects of F-Actin and Myosin Subfragment 1. Biochemistry 1985, 24, 6631–6638. [Google Scholar] [CrossRef]
- Ishii, Y.; Lehrer, S.S. Excimer Fluorescence of Pyrenyliodoacetamide-Labeled Tropomyosin: A Probe of the State of Tropomyosin in Reconstituted Muscle Thin Filaments. Biochemistry 1990, 29, 1160–1166. [Google Scholar] [CrossRef]
- Offer, G. Myosin-Binding Protein-C: Bridging the Gap. J. Mol. Biol. 2015, 427, 231–235. [Google Scholar] [CrossRef]
- Bunch, T.A.; Kanassatega, R.-S.; Lepak, V.C.; Colson, B.A. Human Cardiac Myosin-Binding Protein C Restricts Actin Structural Dynamics in a Cooperative and Phosphorylation-Sensitive Manner. J. Biol. Chem. 2019, 294, 16228–16240. [Google Scholar] [CrossRef] [Green Version]
- Starr, R.; Offer, G. The Interaction of C-Protein with Heavy Meromyosin and Subfragment-2. Biochem. J. 1978, 171, 813–816. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Risi, C.; Belknap, B.; Forgacs-Lonart, E.; Harris, S.P.; Schröder, G.F.; White, H.D.; Galkin, V.E. N-Terminal Domains of Cardiac Myosin Binding Protein C Cooperatively Activate the Thin Filament. Structure 2018, 26, 1604–1611. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cimiotti, D.; Fujita-Becker, S.; Möhner, D.; Smolina, N.; Budde, H.; Wies, A.; Morgenstern, L.; Gudkova, A.; Sejersen, T.; Sjöberg, G.; et al. Infantile Restrictive Cardiomyopathy: CTnI-R170G/W Impair the Interplay of Sarcomeric Proteins and the Integrity of Thin Filaments. PLoS One 2020, 15, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Taylor, R.S.; Weeds, A.G. The Magnesium-Ion-Dependent Adenosine Triphosphatase of Bovine Cardiac Myosin and Its Subfragment-1. Biochem. J. 1976, 159, 301–315. [Google Scholar] [CrossRef] [Green Version]
- Erdmann, C. Charakterisierung von HCM Und DCM Auslösenden Mutationen in Humanem Kardialem α-Aktin. Ph.D. Thesis, Ruhr-University Bochum, Bochum, Germany, 2017. [Google Scholar]
- Carrier, L.; Mearini, G.; Stathopoulou, K.; Cuello, F. Cardiac Myosin-Binding Protein C (MYBPC3) in Cardiac Pathophysiology. Gene 2015, 573, 188–197. [Google Scholar] [CrossRef]
- Trivedi, D.V.; Adhikari, A.S.; Sarkar, S.S.; Ruppel, K.M.; Spudich, J.A. Hypertrophic Cardiomyopathy and the Myosin Mesa: Viewing an Old Disease in a New Light. Biophys. Rev. 2018, 10, 27–48. [Google Scholar] [CrossRef] [Green Version]
- Mogensen, J.; Perrot, A.; Andersen, P.S.; Havndrup, O.; Klausen, I.C.; Christiansen, M.; Bross, P.; Egeblad, H.; Bundgaard, H.; Osterziel, K.J.; et al. Clinical and Genetic Characteristics of Alpha Cardiac Actin Gene Mutations in Hypertrophic Cardiomyopathy. J. Med. Genet. 2004, 41, e10. [Google Scholar] [CrossRef] [Green Version]
- von der Ecken, J.; Müller, M.; Lehman, W.; Manstein, D.J.; Penczek, P.A.; Raunser, S. Structure of the F-Actin-Tropomyosin Complex. Nature 2015, 519, 114–117. [Google Scholar] [CrossRef] [Green Version]
- Wong, W.W.; Doyle, T.C.; Cheung, P.; Olson, T.M.; Reisler, E. Functional Studies of Yeast Actin Mutants Corresponding to Human Cardiomyopathy Mutations. J. Muscle Res. Cell Motil. 2001, 22, 665–674. [Google Scholar] [CrossRef]
- Ohki, T.; Ohno, C.; Oyama, K.; Mikhailenko, S.V.; Ishiwata, S. Purification of Cytoplasmic Actin by Affinity Chromatography Using the C-Terminal Half of Gelsolin. Biochem. Biophys. Res. Commun. 2009, 383, 146–150. [Google Scholar] [CrossRef]
- Song, W.; Dyer, E.; Stuckey, D.; Leung, M.-C.; Memo, M.; Mansfield, C.; Ferenczi, M.; Liu, K.; Redwood, C.; Nowak, K.; et al. Investigation of a Transgenic Mouse Model of Familial Dilated Cardiomyopathy. J. Mol. Cell. Cardiol. 2010, 49, 380–389. [Google Scholar] [CrossRef] [PubMed]
- Bailey, K. Tropomyosin: A New Asymmetric Protein Component of the Muscle Fibril. Biochem. J. 1948, 43, 271–279. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Schmidtmann, A.; Redlich, A.; Westerdorf, B.; Jaquet, K.; Thieleczek, R. Effects of Phosphorylation and Mutation R145G on Human Cardiac Troponin I Function. Biochemistry 2001, 40, 14593–14602. [Google Scholar] [CrossRef] [PubMed]
- Trentham, D.R.; Bardsley, R.G.; Eccleston, J.F.; Weeds, A.G. Elementary Processes of the Magnesium Ion-Dependent Adenosine Triphosphatase Activity of Heavy Meromyosin. A Transient Kinetic Approach to the Study of Kinases and Adenosine Triphosphatases and a Colorimetric Inorganic Phosphate Assay in Situ. Biochem. J. 1972, 126, 635–644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qu, Z.; Fujita-Becker, S.; Ballweber, E.; Ince, S.; Herrmann, C.; Schröder, R.R.; Mannherz, H.G. Interaction of Isolated Cross-Linked Short Actin Oligomers with the Skeletal Muscle Myosin Motor Domain. FEBS J. 2018, 285, 1715–1729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Qu, Z.; Silvan, U.; Jockusch, B.M.; Aebi, U.; Schoenenberger, C.-A.; Mannherz, H.G. Distinct Actin Oligomers Modulate Differently the Activity of Actin Nucleators. FEBS J. 2015, 282, 3824–3840. [Google Scholar] [CrossRef]
- Bradford, M.M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- HESTERKAMP, T.; WEEDS, A.G.; MANNHERZ, H.G. The Actin Monomers in the Ternary Gelsolin: 2 Actin Complex Are in an Antiparallel Orientation. Eur. J. Biochem. 1993, 218, 507–513. [Google Scholar] [CrossRef]
- JAHN, W. Easily Prepared Holey Films for Use in Cryo-Electron Microscopy. J. Microsc. 1995, 179, 333–334. [Google Scholar] [CrossRef]
- Steger, C. An Unbiased Detector of Curvilinear Structures. IEEE Trans. Pattern Anal. Mach. Intell. 1998, 20, 113–125. [Google Scholar] [CrossRef] [Green Version]
- Ishii, Y.; Lehrer, S.S. Two-Site Attachment of Troponin to Pyrene-Labeled Tropomyosin. J. Biol. Chem. 1991, 266, 6894–6903. [Google Scholar] [CrossRef]
- Babu, A.; Su, H.; Ryu, Y.; Gulati, J. Determination of residue specificity in the EF-hand of troponin C for Ca2+ coordination, by genetic engineering. J. Biol. Chem. 1992, 267, 15469–15474. [Google Scholar] [CrossRef]
- Reiffert, S.; Maytum, R.; Geeves, M.; Lohmann, K.; Greis, T.; Blüggel, M.; Meyer, H.E.; Heilmeyer, L.M.G.; Jaquet, K. Characterization of the cardiac holotroponin complex reconstituted from native cardiac troponin T and recombinant I and C. Eur. J. Biochem. 1999, 261, 40–47. [Google Scholar] [CrossRef] [Green Version]
| A. Half Time of Polymerization (t 1/2) in Min. | ||||
| Bovine c-Actin | Recomb. wt c-actin | p.A295S | p.R312H | p.E361G |
| 2.7 ± 0.6 | 2.5 ± 0.3 | 8.5 ± 0.6 | 10.8 ± 0.5 | 1.6 ± 0.2 |
| B. Critical Concentration of Polymerization (Cc) in μM | ||||
| Bovine c-actin | recomb. wt c-actin | p.A295S | p.R312H | p.E361G |
| 0.21 ± 0.1 | 0.2 ± 0.09 | 0.4. ± 0.3 | 0.7 ± 0.12 | 0.2 ± 0.11 |
| c-Actin Variant | Bovine c-Actin | wt Recombinant | p.A295S | p.R312H | p.E361G |
|---|---|---|---|---|---|
| apparent KM (µM) | 0.081 | 0.165 | 0.966 | 0.096 | 0.075 |
| +TM/TN apparent KM (µM) | n.d. | 0.217 | 3.31 | 0.0243 | 0.173 |
| Vmax (µM ATP hydrol/sec/µM S1) | 0.44 | 0.334 | 0.799 | 0.444 | 0.641 |
| +TM/TN Vmax (µM ATP hydrol/sec/µM S1) | n.d. | 0.563 | 4.347 | 0.267 | 0.769 |
| Actin Variant | pCa50 | Hill Coefficient | Min. ATPase µM ATP hyd/µM S1/s | Max. ATPase µM ATP hyd/µMS1/s | Fold Stimulation (pCa Max./Min.) |
|---|---|---|---|---|---|
| Bovine n = 3 R2 = 0.891 | 6.93 ± 0.20 | 1.182 ± 0.54 | 0.086 | 0.44 | 5.625 |
| WT rec n = 4 R2 = 0.894 | 6.84 ± 0.09 | 1.68 ± 0.559 | 0.093 | 0.379 | 4.0 |
| p.A295S n = 3 R2 = 0.933 | 7.06 ± 0.09 | 1.497 ± 0.452 | 0.078 | 0.387 | 4.96 |
| p.R312H n = 3 R2= 0.789 | 6.92 ± 0.19 | 1.342 ± 0.824 | 0.054 | 0.346 | 5.83 |
| p.E361G n = 3 R2 = 0.927 | 6.84 ± 0.11 | 1.021 ± 0.256 | 0.057 | 0.301 | 5.0 |
| p.R312H | p.R312H + Levo | WTc-Actin | p.E361G | p.E361G + Levo | WT c-Actin | |
|---|---|---|---|---|---|---|
| pCa50 | 6.923 ± 0.196 | 7.19 ± 0.20 | 6.84 ± 0.09 | 6.84 ± 0.11 | 7.44 ± 0.34 | 6.84 ± 0.09 |
| Hill coefficient | 1.342 ± 0.824 | 1.9 ± 1.92 | 1.68 ± 0.55 | 1.02 ±0.25 | 1.75 ±1.36 | 1.68 ± 0.55 |
| Bovine c-Actin | Recomb. wt c-Actin | |||||
| pCa50 | Hill | pCa50 | Hill | |||
| +cTm/cTn | 7.32 ± 0.14 (n = 9; R2 = 0.724) | 1.03 ± 0.27 | 7.55 ± 0.21 (n = 4; R2 = 0.797) | 0.88 ± 0.31 | ||
| cTm/cTn + S1 | 7.53 ± 0.12 (n = 6; R2 = 0.812) | 1.22 ± 0.31 | 7.45 ± 0.13 (n = 4; R2 = 0,871) | 1.31 ± 0.36 | ||
| cTm/cTn + MyBP-C | 7.15 ± 0.14 (n = 2; R2 = 0.90) | 1.52 ± 0.67 | 7.48 ± 0.17 (n = 3; R2 = 0.913) | 1.74 ± 0.63 | ||
| cTm/cTn + S1 + MyBP-C | 7.47 ± 0.2 (n = 3; R2 = 0.789) | 1.1 ± 0.42 | 7.32 ± 0.06 (n = 3; R2 = 0.977) | 1.58 ± 0.26 | ||
| p.A295S | p.R312H | p.E361G | ||||
| pCa50 | Hill | pCa50 | Hill | pCa50 | Hill | |
| +cTm/cTn | 7.32 ± 0.1 (n = 9; R2 = 0.873) | 1.55 ± 0.43 | 7.1 ± 0.03 (n = 3; R2 = 0.950) | 3.9 ± 2.4 | 7.6 ± 0.11 (n = 6; R2 = 0.90) | 1.05 ± 0.2 |
| cTm/cTn + S1 | 7.26 ± 0.61 (n = 4; R2 = 0.8638) | 1.74 ± 0.61 | 6.76 ± 0.21 (n = 5; R2 = 0.817) | 1.28 ± 0.26 | 7.07 ± 0.07 (n = 6; R2 = 0.761) | 2.77 ± 1.56 |
| cTm/cTn +MyBP-C | 7.37 ± 0.1 (n = 5; R2 = 0.936) | 0.85 ± 0.13 | 7.1 ± 0.09 (n = 3; R2 = 0.936 | 1.42 ± 0.37 | 7.38 ± 0.2 (n = 3; R2 = 0.879) | 0.86 ± 0.27 |
| cTm/cTn +S1 + MyBP-C | 7.22 ± 0.16 (n = 5; R2 = 0.842) | 0.96 ± 0.25 | 7.5 ± 0.17 (n = 3; R2 = 0.893) | 1.69 ± 0.65 | 7.13 ± 0.14 (n = 3; R2 = 0.878 | 1.33 ± 0.49 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hassoun, R.; Erdmann, C.; Schmitt, S.; Fujita-Becker, S.; Mügge, A.; Schröder, R.R.; Geyer, M.; Borbor, M.; Jaquet, K.; Hamdani, N.; et al. Functional Characterization of Cardiac Actin Mutants Causing Hypertrophic (p.A295S) and Dilated Cardiomyopathy (p.R312H and p.E361G). Int. J. Mol. Sci. 2022, 23, 4465. https://doi.org/10.3390/ijms23084465
Hassoun R, Erdmann C, Schmitt S, Fujita-Becker S, Mügge A, Schröder RR, Geyer M, Borbor M, Jaquet K, Hamdani N, et al. Functional Characterization of Cardiac Actin Mutants Causing Hypertrophic (p.A295S) and Dilated Cardiomyopathy (p.R312H and p.E361G). International Journal of Molecular Sciences. 2022; 23(8):4465. https://doi.org/10.3390/ijms23084465
Chicago/Turabian StyleHassoun, Roua, Constanze Erdmann, Sebastian Schmitt, Setsuko Fujita-Becker, Andreas Mügge, Rasmus R. Schröder, Matthias Geyer, Mina Borbor, Kornelia Jaquet, Nazha Hamdani, and et al. 2022. "Functional Characterization of Cardiac Actin Mutants Causing Hypertrophic (p.A295S) and Dilated Cardiomyopathy (p.R312H and p.E361G)" International Journal of Molecular Sciences 23, no. 8: 4465. https://doi.org/10.3390/ijms23084465
APA StyleHassoun, R., Erdmann, C., Schmitt, S., Fujita-Becker, S., Mügge, A., Schröder, R. R., Geyer, M., Borbor, M., Jaquet, K., Hamdani, N., & Mannherz, H. G. (2022). Functional Characterization of Cardiac Actin Mutants Causing Hypertrophic (p.A295S) and Dilated Cardiomyopathy (p.R312H and p.E361G). International Journal of Molecular Sciences, 23(8), 4465. https://doi.org/10.3390/ijms23084465







