Systematic Analysis and Identification of Drought-Responsive Genes of the CAMTA Gene Family in Wheat (Triticum aestivum L.)
Abstract
:1. Introduction
2. Results
2.1. Identification and Structural Analyses of TaCAMTA Genes in the Wheat Genome
2.2. Chromosomal Location of TaCAMTAs
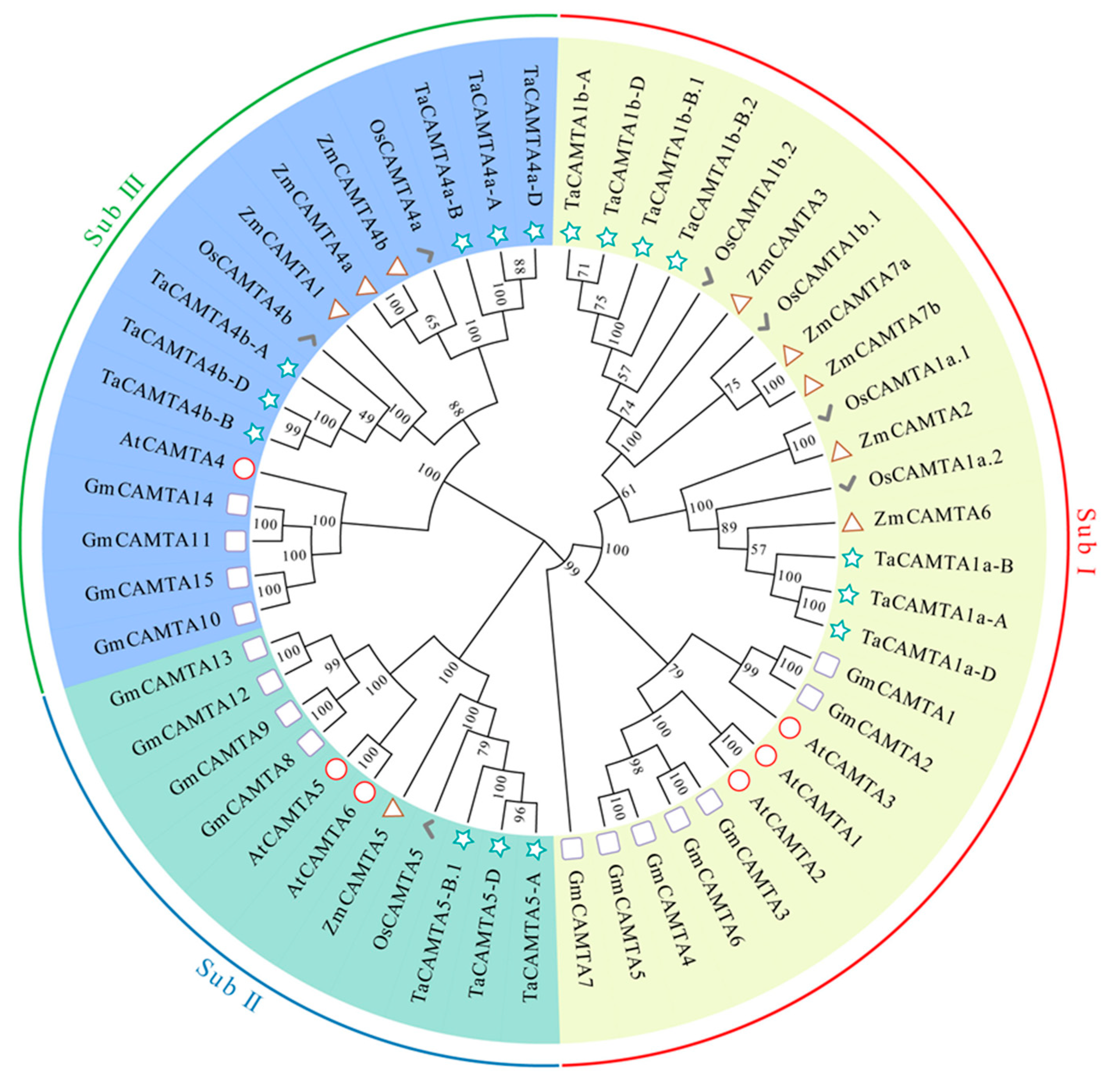
2.3. Phylogenetic Analysis of TaCAMTAs
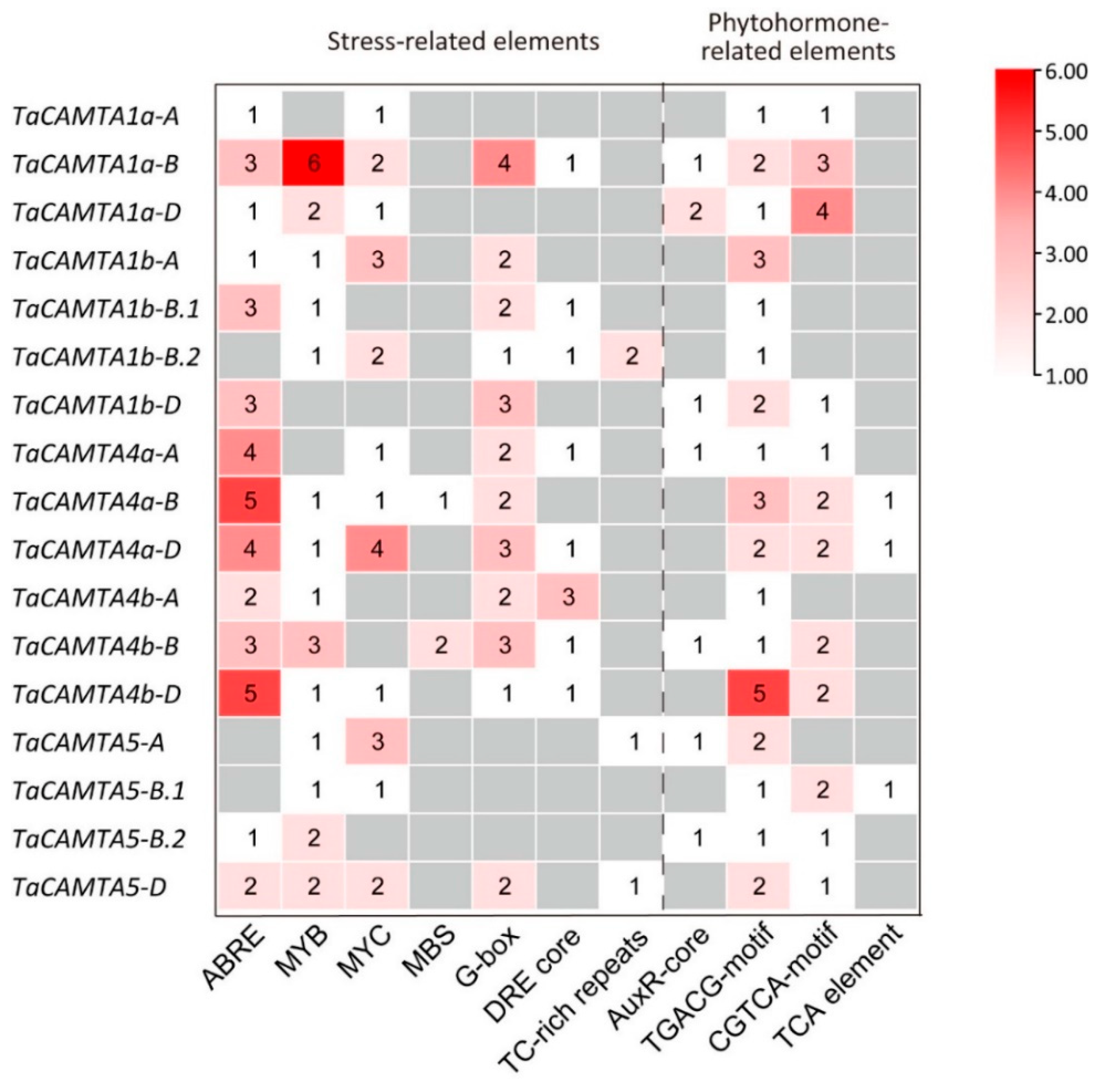
2.4. Cis-Acting Regulatory Elements in the Promoter Regions of TaCAMTAs
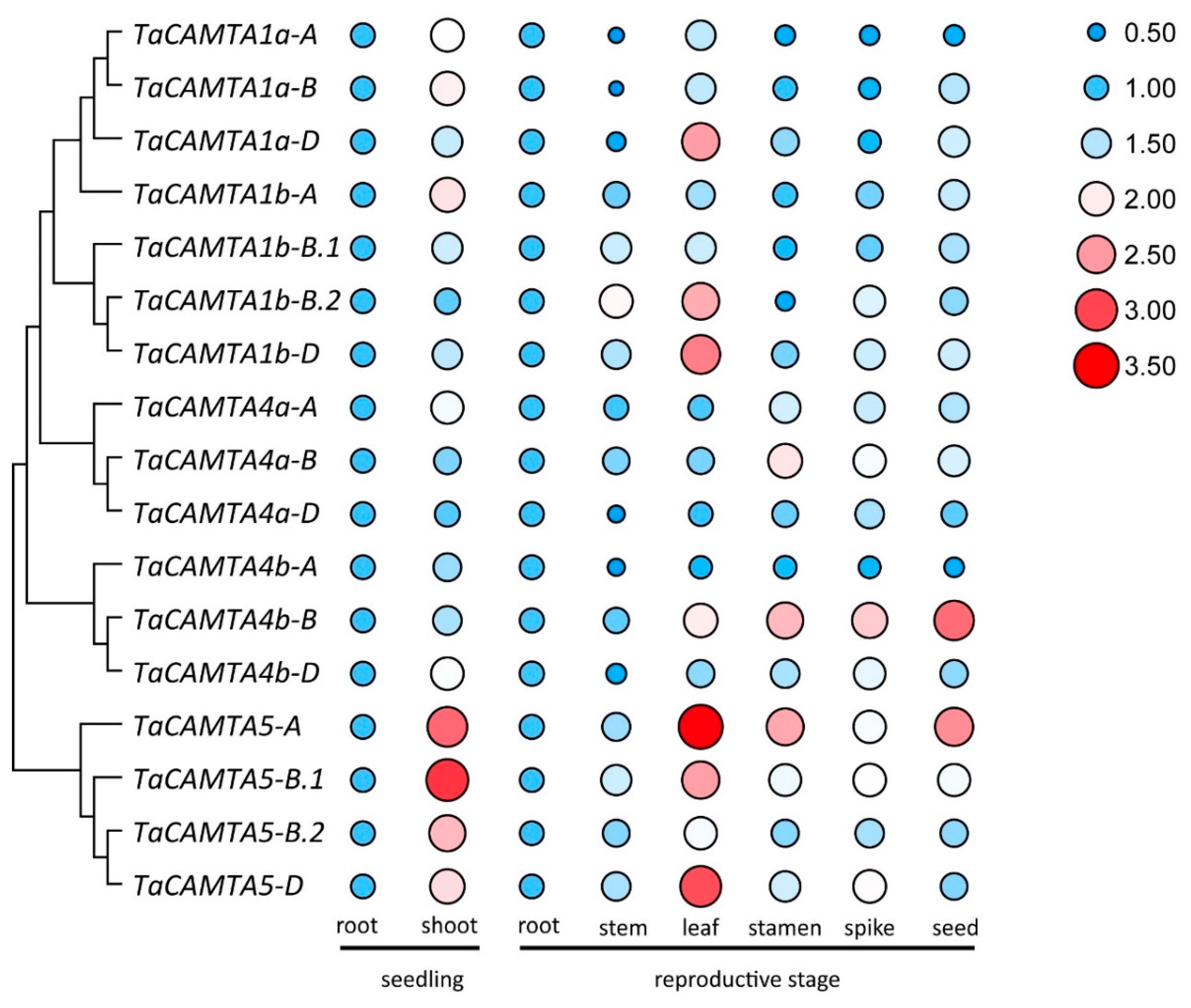
2.5. Stage- and Tissue-Specific Expression Levels of TaCAMTA Genes
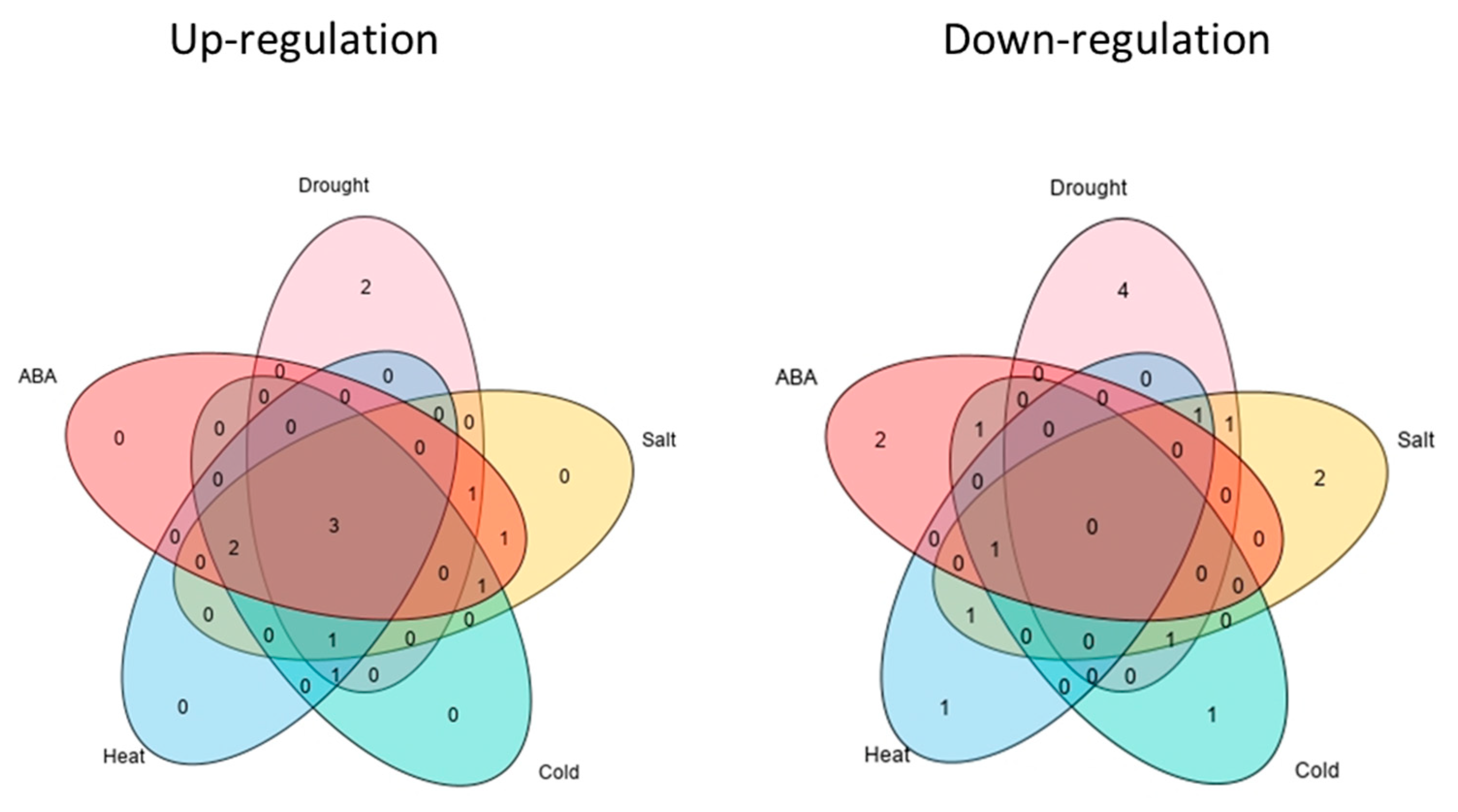
2.6. Expression Profiles of Predicted CAMTA Sequences during Abiotic Stress
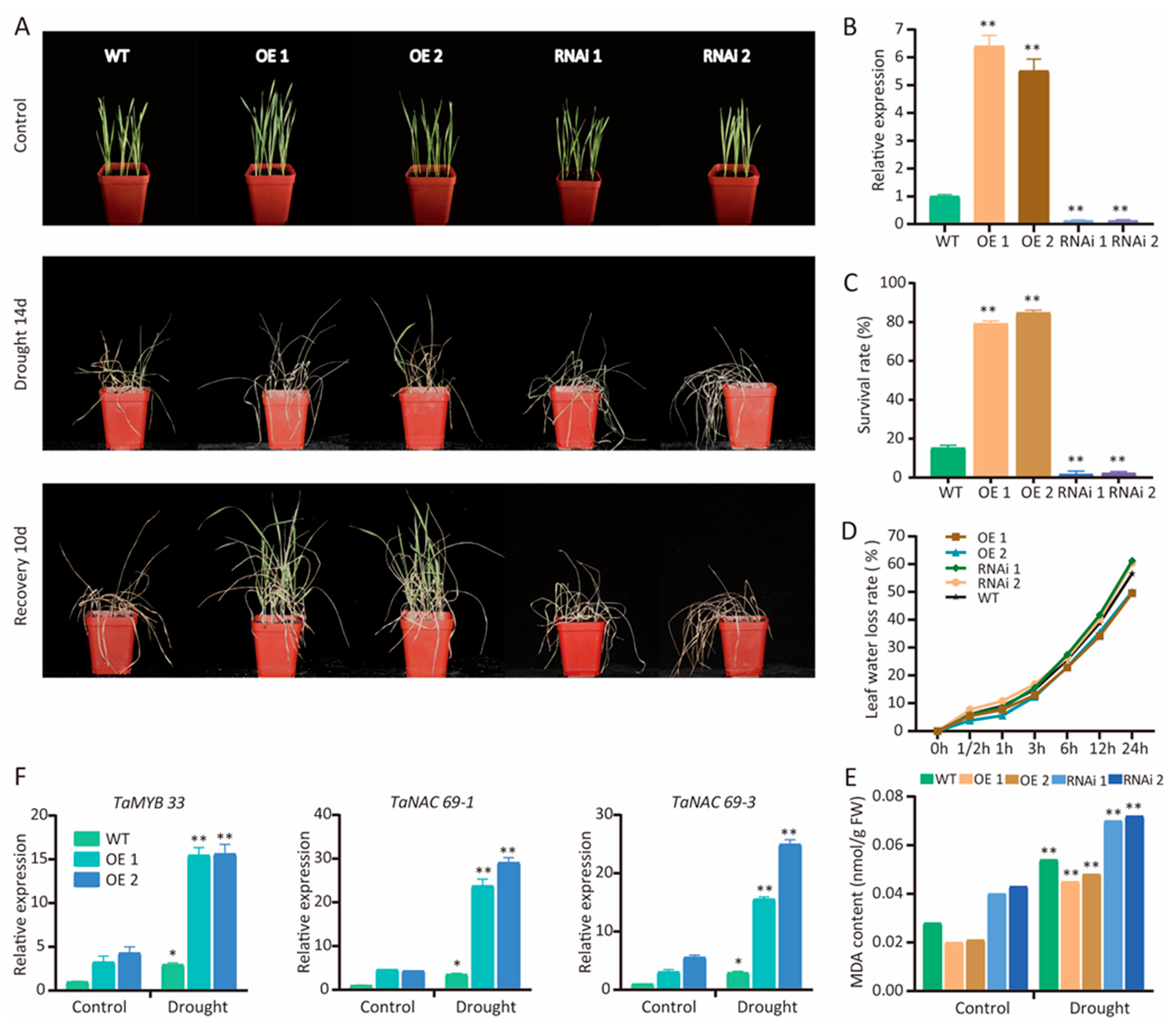
2.7. TaCAMTA1b-B.1 Regulates Prominent Drought-Responsive Genes
3. Discussion
3.1. Molecular Mechanisms of CAMTA Duplication
3.2. Basic Characteristics of CAMTAs in Wheat
3.3. TaCAMTA1b-B.1 Regulates the Expression of Stress-Associated Genes in Response to Drought
4. Materials and Methods
4.1. Identification of CAMTA Genes in Wheat
4.2. Chromosomal Location and Sequence Analysis of CAMTA Genes
4.3. Multiple Sequence Alignment and Phylogenetic Analysis of CAMTA
4.4. Gene Structure and Conserved Motif Analysis of Wheat CAMTAs
4.5. Vector Construction and Plant Transformation
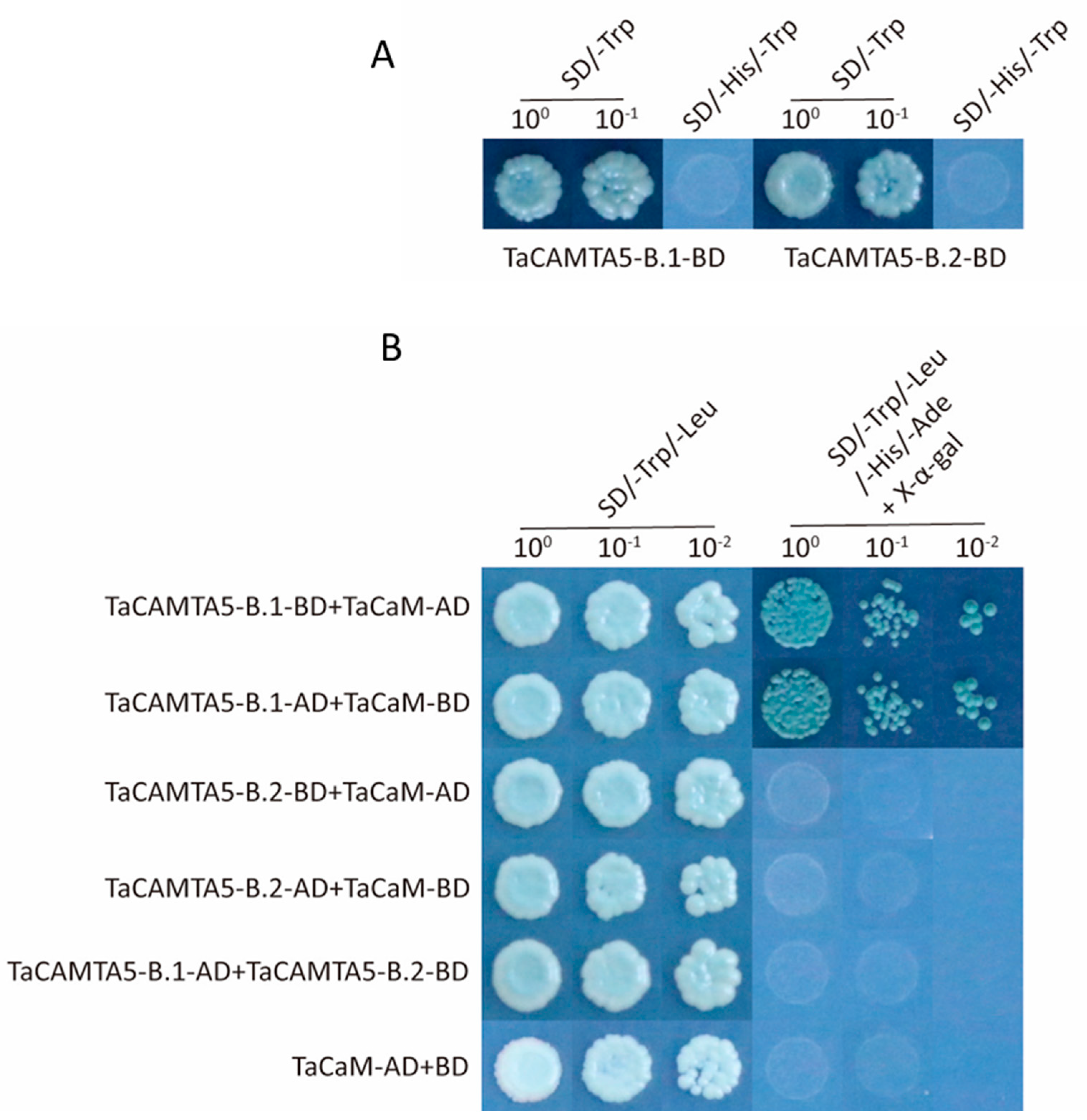
4.6. Yeast Two-Hybrid Assay
4.7. Plant Materials and Stress Treatments
4.8. RNA Isolation and Quantitative Real-Time RT-PCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kudla, J.; Batistic, O.; Hashimoto, K. Calcium signals: The lead currency of plant information processing. Plant Cell. 2010, 22, 541–563. [Google Scholar] [CrossRef] [PubMed]
- Ikura, M.; Osawa, M.; Ames, J.B. The role of calcium-binding proteins in the control of transcription: Structure to function. Bioessays 2002, 24, 625–636. [Google Scholar] [CrossRef] [PubMed]
- Reddy, A.S. Calcium: Silver bullet in signaling. Plant Sci. 2001, 160, 381–404. [Google Scholar] [CrossRef]
- Poovaiah, B.W.; Du, L.; Wang, H.; Yang, T. Recent advances in calcium/calmodulin-mediated signaling with an emphasis on plant-microbe interactions. Plant Physiol. 2013, 163, 531–542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bouché, N.; Scharlat, A.; Snedden, W.; Bouchez, D.; Fromm, H. A Novel Family of Calmodulin-binding Transcription Activators in Multicellular Organisms. J. Biol. Chem. 2002, 277, 21851–21861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, T.; Poovaiah, B.W. An early ethylene up-regulated gene encoding a calmodulin-binding protein involved in plant senescence and death. J. Biol. Chem. 2000, 275, 38467–38473. [Google Scholar] [CrossRef] [Green Version]
- Iqbal, Z.; Shariq Iqbal, M.; Singh, S.P.; Buaboocha, T. Ca2+/Calmodulin Complex Triggers CAMTA Transcriptional Machinery Under Stress in Plants: Signaling Cascade and Molecular Regulation. Front. Plant Sci. 2020, 11, 598327. [Google Scholar] [CrossRef]
- Perrino, E.V.; Wagensommer, R.P. Crop Wild Relatives (CWRs) Threatened and Endemic to Italy: Urgent Actions for Protection and Use. Biology 2022, 11, 193. [Google Scholar] [CrossRef]
- Abenavoli, L.; Milanovic, M.; Procopio, A.C.; Spampinato, G.; Maruca, G.; Perrino, E.V.; Mannino, G.C.; Fagoonee, S.; Luzza, F.; Musarella, C.M. Ancient wheats: Beneficial effects on insulin resistance. Minerva Med. 2021, 112, 641–650. [Google Scholar] [CrossRef]
- Galon, Y.; Finkler, A.; Fromm, H. Calcium-regulated transcription in plants. Mol. Plant 2010, 3, 653–669. [Google Scholar] [CrossRef]
- Doherty, C.J.; Van Buskirk, H.A.; Myers, S.J.; Thomashow, M.F. Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell 2009, 21, 972–984. [Google Scholar] [CrossRef] [Green Version]
- Du, L.; Ali, G.S.; Simons, K.A.; Hou, J.; Yang, T.; Reddy, A.S.; Poovaiah, B.W. Ca2+/calmodulin regulates salicylic-acid-mediated plant immunity. Nature 2009, 457, 1154–1158. [Google Scholar] [CrossRef]
- Galon, Y.; Nave, R.; Boyce, J.M.; Nachmias, D.; Knight, M.R.; Fromm, H. Calmodulin-binding transcription activator (CAMTA) 3 mediates biotic defense responses in Arabidopsis. FEBS Lett. 2008, 582, 943–948. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.; Park, S.; Gilmour, S.J.; Thomashow, M.F. Roles of CAMTA transcription factors and salicylic acid in configuring the low-temperature transcriptome and freezing tolerance of Arabidopsis. Plant J. 2013, 75, 364–376. [Google Scholar] [CrossRef]
- Koo, S.C.; Choi, M.S.; Chun, H.J.; Shin, D.B.; Park, B.S.; Kim, Y.H.; Park, H.M.; Seo, H.S.; Song, J.T.; Kang, K.Y.; et al. The calmodulin-binding transcription factor OsCBT suppresses defense responses to pathogens in rice. Mol. Cells 2009, 27, 563–570. [Google Scholar] [CrossRef] [PubMed]
- Laluk, K.; Prasad, K.V.; Savchenko, T.; Celesnik, H.; Dehesh, K.; Levy, M.; Mitchell-Olds, T.; Reddy, A.S. The calmodulin-binding transcription factor SIGNAL RESPONSIVE1 is a novel regulator of glucosinolate metabolism and herbivory tolerance in Arabidopsis. Plant Cell Physiol. 2012, 53, 2008–2015. [Google Scholar] [CrossRef]
- Nie, H.; Zhao, C.; Wu, G.; Wu, Y.; Chen, Y.; Tang, D. SR1, a calmodulin-binding transcription factor, modulates plant defense and ethylene-induced senescence by directly regulating NDR1 and EIN3. Plant Physiol. 2012, 158, 1847–1859. [Google Scholar] [CrossRef] [Green Version]
- Qiu, Y.; Xi, J.; Du, L.; Suttle, J.C.; Poovaiah, B.W. Coupling calcium/calmodulin-mediated signaling and herbivore-induced plant response through calmodulin-binding transcription factor AtSR1/CAMTA3. Plant Mol. Biol. 2012, 79, 89–99. [Google Scholar] [CrossRef]
- Büyük, İ.; İlhan, E.; Şener, D.; Özsoy, A.U.; Aras, S. Genome-wide identification of CAMTA gene family members in Phaseolus vulgaris L. and their expression profiling during salt stress. Mol. Biol. Rep. 2019, 46, 2721–2732. [Google Scholar] [CrossRef]
- Pant, P.; Iqbal, Z.; Pandey, B.K.; Sawant, S.V. Genome-wide comparative and evolutionary analysis of Calmodulin-binding Transcription Activator (CAMTA) family in Gossypium species. Sci Rep. 2018, 8, 5573. [Google Scholar] [CrossRef]
- Noman, M.; Aysha, J.; Ketehouli, T.; Yang, J.; Du, L.; Wang, F.; Li, H. Calmodulin binding transcription activators: An interplay between calcium signalling and plant stress tolerance. J. Plant Physiol. 2021, 256, 153327. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Poovaiah, B.W. A calmodulin-binding/CGCG box DNA-binding protein family involved in multiple signaling pathways in plants. J. Biol. Chem. 2002, 277, 45049–45058. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, Y.S.; An, C.; Park, S.; Gilmour, S.J.; Wang, L.; Renna, L.; Brandizzi, F.; Grumet, R.; Thomashow, M.F. CAMTA-Mediated Regulation of Salicylic Acid Immunity Pathway Genes in Arabidopsis Exposed to Low Temperature and Pathogen Infection. Plant Cell 2017, 29, 2465–2477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pandey, N.; Ranjan, A.; Pant, P.; Tripathi, R.K.; Ateek, F.; Pandey, H.P.; Patre, U.V.; Sawant, S.V. CAMTA 1 regulates drought responses in Arabidopsis thaliana. BMC Genom. 2013, 14, 216. [Google Scholar] [CrossRef] [Green Version]
- Shkolnik, D.; Finkler, A.; Pasmanik-Chor, M.; Fromm, H. CALMODULIN-BINDING TRANSCRIPTION ACTIVATOR 6: A Key Regulator of Na+ Homeostasis during Germination. Plant Physiol. 2019, 180, 1101–1118. [Google Scholar] [CrossRef] [Green Version]
- Tokizawa, M.; Kobayashi, Y.; Saito, T.; Kobayashi, M.; Iuchi, S.; Nomoto, M.; Tada, Y.; Yamamoto, Y.Y.; Koyama, H. Sensitive to proton rhizotoxicity1, calmodulin binding transcription activator2, and other transcription factors are involved in aluminum-activated malate transporter1 expression. Plant Physiol. 2015, 167, 991–1003. [Google Scholar] [CrossRef] [Green Version]
- Wu, L.; Sadhukhan, A.; Kobayashi, Y.; Ogo, N.; Tokizawa, M.; Agrahari, R.K.; Ito, H.; Iuchi, S.; Kobayashi, M.; Asai, A.; et al. Involvement of phosphatidylinositol metabolism in aluminum-induced malate secretion in Arabidopsis. J. Exp. Bot. 2019, 70, 3329–3342. [Google Scholar] [CrossRef]
- Mitsuda, N.; Isono, T.; Sato, M.H. Arabidopsis CAMTA family proteins enhance V-PPase expression in pollen. Plant Cell Physiol. 2003, 44, 975–981. [Google Scholar] [CrossRef] [Green Version]
- Yuan, P.; Tanaka, K.; Poovaiah, B.W. Calmodulin-binding transcription activator AtSR1/CAMTA3 fine-tunes plant immune response by transcriptional regulation of the salicylate receptor NPR1. Plant Cell Environ. 2021, 44, 3140–3154. [Google Scholar] [CrossRef]
- Galon, Y.; Aloni, R.; Nachmias, D.; Snir, O.; Feldmesser, E.; Scrase-Field, S.; Boyce, J.M.; Bouche, N.; Knight, M.R.; Fromm, H. Calmodulin-binding transcription activator 1 mediates auxin signaling and responds to stresses in Arabidopsis. Planta 2010, 232, 165–178. [Google Scholar] [CrossRef]
- Wei, M.; Xu, X.; Li, C. Identification and expression of CAMTA genes in Populus trichocarpa under biotic and abiotic stress. Sci Rep. 2017, 7, 17910. [Google Scholar] [CrossRef] [Green Version]
- Kakar, K.U.; Nawaz, Z.; Cui, Z.; Cao, P.; Jin, J.; Shu, Q.; Ren, X. Evolutionary and expression analysis of CAMTA gene family in Nicotiana tabacum yielded insights into their origin, expansion and stress responses. Sci. Rep. 2018, 8, 10322. [Google Scholar] [CrossRef]
- Saeidi, K.; Zare, N.; Baghizadeh, A.; Asghari-Zakaria, R. Phaseolus vulgaris genome possesses CAMTA genes, and phavuCAMTA1 contributes to the drought tolerance. J. Genet. 2019, 98, 31. [Google Scholar] [CrossRef]
- Yue, R.; Lu, C.; Sun, T.; Peng, T.; Han, X.; Qi, J.; Yan, S.; Tie, S. Identification and expression profiling analysis of calmodulin-binding transcription activator genes in maize (Zea mays L.) under abiotic and biotic stresses. Front. Plant Sci. 2015, 6, 576. [Google Scholar] [CrossRef] [Green Version]
- Leng, X.; Han, J.; Wang, X.; Zhao, M.; Sun, X.; Wang, C.; Fang, J. Characterization of a Calmodulin-binding Transcription Factor from Strawberry (Fragaria x ananassa). Plant Genome. 2015, 8. [Google Scholar] [CrossRef]
- Zhang, J.; Pan, X.; Ge, T.; Yi, S.; Lv, Q.; Zheng, Y.; Ma, Y.; Liu, X.; Xie, R. Genome-wide identification of citrus CAMTA genes and their expression analysis under stress and hormone treatments. J. Hortic. Sci. Biotechnol. 2018, 94, 331–340. [Google Scholar] [CrossRef]
- Ali, E.; Raza, M.A.; Cai, M.; Hussain, N.; Shahzad, A.N.; Hussain, M.; Ali, M.; Bukhari, S.A.H.; Sun, P. Calmodulin-binding transcription activator (CAMTA) genes family: Genome-wide survey and phylogenetic analysis in flax (Linum usitatissimum). PLoS ONE 2020, 15, e0236454. [Google Scholar] [CrossRef]
- Yang, T.; Peng, H.; Whitaker, B.D.; Jurick, W.M. Differential expression of calcium/calmodulin-regulated SlSRs in response to abiotic and biotic stresses in tomato fruit. Physiol. Plant. 2013, 148, 445–455. [Google Scholar] [CrossRef]
- Li, X.; Huang, L.; Zhang, Y.; Ouyang, Z.; Hong, Y.; Zhang, H.; Li, D.; Song, F. Tomato SR/CAMTA transcription factors SlSR1 and SlSR3L negatively regulate disease resistance response and SlSR1L positively modulates drought stress tolerance. BMC Plant Biol. 2014, 14, 286. [Google Scholar] [CrossRef]
- Wang, G.; Zeng, H.; Hu, X.; Zhu, Y.; Chen, Y.; Shen, C.; Wang, H.; Poovaiah, B.W.; Du, L. Identification and expression analyses of calmodulin-binding transcription activator genes in soybean. Plant and Soil. 2014, 386, 205–221. [Google Scholar] [CrossRef]
- Noman, M.; Jameel, A.; Qiang, W.D.; Ahmad, N.; Liu, W.C.; Wang, F.W.; Li, H.Y. Overexpression of GmCAMTA12 Enhanced Drought Tolerance in Arabidopsis and Soybean. Int. J. Mol. Sci. 2019, 20, 4849. [Google Scholar] [CrossRef] [Green Version]
- Rahman, H.; Xu, Y.P.; Zhang, X.R.; Cai, X.Z. Brassica napus Genome Possesses Extraordinary High Number of CAMTA Genes and CAMTA3 Contributes to PAMP Triggered Immunity and Resistance to Sclerotinia sclerotiorum. Front. Plant Sci. 2016, 7, 581. [Google Scholar] [CrossRef] [Green Version]
- Araus, J.L.; Slafer, G.A.; Reynolds, M.P.; Royo, C. Plant breeding and drought in C3 cereals: What should we breed for? Ann Bot. 2002, 89, 925–940. [Google Scholar] [CrossRef]
- Zhang, H.; Zhu, J.; Gong, Z.; Zhu, J.K. Abiotic stress responses in plants. Nat. Rev. Genet. 2022, 23, 104–119. [Google Scholar] [CrossRef]
- Pfeiffer, W.; Trethowan, R.M.; van Ginkel, M.; Ortiz-Monasterio, I.; Rajaram, S. Breeding for Abiotic Stress Tolerance in Wheat. In Abiotic Stresses Plant Resistance through Breeding and Molecular Approaches; CIMMTY: Mexico City, Mexico, 2005; pp. 401–489. [Google Scholar]
- Lesk, C.; Rowhani, P.; Ramankutty, N. Influence of extreme weather disasters on global crop production. Nature 2016, 529, 84–87. [Google Scholar] [CrossRef]
- Alotaibi, F.; Alharbi, S.; Alotaibi, M.; Mosallam, M.A.; Alrajhi, A. Wheat Omics: Classical Breeding to New Breeding Technologies. Saudi J. Biol. Sci. 2020, 28, 1433–1444. [Google Scholar] [CrossRef]
- Valkoun, J.J. Wheat Pre-Breeding Using Wild Progenitors. Euphytica 2001, 119, 17–23. [Google Scholar] [CrossRef]
- Ashraf, M. Inducing drought tolerance in plants: Recent advances. Biotechnol. Adv. 2010, 28, 169–183. [Google Scholar] [CrossRef]
- Borrill, P.; Adamski, N.; Uauy, C. Genomics as the key to unlocking the polyploid potential of wheat. New Phytol. 2015, 208, 1008–1022. [Google Scholar] [CrossRef]
- He, L.; Chen, X.; Xu, M.; Liu, T.; Zhang, T.; Li, J.; Yang, J.; Chen, J.; Zhong, K. Genome-Wide Identification and Characterization of the Cystatin Gene Family in Bread Wheat (Triticum aestivum L.). Int J. Mol. Sci. 2021, 22, 10264. [Google Scholar] [CrossRef]
- Yang, F.; Dong, F.S.; Hu, F.H.; Liu, Y.W.; Chai, J.F.; Zhao, H.; Lv, M.Y.; Zhou, S. Genome-wide identification and expression analysis of the calmodulin-binding transcription activator (CAMTA) gene family in wheat (Triticum aestivum L.). BMC Genet. 2020, 21, 105. [Google Scholar] [CrossRef] [PubMed]
- Adhikari, L.; Raupp, J.; Wu, S.; Wilson, D.; Evers, B.; Koo, D.H.; Singh, N.; Friebe, B.; Poland, J. Genetic characterization and curation of diploid A-genome wheat species. Plant Physiol. 2022, 188, 2101–2114. [Google Scholar] [CrossRef] [PubMed]
- Perrino, E.V.; Wagensommer, R.P.; Medagli, P. Aegilops (Poaceae) in Italy: Taxonomy, geographical distribution, ecology, vulnerability and conservation. Syst. Biodivers. 2014, 12, 331–349. [Google Scholar] [CrossRef]
- Yang, Y.; Sun, T.; Xu, L.; Pi, E.; Wang, S.; Wang, H.; Shen, C. Genome-wide identification of CAMTA gene family members in Medicago truncatula and their expression during root nodule symbiosis and hormone treatments. Front. Plant Sci. 2015, 6, 459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vogel, C.; Teichmann, S.A.; Pereira-Leal, J. The relationship between domain duplication and recombination. J. Mol. Biol. 2005, 346, 355–365. [Google Scholar] [CrossRef] [PubMed]
- Li, W.H.; Makova, K.D. Domain Duplication and Gene Elongation. eLS 2006. [Google Scholar] [CrossRef]
- Toll-Riera, M.; Laurie, S.; Rad-Trilla, N.; Alba, M.M. Partial Gene Duplication and the Formation of Novel Genes. In Gene Duplication; IntechOpen: London, UK, 2011. [Google Scholar] [CrossRef] [Green Version]
- Samonte, R.V.; Eichler, E.E. Segmental duplications and the evolution of the primate genome. Nat. Rev. Genet. 2002, 3, 65–72. [Google Scholar] [CrossRef]
- Glover, N.M.; Redestig, H.; Dessimoz, C. Homoeologs: What Are They and How Do We Infer Them? Trends Plant Sci. 2016, 21, 609–621. [Google Scholar] [CrossRef] [Green Version]
- Rahman, H.; Yang, J.; Xu, Y.P.; Munyampundu, J.P.; Cai, X.Z. Phylogeny of Plant CAMTAs and Role of AtCAMTAs in Nonhost Resistance to Xanthomonas oryzae pv. oryzae. Front. Plant Sci. 2016, 7, 177. [Google Scholar] [CrossRef]
- Xiao, P.; Feng, J.W.; Zhu, X.T.; Gao, J. Evolution Analyses of CAMTA Transcription Factor in Plants and Its Enhancing Effect on Cold-tolerance. Front. Plant Sci. 2021, 12, 758187. [Google Scholar] [CrossRef]
- Qin, Y.; Wang, M.; Tian, Y.; He, W.; Han, L.; Xia, G. Over-expression of TaMYB33 encoding a novel wheat MYB transcription factor increases salt and drought tolerance in Arabidopsis. Mol. Biol. Rep. 2012, 39, 7183–7192. [Google Scholar] [CrossRef]
- Xue, G.P.; Way, H.M.; Richardson, T.; Drenth, J.; Joyce, P.A.; McIntyre, C.L. Overexpression of TaNAC69 leads to enhanced transcript levels of stress up-regulated genes and dehydration tolerance in bread wheat. Mol. Plant 2011, 4, 697–712. [Google Scholar] [CrossRef]
- Wang, K.; Liu, H.; Du, L.; Ye, X. Generation of marker-free transgenic hexaploid wheat via an Agrobacterium-mediated co-transformation strategy in commercial Chinese wheat varieties. Plant. Biotechnol. J. 2017, 15, 614–623. [Google Scholar] [CrossRef]
- Wang, K.; Shi, L.; Liang, X.; Zhao, P.; Wang, W.; Liu, J.; Chang, Y.; Hiei, Y.; Yanagihara, C.; Du, L.; et al. The gene TaWOX5 overcomes genotype dependency in wheat genetic transformation. Nat. Plants 2022, 8, 110–117. [Google Scholar] [CrossRef]
- Xu, L.; Tang, Y.; Gao, S.; Su, S.; Hong, L.; Wang, W.; Fang, Z.; Li, X.; Ma, J.; Quan, W.; et al. Comprehensive analyses of the annexin gene family in wheat. BMC Genom. 2016, 17, 415. [Google Scholar] [CrossRef] [Green Version]
- Dandan, Q.; Songchao, X.; Gang, L.; Zhongfu, N.; Yingyin, Y.; Qixin, S.; Huiru, P. Isolation and Functional Characterization of Heat-stress-responsive Gene TaWTF1 from Wheat. Chin. Bull. Bot. 2013, 48, 34–41. [Google Scholar] [CrossRef]
- Liang, Y.K.; Xie, X.; Lindsay, S.E.; Wang, Y.B.; Masle, J.; Williamson, L.; Leyser, O.; Hetherington, A.M. Cell wall composition contributes to the control of transpiration efficiency in Arabidopsis thaliana. Plant J. 2010, 64, 679–686. [Google Scholar] [CrossRef]
- Heath, R.L.; Packer, L. Photoperoxidation in isolated chloroplasts. I. Kinetics and stoichiometry of fatty acid peroxidation. Arch. Biochem. Biophys. 1968, 125, 189–198. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, D.; Wu, X.; Gao, S.; Zhang, S.; Wang, W.; Fang, Z.; Liu, S.; Wang, X.; Zhao, C.; Tang, Y. Systematic Analysis and Identification of Drought-Responsive Genes of the CAMTA Gene Family in Wheat (Triticum aestivum L.). Int. J. Mol. Sci. 2022, 23, 4542. https://doi.org/10.3390/ijms23094542
Wang D, Wu X, Gao S, Zhang S, Wang W, Fang Z, Liu S, Wang X, Zhao C, Tang Y. Systematic Analysis and Identification of Drought-Responsive Genes of the CAMTA Gene Family in Wheat (Triticum aestivum L.). International Journal of Molecular Sciences. 2022; 23(9):4542. https://doi.org/10.3390/ijms23094542
Chicago/Turabian StyleWang, Dezhou, Xian Wu, Shiqin Gao, Shengquan Zhang, Weiwei Wang, Zhaofeng Fang, Shan Liu, Xiaoyan Wang, Changping Zhao, and Yimiao Tang. 2022. "Systematic Analysis and Identification of Drought-Responsive Genes of the CAMTA Gene Family in Wheat (Triticum aestivum L.)" International Journal of Molecular Sciences 23, no. 9: 4542. https://doi.org/10.3390/ijms23094542






