Probing and Manipulating the Lateral Pressure Profile in Lipid Bilayers Using Membrane-Active Peptides—A Solid-State 19F NMR Study
Abstract
:1. Introduction
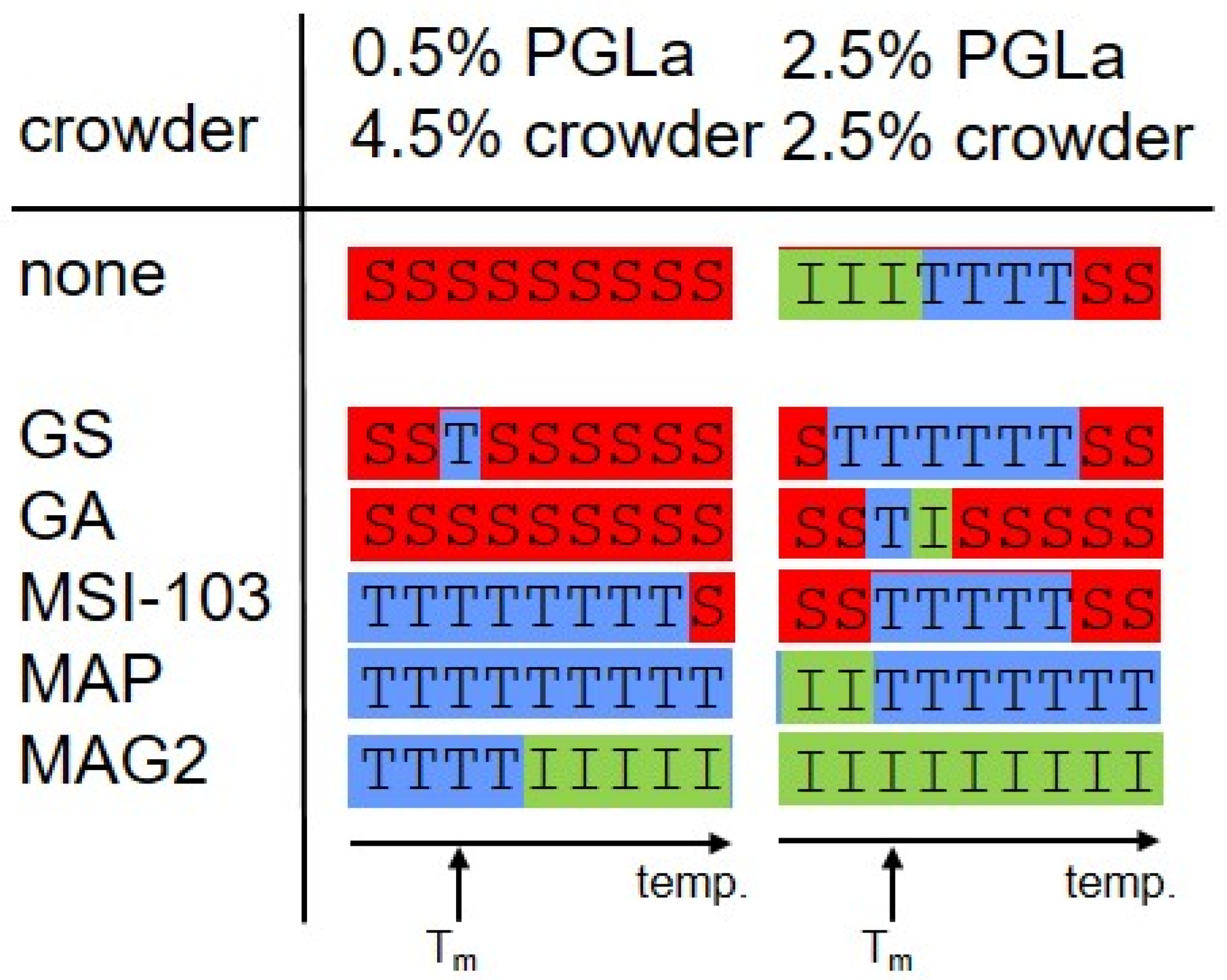
2. Results
3. Discussion
4. Materials and Methods
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
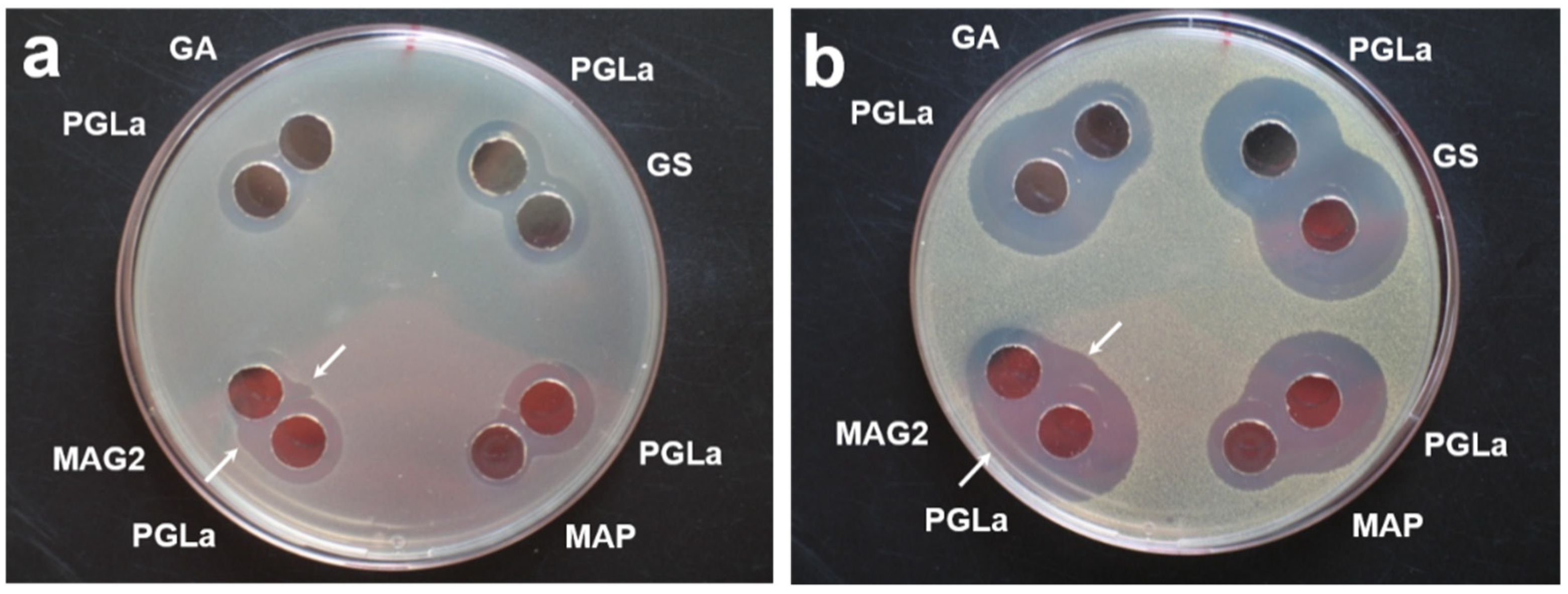
Appendix A. Antimicrobial Activity
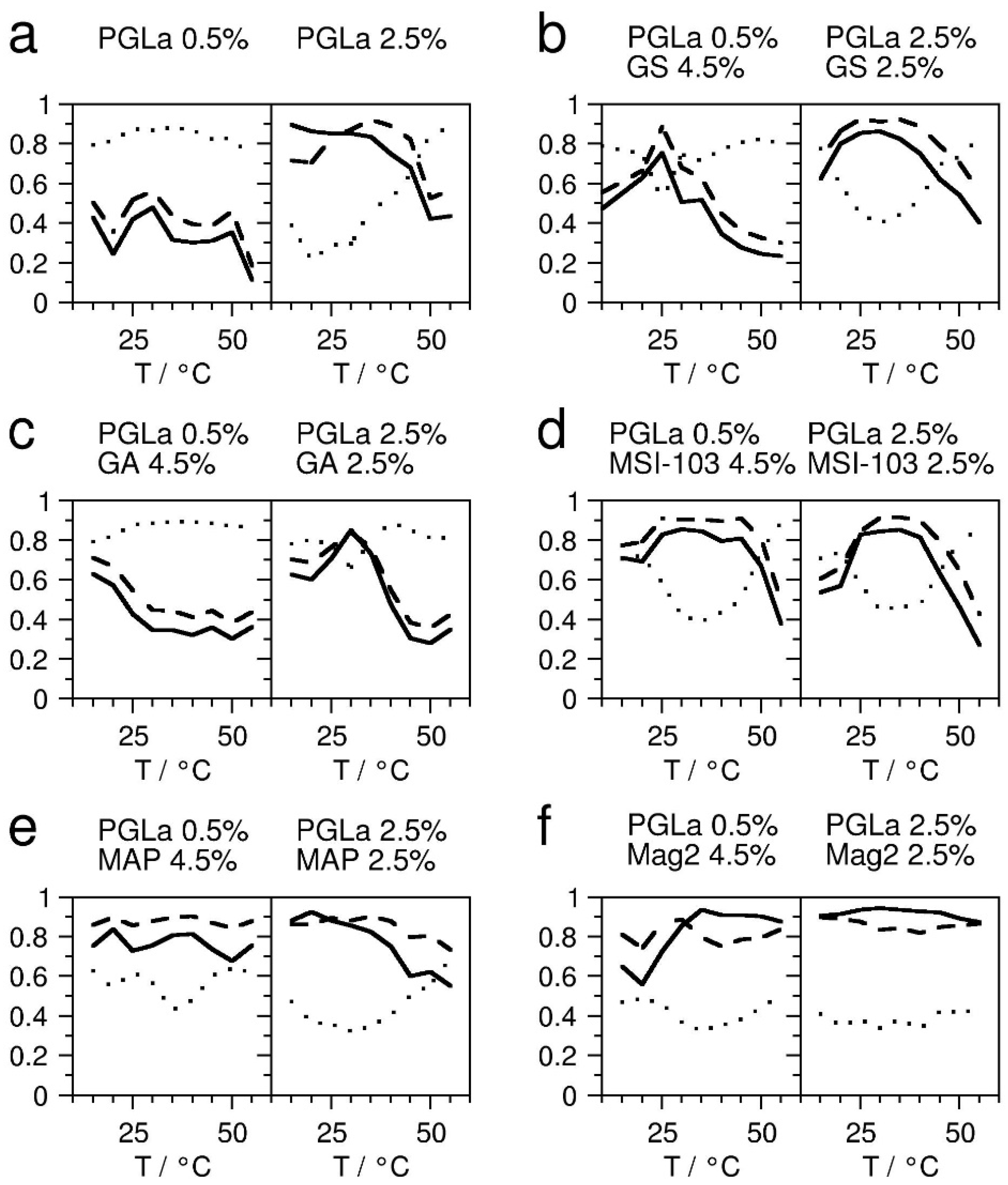
Appendix B. Determination of the Peptide Insertion States
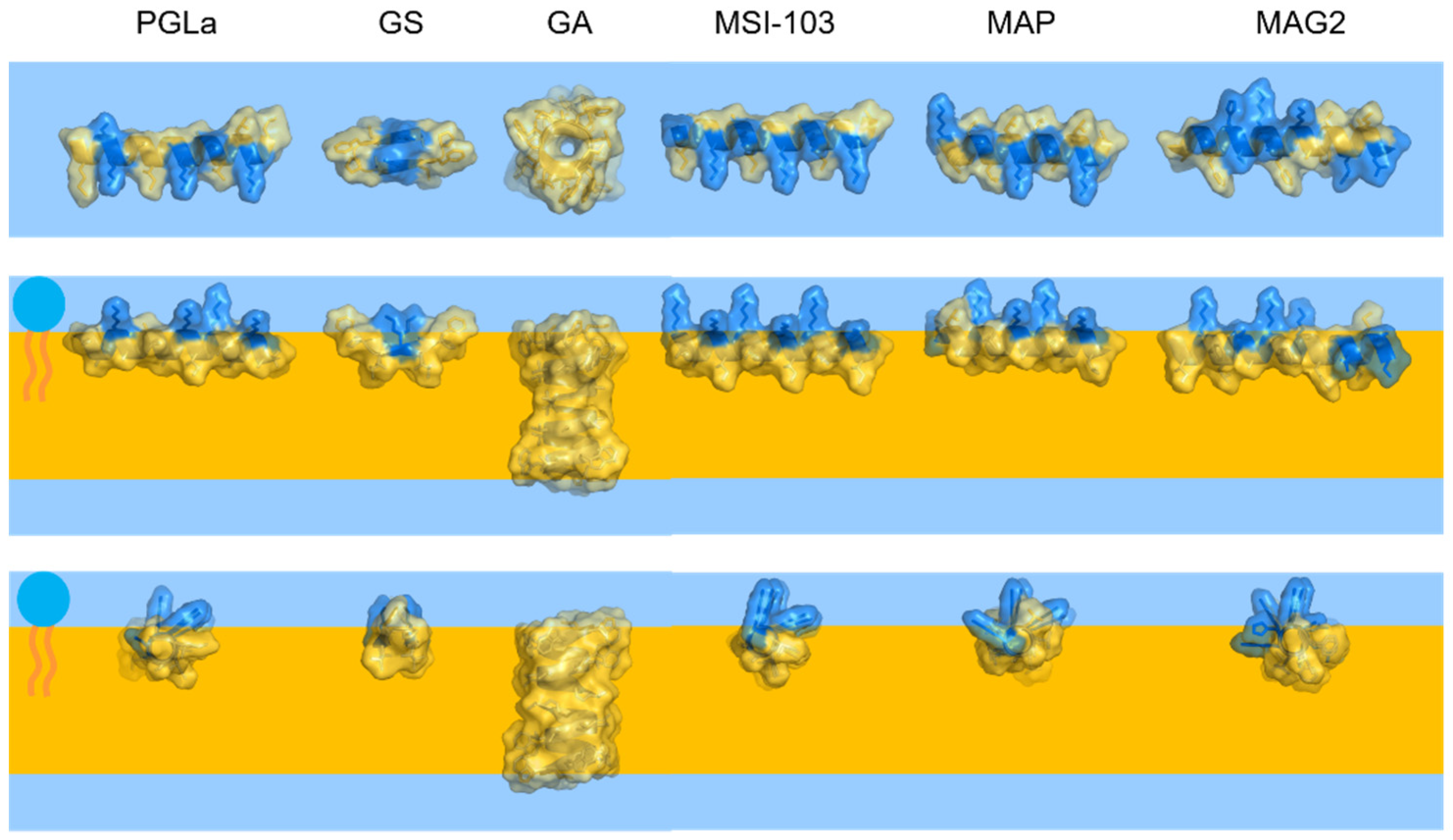
Appendix C. Comparison of Peptide Dimensions
References
- Cantor, R.S. Lateral pressures in cell membranes: A mechanism for modulation of protein function. J. Phys. Chem. B 1997, 101, 1723–1725. [Google Scholar] [CrossRef]
- Marsh, D. Lateral pressure profile, spontaneous curvature frustration, and the incorporation and conformation of proteins in membranes. Biophys. J. 2007, 93, 3884–3899. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perozo, E.; Cortes, D.M.; Sompornpisut, P.; Kloda, A.; Martinac, B. Open channel structure of MscL and the gating mechanism of mechanosensitive channels. Nature 2002, 418, 942–948. [Google Scholar] [CrossRef] [PubMed]
- Perozo, E.; Kloda, A.; Cortes, D.M.; Martinac, B. Physical principles inderlying the transduction of bilayer deformation forces during mechanosensitive channel gating. Nat. Struct. Biol. 2002, 9, 696–703. [Google Scholar] [CrossRef] [PubMed]
- Grage, S.L.; Keleshian, A.M.; Turdzeladze, T.; Battle, A.R.; Tay, W.C.; May, R.P.; Holt, S.A.; Contera, S.A.; Moulin, M.; Pal, P.; et al. Bilayer-mediated clustering and functional interaction of MscL channels. Biophys. J. 2011, 100, 1252–1260. [Google Scholar] [CrossRef]
- Battle, A.; Ridone, P.; Bavi, N.; Nakayama, Y.; Nikolaev, Y.; Martinac, B. Lipid-protein interactions: Lessons learned from stress. Biochim. Biophys. Acta-Rev. Biomembr. 2015, 1848, 1744–1756. [Google Scholar] [CrossRef] [PubMed]
- Ridone, P.; Grage, S.L.; Patkunarajah, A.; Battle, A.R.; Ulrich, A.S.; Martinac, B. “Force-from-lipids” gating of mechanosensitive channels modulated by PUFAs. J. Mechanical Behav. Biomed. Mat. 2018, 79, 158–167. [Google Scholar] [CrossRef] [PubMed]
- Henderson, J.M.; Lee, K.Y.C. Promising antimicrobial agents designed from natural peptide templates. Curr. Opin. Solid State Mater. Sci. 2013, 17, 175–192. [Google Scholar] [CrossRef]
- Shai, Y. Mechanism of the binding, insertion and destabilization of phospholipid bilayer membranes by α-helical antimicrobial peptides and cell non-selective membrane-lytic peptides. Biochim. Biophys. Acta-Rev. Biomembr. 1999, 1462, 55–70. [Google Scholar] [CrossRef] [Green Version]
- Wimley, W. Describing the mechanism of antimicrobial peptide action with the interfacial activity model. ACS Chem. Biol. 2010, 5, 905–917. [Google Scholar] [CrossRef] [Green Version]
- Strandberg, E.; Tiltak, D.; Ehni, S.; Wadhwani, P.; Ulrich, A.S. Lipid shape is a key factor for membrane interactions of amphipathic helical peptides. Biochim. Biophys. Acta-Rev. Biomembr. 2012, 1818, 1764–1776. [Google Scholar] [CrossRef] [Green Version]
- Strandberg, E.; Bentz, D.; Wadhwani, P.; Ulrich, A.S. Chiral supramolecular architecture of stable transmembrane pores formed by an α-helical antibiotic peptide in the presence of lyso-lipids. Sci. Rep. 2020, 10, 4710. [Google Scholar] [CrossRef] [PubMed]
- Cantor, R.S. Lipid composition and the lateral pressure profile in bilayers. Biophys. J. 1999, 76, 2625–2639. [Google Scholar] [CrossRef] [Green Version]
- Israelachvili, J.N.; Mitchell, D.J.; Ninham, B.W. Theory of self-assembly of lipid bilayers and vesicles. Biochim. Biophys. Acta-Rev. Biomembr. 1977, 470, 185–201. [Google Scholar] [CrossRef]
- Cullis, P.R.; de Kruijff, B. Lipid polymorphism and the functional roles of lipids in biological membranes. Biochim. Biophys. Acta-Rev. Biomembr. 1979, 559, 399–420. [Google Scholar] [CrossRef]
- Kollmitzer, B.; Heftberger, P.; Rappolt, M.; Pabst, G. Monolayer spontaneous curvature of raft-forming membrane lipids. Soft Matter 2013, 9, 10877–10884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lohner, K.; Prenner, E.J. Differential scanning calorimetry and X-ray diffraction studies of the specificity of the interaction of antimicrobial peptides with membrane-mimetic systems. Biochim. Biophys. Acta-Biomembr. 1999, 1462, 141–156. [Google Scholar] [CrossRef] [Green Version]
- Bechinger, B.; Lohner, K. Detergent-like actions of linear amphipathic cationic antimicrobial peptides. Biochim. Biophys. Acta-Rev. Biomembr. 2006, 1758, 1529–1539. [Google Scholar] [CrossRef] [Green Version]
- Koller, D.; Lohner, K. The role of spontaneous lipid curvature in the interaction of interfacially active peptides with membranes. Biochim. Biophys. Acta-Rev. Biomembr. 2014, 1838, 2250–2259. [Google Scholar] [CrossRef] [Green Version]
- Grage, S.L.; Afonin, S.; Ulrich, A.S. Dynamic transitions of membrane-active peptides. Methods Mol. Biol. 2008, 618, 183–207. [Google Scholar]
- Strandberg, E.; Ulrich, A.S. Solid-state 19F-NMR analysis of peptides in oriented biomembranes. In Modern Magnetic Resonance; Webb, G.A., Ed.; Springer International Publishing: Berlin/Heidelberg, Germany, 2017; pp. 1–18. [Google Scholar]
- Afonin, S.; Glaser, R.W.; Sachse, C.; Salgado, J.; Wadhwani, P.; Ulrich, A.S. 19F NMR screening of unrelated antimicrobial peptides shows that membrane interactions are largely governed by lipids. Biochim. Biophys. Acta-Rev. Biomembr. 2014, 1838, 2260–2268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grage, S.L.; Afonin, S.; Ulrich, A.S. 19F NMR in biomembranes. In Solid-State NMR: Applications in Biomembrane Structure; Separovic, F., Sani, M.-A., Eds.; IOP Publishing Ltd.: Bristol, UK, 2020. [Google Scholar]
- Glaser, R.W.; Sachse, C.; Dürr, U.H.N.; Wadhwani, P.; Ulrich, A.S. Orientation of the antimicrobial peptide PGLa in lipid membranes determined from 19F-NMR dipolar couplings of 4-CF3-phenylglycine labels. J. Magn. Reson. 2004, 168, 153–163. [Google Scholar] [CrossRef] [PubMed]
- Glaser, R.W.; Sachse, C.; Dürr, U.H.N.; Wadhwani, P.; Afonin, S.; Strandberg, E.; Ulrich, A.S. Concentration-dependent realignment of the antimicrobial peptide PGLa in lipid membranes observed by solid-state 19F-NMR. Biophys. J. 2005, 88, 3392–3397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Afonin, S.; Grage, S.L.; Ieronimo, M.; Wadhwani, P.; Ulrich, A.S. Temperature-dependent transmembrane insertion of the amphiphilic peptide PGLa in lipid bilayers observed by solid state 19F NMR spectroscopy. J. Am. Chem. Soc. 2008, 130, 16512–16514. [Google Scholar] [CrossRef] [PubMed]
- Strandberg, E.; Zerweck, J.; Wadhwani, P.; Ulrich, A.S. Synergistic insertion of antimicrobial magainin-family peptides in membranes depends on the lipid spontaneous curvature. Biophys. J. 2013, 104, L9–L11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salnikov, E.S.; Bechinger, B. Lipid-controlled peptide topology and interactions in bilayers: Structural insights into the synergistic enhancement of the antimicrobial activities of PGLa and Magainin 2. Biophys. J. 2011, 100, 1473–1480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsuzaki, K.; Mitani, Y.; Akada, K.; Murase, O.; Yoneyama, S.; Zasloff, M.; Miyajima, K. Mechanism of synergism between antimicrobial peptides magainin 2 and PGLa. Biochemistry 1998, 37, 15144–15153. [Google Scholar] [CrossRef]
- Strandberg, E.; Tremouilhac, P.; Wadhwani, P.; Ulrich, A.S. Synergistic transmembrane insertion of the heterodimeric PGLa/magainin 2 complex studied by solid-state NMR. Biochim. Biophys. Acta-Biomembr. 2009, 1788, 1667–1679. [Google Scholar] [CrossRef] [Green Version]
- Afonin, S.; Dürr, U.H.N.; Wadhwani, P.; Salgado, J.; Ulrich, A.S. Solid state NMR structure analysis of the antimicrobial peptide gramicidin S in lipid membranes: Concentration-dependent re-alignment and self-assembly as a β-barrel. Top. Curr. Chem. 2008, 273, 139–154. [Google Scholar]
- Salgado, J.; Grage, S.L.; Kondejewski, L.H.; Hodges, R.S.; McElhaney, R.N.; Ulrich, A.S. Membrane-bound structure and alignment of the antimicrobial β-sheet. J. Biomol. NMR 2001, 21, 191–208. [Google Scholar] [CrossRef]
- Fu, R.; Cotten, M.; Cross, T.A. Inter- and intramolecular distance measurements by solid-state MAS NMR: Determination of gramicidin A channel dimer structure in hydrated phospholipid bilayers. J. Biomol. NMR 2000, 16, 261–268. [Google Scholar] [CrossRef] [PubMed]
- Maloy, W.L.; Kari, U.P. Structure-activity studies on magainins and other host defense peptides. Biopolymers 1995, 37, 105–122. [Google Scholar] [CrossRef] [PubMed]
- Langel, U. Cell-Penetrating Peptides: Processes and Applications, 1st ed.; CRC Press: Boca Raton, FL, USA, 2002. [Google Scholar]
- Bürck, J.; Roth, S.; Wadhwani, P.; Afonin, S.; Kanithasen, N.; Strandberg, E.; Ulrich, A.S. Conformation and membrane orientation of amphiphilic helical peptides by oriented circular dichroism. Biophys. J. 2008, 95, 3872–3881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strandberg, E.; Kanithasen, N.; Tiltak, D.; Bürck, J.; Wadhwani, P.; Zwernemann, O.; Ulrich, A.S. Solid-state NMR analysis comparing the designer-made antibiotic MSI-103 with its parent peptide PGLa in lipid bilayers. Biochemistry 2008, 47, 2601–2616. [Google Scholar] [CrossRef]
- Strandberg, E.; Horn, D.; Reißer, S.; Zerweck, J.; Wadhwani, P.; Ulrich, A.S. 2H-NMR and MD simulations reveal membrane-bound conformation of Magainin 2 and its synergy with PGLa. Biophys. J. 2016, 111, 2149–2161. [Google Scholar] [CrossRef] [Green Version]
- Zerweck, J.; Strandberg, E.; Kukharenko, O.; Reichert, J.; Bürck, J.; Wadhwani, P.; Ulrich, A.S. Molecular mechanism of synergy between the antimicrobial peptides PGLa and magainin 2. Sci. Rep. 2017, 7, 13153. [Google Scholar] [CrossRef]
- Dymond, K.M. Lipid monolayer spontaneous curvatures: A collection of published values. Chem. Phys. Lipids 2021, 239, 105117. [Google Scholar] [CrossRef]
- Strandberg, E.; Wadhwani, P.; Tremouilhac, P.; Dürr, U.H.N.; Ulrich, A.S. Solid-state NMR analysis of the PGLa peptide orientation in DMPC bilayers: Structural fidelity of 2H-labels versus high sensitivity of 19F-NMR. Biophys. J. 2006, 90, 1676–1686. [Google Scholar] [CrossRef] [Green Version]
- Gullingsrud, J.; Schulten, K. Lipid bilayer pressure profiles and mechanosensitive channel gating. Biophys. J. 2004, 86, 3496–3509. [Google Scholar] [CrossRef] [Green Version]
- Tremouilhac, P.; Strandberg, E.; Wadhwani, P.; Ulrich, A.S. Synergistic Transmembrane Alignment of the Antimicrobial Heterodimer PGLa/Magainin. J. Biol. Chem. 2006, 281, 32089–32094. [Google Scholar] [CrossRef] [Green Version]
- Pujals, S.; Miyamae, H.; Afonin, S.; Murayama, T.; Hirose, H.; Nakase, I.; Taniuchi, K.; Umeda, M.; Sakamoto, K.; Ulrich, A.S.; et al. Curvature engineering: Positive membrane curvature induced by Epsin N-terminal peptide boosts internalization of octaarginine. ACS Chem. Biol. 2013, 8, 1894–1899. [Google Scholar] [CrossRef] [PubMed]
- Berditsch, M.; Afonin, S.; Ulrich, A.S. The ability of Aneurinibacillus migulanus (Bacillus brevis) to produce the antibiotic gramicidin S is correlated with phenotype variation. Appl. Environ. Microbiol. 2007, 73, 6620–6628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, C.D.; Meienhofer, J. Solid-phase peptide-synthesis using mild base cleavage of N-alpha-fluorenylmethyloxycarbonyl-amino acids, exemplified by a synthesis of dihydrosomatostatin. Int. J. Peptide Protein Res. 1978, 11, 246–249. [Google Scholar] [CrossRef] [PubMed]
- Fields, G.B.; Noble, R.L. Solid-phase peptide-synthesis utilizing 9-fluorenylmethoxycarbonyl amino-acids. Int. J. Pept. Protein Res. 1990, 35, 161–214. [Google Scholar] [CrossRef]
- Afonin, S.; Glaser, R.W.; Berditchevskaia, M.; Wadhwani, P.; Gührs, K.-H.; Möllmann, U.; Perner, A.; Ulrich, A.S.; Guehrs, K.-H. 4-fluorophenylglycine as a label for 19F NMR structure analysis of membrane-associated peptides. ChemBioChem 2003, 4, 1151–1163. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grage, S.L.; Afonin, S.; Ieronimo, M.; Berditsch, M.; Wadhwani, P.; Ulrich, A.S. Probing and Manipulating the Lateral Pressure Profile in Lipid Bilayers Using Membrane-Active Peptides—A Solid-State 19F NMR Study. Int. J. Mol. Sci. 2022, 23, 4544. https://doi.org/10.3390/ijms23094544
Grage SL, Afonin S, Ieronimo M, Berditsch M, Wadhwani P, Ulrich AS. Probing and Manipulating the Lateral Pressure Profile in Lipid Bilayers Using Membrane-Active Peptides—A Solid-State 19F NMR Study. International Journal of Molecular Sciences. 2022; 23(9):4544. https://doi.org/10.3390/ijms23094544
Chicago/Turabian StyleGrage, Stephan L., Sergii Afonin, Marco Ieronimo, Marina Berditsch, Parvesh Wadhwani, and Anne S. Ulrich. 2022. "Probing and Manipulating the Lateral Pressure Profile in Lipid Bilayers Using Membrane-Active Peptides—A Solid-State 19F NMR Study" International Journal of Molecular Sciences 23, no. 9: 4544. https://doi.org/10.3390/ijms23094544
APA StyleGrage, S. L., Afonin, S., Ieronimo, M., Berditsch, M., Wadhwani, P., & Ulrich, A. S. (2022). Probing and Manipulating the Lateral Pressure Profile in Lipid Bilayers Using Membrane-Active Peptides—A Solid-State 19F NMR Study. International Journal of Molecular Sciences, 23(9), 4544. https://doi.org/10.3390/ijms23094544






