Mutant lpa1 Analysis of ZmLPA1 Gene Regulates Maize Leaf-Angle Development through the Auxin Pathway
Abstract
:1. Introduction
2. Results
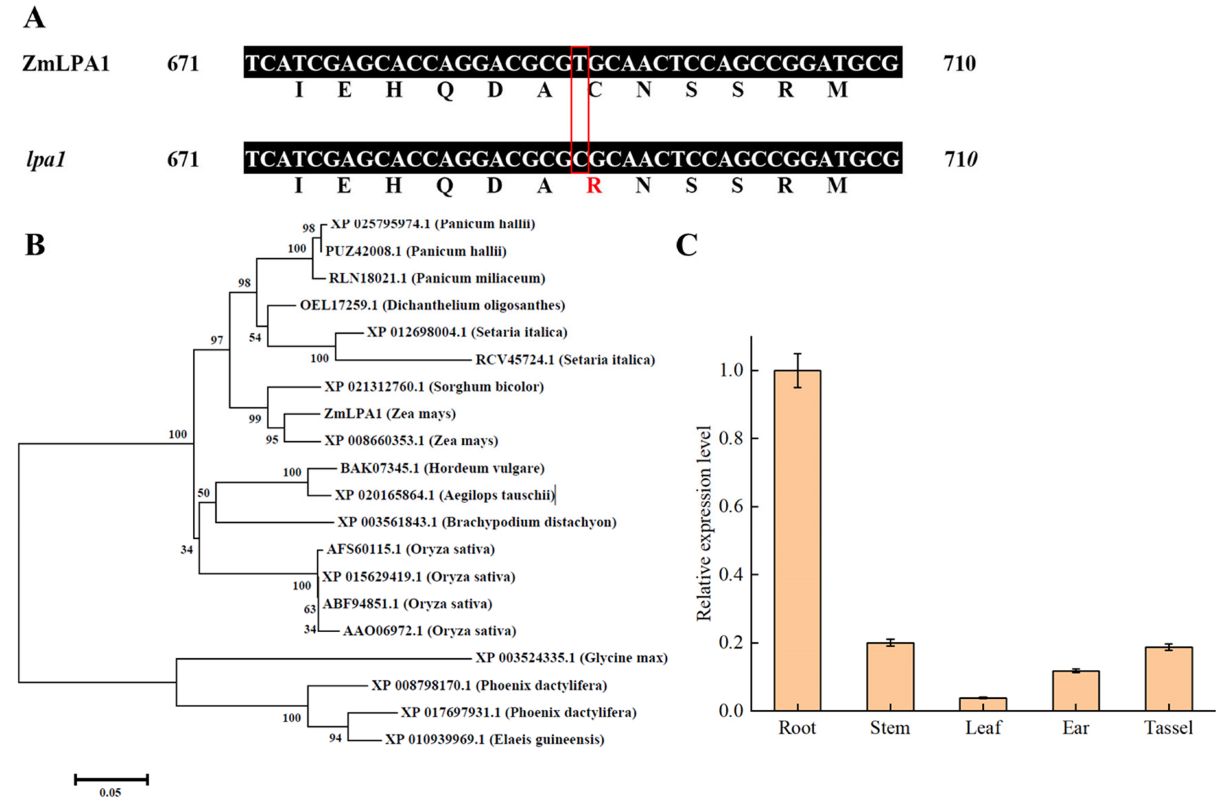
2.1. Identification of ZmLPA1 Gene
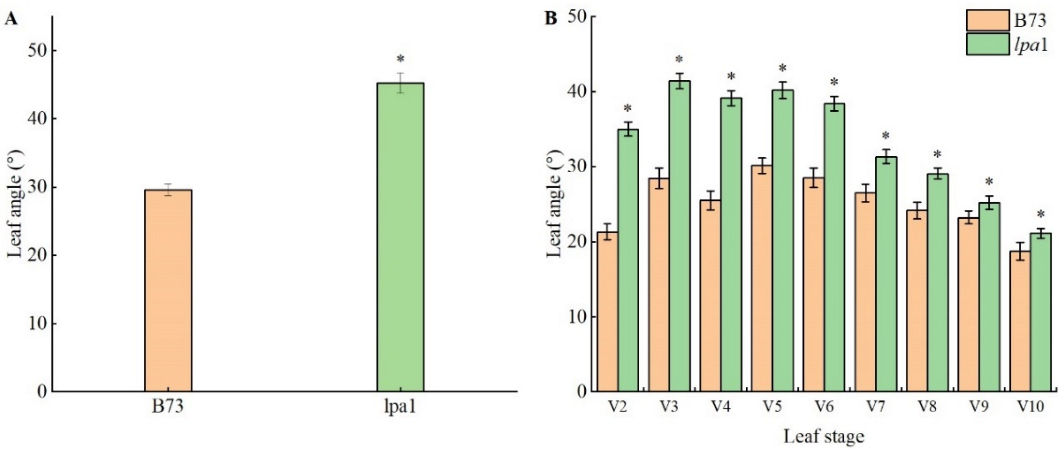
2.2. Phenotype Identification of Maize Mutant lpa1
2.3. Analysis of Agronomic Characters of Maize Mutant lpa1
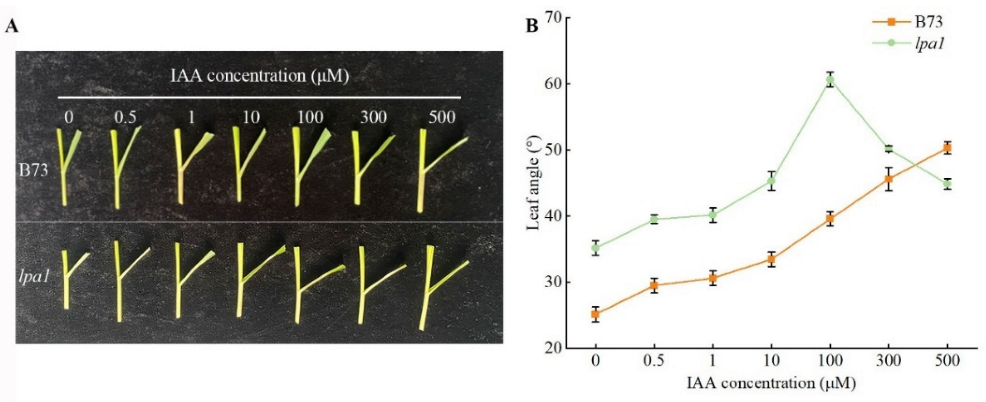
2.4. Screening of the Optimal Concentration of Exogenous Hormones
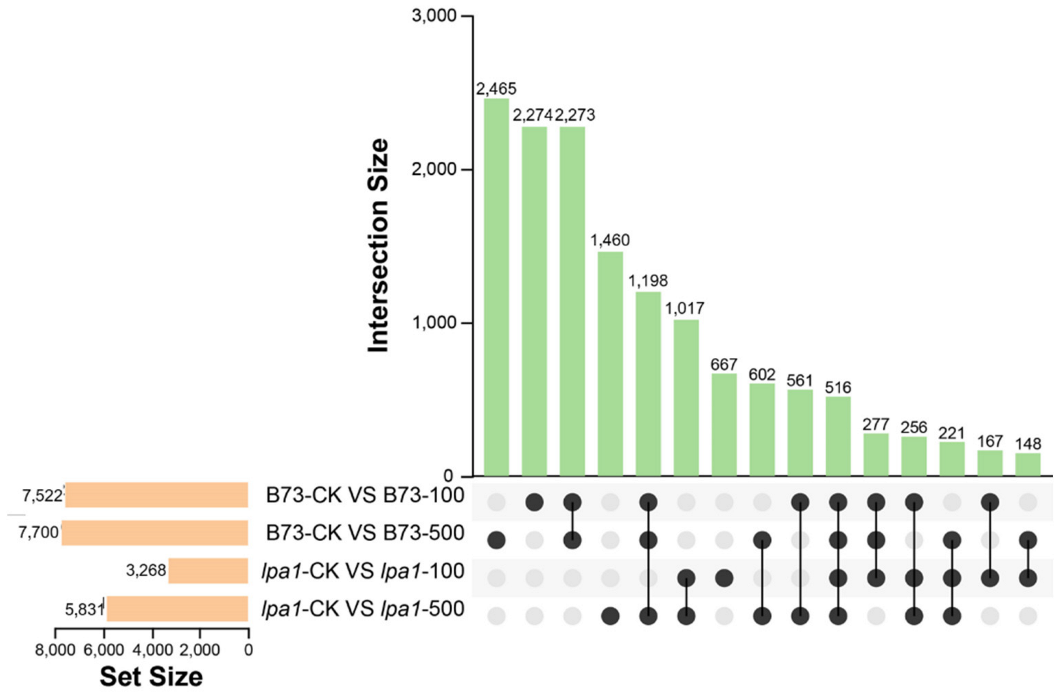
2.5. Upset Plot Analysis of DEGs in B73 and lpa1 under Different Concentrations of IAA
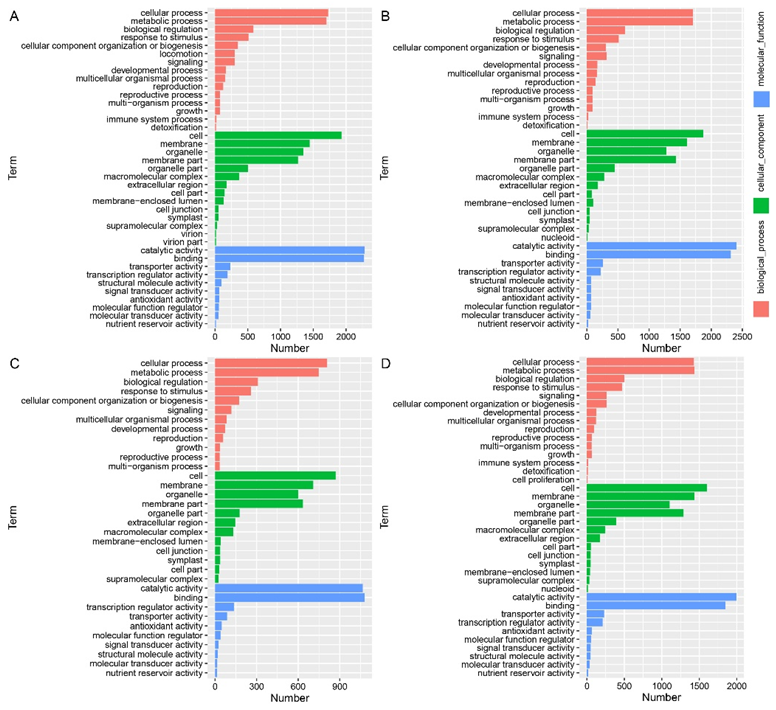
2.6. Gene-Ontology Classification of DEGs in the Leaf Angle between B73 and lpa1 under Different IAA Concentrations
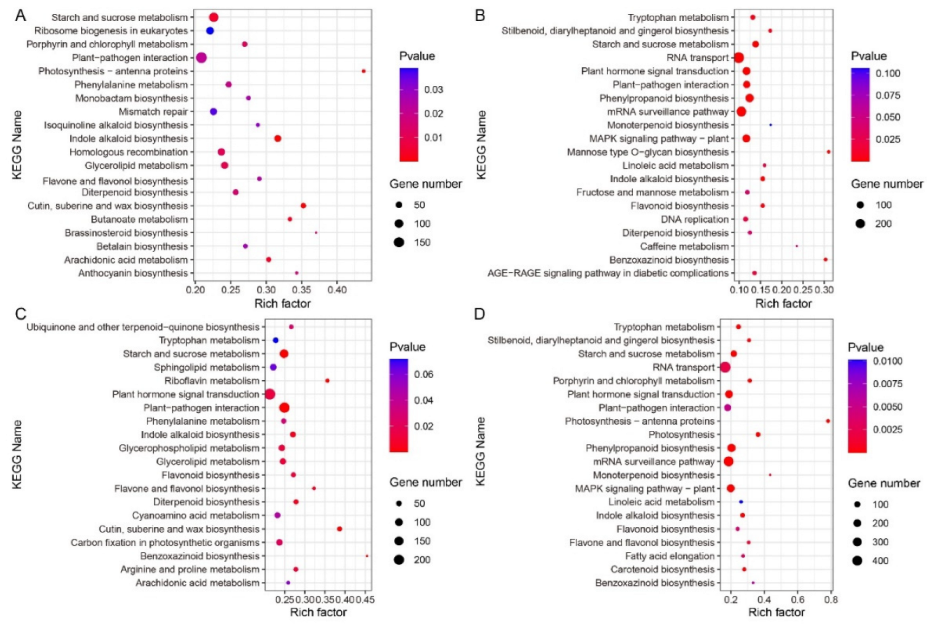
2.7. Pathway Enrichment Analysis of B73 and lpa1 at Different IAA Concentrations
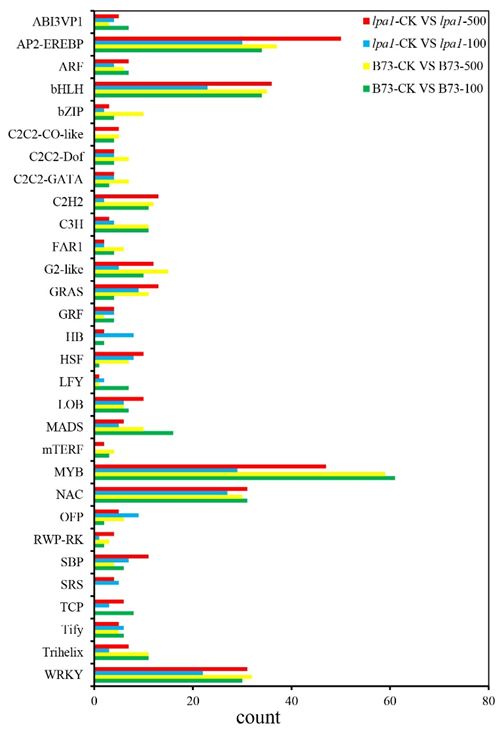
2.8. Transcription-Factor Family Analysis
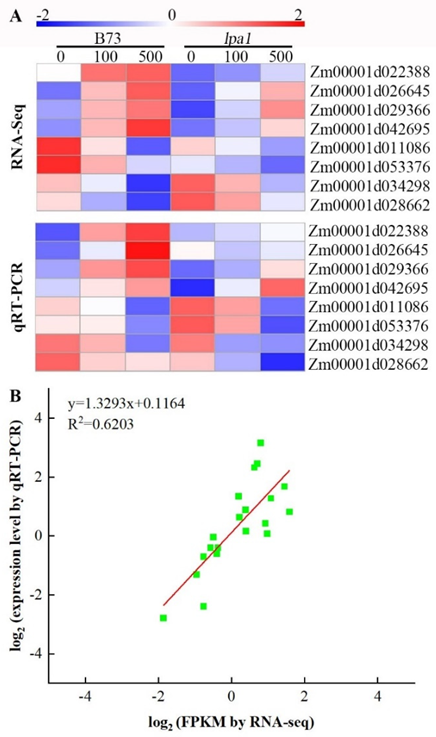
2.9. Validation of DEGs by qRT-PCR Analysis
3. Discussion
3.1. Functional Study of ZmLPA1 in Maize
3.2. Plant-Type Characteristics of Mutant lpa1
3.3. The Effect of Exogenous IAA on Maize Leaf Angle
3.4. Differentially Expressed Genes Comparison and GO Function Analysis
3.5. Analysis of Differential Genes Related to Maize Leaf Angle under Different Concentrations of Exogenous IAA Treatment
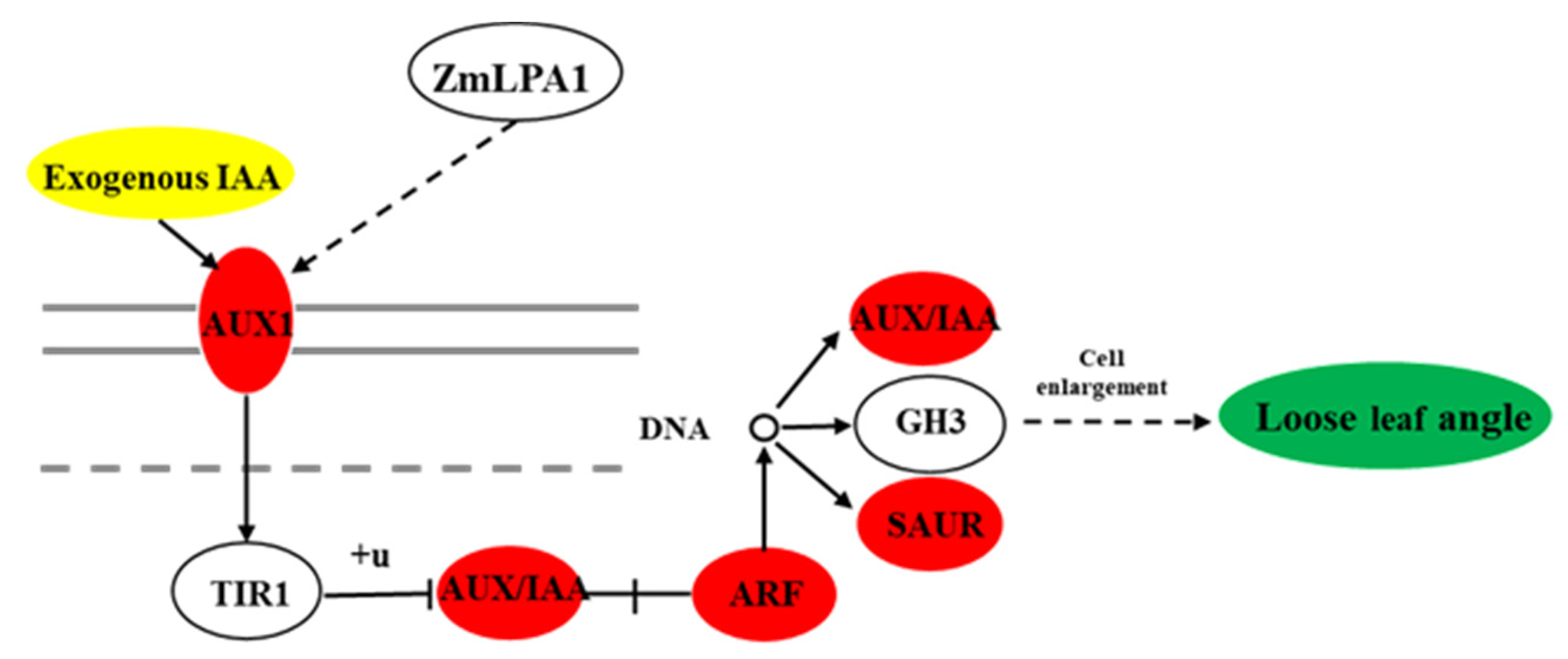
3.6. The Effect of ZmLPA1 on Maize Auxin Signal-Transduction-Related Genes
4. Materials and Methods
4.1. Plant Materials and Planting Conditions
4.2. Mutant Phenotype Identification
4.3. Data Analysis
4.4. Extraction of Total RNA and Synthesis of First Strand cDNA
4.5. PCR Amplification of ZmLPA1 and Sequencing of the Product
4.6. Bioinformatics Analysis of ZmLPA1
4.7. Material Hormone Treatment
4.8. Total RNA Extraction and Detection
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, J.; Fernie, A.R.; Yan, J. The Past, Present, and Future of Maize Improvement: Domestication, Genomics, and Functional Genomic Routes toward Crop Enhancement. Plant Commun. 2020, 1, 100010. [Google Scholar] [CrossRef] [PubMed]
- Ciampitti, I.; Vyn, T.J. Physiological perspectives of changes over time in maize yield dependency on nitrogen uptake and associated nitrogen efficiencies: A review. Field Crops Res. 2012, 133, 48–67. [Google Scholar] [CrossRef]
- Messmer, R.; Fracheboud, Y.; Banziger, M.; Vargas, M.; Stamp, P.; Ribaut, J.M. Drought stress and tropical maize: QTL-by-environment interactions and stability of QTLs across environments for yield components and secondary traits. Theor. Appl. Genet. 2009, 119, 913–930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chern, M.; Canlas, P.E.; Fitzgerald, H.A.; Ronald, P.C. Rice NRR, a negative regulator of disease resistance, interacts with Arabidopsis NPR1 and rice NH1. Plant J. 2005, 43, 623–635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colasanti, J.; Tremblay, R.; Wong, A.Y.; Coneva, V.; Kozaki, A.; Mable, B.K. The maize INDETERMINATE1 flowering time regulator defines a highly conserved zinc finger protein family in higher plants. BMC Genom. 2006, 7, 158. [Google Scholar] [CrossRef] [Green Version]
- Correll, M.J.; Kiss, J.Z. Interactions between gravitropism and phototropism in plants. J. Plant Growth Regul. 2002, 21, 89–101. [Google Scholar] [CrossRef]
- Duan, K.; Li, L.; Hu, P.; Xu, S.P.; Xu, Z.H.; Xue, H.W. A brassinolide-suppressed rice MADS-box transcription factor, OsMDP1, has a negative regulatory role in BR signaling. Plant J. Cell Mol. Biol. 2006, 47, 519–531. [Google Scholar] [CrossRef]
- Vazin, F.; Hassanzadeh, M.; Madani, A.; Nassiri-Mahallati, M.; Nasri, M. Modeling light interception and distribution in mixed canopy of common cocklebur (Xanthium stramarium) in competition with corn. Planta Daninha 2010, 28, 455–462. [Google Scholar] [CrossRef] [Green Version]
- Donald, C.M. The breeding of crop ideotypes. Euphytica 1968, 17, 385–403. [Google Scholar] [CrossRef]
- Buckler, E.S.; Tian, F.; Bradbury, P.J.; Brown, P.J.; Hung, H.; Sun, Q.; Holland, J.B. Genome-wide association study of leaf architecture in the maize nested association mapping population. Nat. Genet. 2011, 43, 159–162. [Google Scholar]
- Stewart, D.W.; Costa, C.; Dwyer, L.M.; Smith, D.L.; Hamilton, R.I.; Ma, B.L. Canopy Structure, Light Interception, and Photosynthesis in Maize. Agron. J. 2003, 95, 1465–1474. [Google Scholar] [CrossRef]
- Mattsson, J.; Ckurshumova, W.; Berleth, T. Auxin signaling in Arabidopsis leaf vascular development. Plant Physiol. 2003, 131, 1327–1339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galli, M.; Liu, Q.; Moss, B.L.; Malcomber, S.; Li, W.; Gaines, C.; Gallavotti, A. Auxin signaling modules regulate maize inflorescence architecture. Proc. Natl. Acad. Sci. USA 2015, 112, 13372–13377. [Google Scholar] [CrossRef] [Green Version]
- Reinhardt, D.; Mandel, T.; Kuhlemeier, C. Auxin regulates the initiation and radial position of plant lateral organs. Plant Cell 2000, 12, 507–518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Terol, J.; Domingo, C.; Talon, M. The GH3 family in plants: Genome wide analysis in rice and evolutionary history based on EST analysis. Gene 2006, 371, 279–290. [Google Scholar] [CrossRef]
- Jain, M.; Kaur, N.; Tyagi, A.K.; Khurana, J.P. The auxin-responsive GH3 gene family in rice (Oryza sativa). Funct. Integr. Genom. 2006, 6, 36–46. [Google Scholar] [CrossRef]
- Yang, J.H.; Han, S.J.; Yoon, E.K.; Lee, W.S. Evidence of an auxin signal pathway, microRNA167-ARF8-GH3, and its response to exogenous auxin in cultured rice cells. Nucleic Acids Res. 2006, 34, 1892–1899. [Google Scholar] [CrossRef]
- Zhao, S.Q.; Xiang, J.J.; Xue, H.W. Studies on the rice LEAF INCLINATION1 (LC1), an IAA-amido synthetase, reveal the effects of auxin in leaf inclination control. Mol. Plant 2013, 6, 174–187. [Google Scholar] [CrossRef] [Green Version]
- Yoshihara, T.; Spalding, E.P. LAZY Genes Mediate the Effects of Gravity on Auxin Gradients and Plant Architecture. Plant Physiol. 2017, 175, 959–969. [Google Scholar] [CrossRef] [Green Version]
- Yoshihara, T.; Spalding, E.P.; Iino, M. AtLAZY1 is a signaling component required for gravitropism of the Arabidopsis thaliana inflorescence. Plant J. 2013, 74, 267–279. [Google Scholar] [CrossRef]
- Band, L.R.; Wells, D.M.; Larrieu, A.; Sun, J.; Middleton, A.M.; French, A.P.; Bennett, M.J. Root gravitropism is regulated by a transient lateral auxin gradient controlled by a tipping-point mechanism. Proc. Natl. Acad. Sci. USA 2012, 109, 4668–4673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ku, L.; Wei, X.; Zhang, S.; Zhang, J.; Guo, S.; Chen, Y. Cloning and characterization of a putative TAC1 ortholog associated with leaf angle in maize (Zea mays L.). PLoS ONE 2011, 6, e20621. [Google Scholar] [CrossRef] [PubMed]
- Juarez, M.T.; Twigg, R.W.; Timmermans, M.C. Specification of adaxial cell fate during maize leaf development. Development 2004, 131, 4533–4544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Ku, L.X.; Han, Z.P.; Guo, S.L.; Liu, H.J.; Zhang, Z.Z.; Chen, Y.H. The ZmCLA4 gene in the qLA4-1 QTL controls leaf angle in maize (Zea mays L.). J. Exp. Bot. 2014, 65, 5063–5076. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strable, J.; Wallace, J.G.; Unger-Wallace, E.; Briggs, S.; Bradbury, P.J.; Buckler, E.S.; Vollbrecht, E. Maize YABBY Genes drooping leaf1 and drooping leaf2 Regulate Plant Architecture. Plant Cell 2017, 29, 1622–1641. [Google Scholar] [CrossRef] [Green Version]
- Wu, X.; Tang, D.; Li, M.; Wang, K.; Cheng, Z. Loose Plant Architecture1, an INDETERMINATE DOMAIN protein involved in shoot gravitropism, regulates plant architecture in rice. Plant Physiol. 2013, 161, 317–329. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.M.; Park, S.J.; Huang, J.; Lee, E.J.; Xuan, Y.H.; Je, B.I.; Han, C.D. Loose Plant Architecture1 (LPA1) determines lamina joint bending by suppressing auxin signalling that interacts with C-22-hydroxylated and 6-deoxo brassinosteroids in rice. J. Exp. Bot. 2016, 67, 1883–1895. [Google Scholar] [CrossRef] [Green Version]
- Ingkasuwan, P.; Netrphan, S.; Prasitwattanaseree, S.; Tanticharoen, M.; Bhumiratana, S.; Meechai, A.; Cheevadhanarak, S. Inferring transcriptional gene regulation network of starch metabolism in Arabidopsis thaliana leaves using graphical Gaussian model. BMC Syst. Biol. 2012, 6, 100. [Google Scholar] [CrossRef] [Green Version]
- Seo, P.J.; Ryu, J.; Kang, S.K.; Park, C.M. Modulation of sugar metabolism by an INDETERMINATE DOMAIN transcription factor contributes to photoperiodic flowering in Arabidopsis. Plant J. 2011, 65, 418–429. [Google Scholar] [CrossRef]
- Feurtado, J.A.; Huang, D.; Wicki-Stordeur, L.; Hemstock, L.E.; Potentier, M.S.; Tsang, E.W.; Cutler, A.J. The Arabidopsis C2H2 zinc finger INDETERMINATE DOMAIN1/ENHYDROUS promotes the transition to germination by regulating light and hormonal signaling during seed maturation. Plant Cell 2011, 23, 1772–1794. [Google Scholar] [CrossRef] [Green Version]
- Tanimoto, M.; Tremblay, R.; Colasanti, J. Altered gravitropic response, amyloplast sedimentation and circumnutation in the Arabidopsis shoot gravitropism 5 mutant are associated with reduced starch levels. Plant Mol. Biol. 2008, 67, 57–69. [Google Scholar] [CrossRef] [PubMed]
- Matsubara, K.; Yamanouchi, U.; Wang, Z.; Minobe, Y.; Izawa, T.; Yano, M. Ehd2, a Rice Ortholog of the Maize INDETERMINATE1 Gene, Promotes Flowering by Up-Regulating Ehd11[C][W]. Plant Physiol. 2008, 148, 1425–1435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, S.J.; Kim, S.L.; Lee, S.; Je, B.I.; Piao, H.L.; Park, S.H.; Han, C.D. Rice Indeterminate 1 (OsId1) is necessary for the expression of Ehd1 (Early heading date 1) regardless of photoperiod. Plant J. 2008, 56, 1018–1029. [Google Scholar] [CrossRef] [PubMed]
- Ning, J.; Zhang, B.; Wang, N.; Zhou, Y.; Xiong, L. Increased leaf angle1, a Raf-like MAPKKK that interacts with a nuclear protein family, regulates mechanical tissue formation in the Lamina joint of rice. Plant Cell 2011, 23, 4334–4347. [Google Scholar] [CrossRef] [Green Version]
- Tang, H.; Zhang, B.; Tan, J.; Tian, Y.; Kang, J.; Ye, G. Grey correlation degree analysis on yield and photosynthetic characteristics of ear leaf on hybrids of corn. Chin. Agric. Sci. Bull. 2011, 27, 69–73. [Google Scholar]
- Yuan, L.M. Mining Key Genes Involved in the Regulation of Ligular Region Development and Leaf Angle (LA) Formation in Maize. Master’s Thesis, Shandong Agricultural University, Tai’an, China, 2016. [Google Scholar]
- Hornitschek, P.; Kohnen, M.V.; Lorrain, S.; Rougemont, J.; Ljung, K.; Lopez-Vidriero, I.; Fankhauser, C. Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J. 2012, 71, 699–711. [Google Scholar] [CrossRef] [Green Version]
- Marchant, A.; Bhalerao, R.; Casimiro, I.; Eklof, J.; Casero, P.J.; Bennett, M.; Sandberg, G. AUX1 promotes lateral root formation by facilitating indole-3-acetic acid distribution between sink and source tissues in the Arabidopsis seedling. Plant Cell 2002, 14, 589–597. [Google Scholar] [CrossRef] [Green Version]
- Mashiguchi, K.; Tanaka, K.; Sakai, T.; Sugawara, S.; Kawaide, H.; Natsume, M.; Kasahara, H. The main auxin biosynthesis pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 18512–18517. [Google Scholar] [CrossRef] [Green Version]
- Stepanova, A.N.; Robertson-Hoyt, J.; Yun, J.; Benavente, L.M.; Xie, D.; Doležal, K.; Alonso, J.M. TAA1-Mediated Auxin Biosynthesis Is Essential for Hormone Crosstalk and Plant Development. Cell 2008, 133, 177–191. [Google Scholar] [CrossRef] [Green Version]
- Cheng, Y.; Dai, X.; Zhao, Y. Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 2006, 20, 1790–1799. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Hammes, U.Z.; Taylor, C.G.; Schachtman, D.P.; Nielsen, E. High-affinity auxin transport by the AUX1 influx carrier protein. Curr. Biol. 2006, 16, 1123–1127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, S.; Xie, Z.; Hu, C.; Zhang, J. A Review of Auxin Response Factors (ARFs) in Plants. Front. Plant Sci. 2016, 7, 47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spartz, A.K.; Lee, S.H.; Wenger, J.P.; Gonzalez, N.; Itoh, H.; Inze, D.; Gray, W.M. The SAUR19 subfamily of SMALL AUXIN UP RNA genes promote cell expansion. Plant J. 2012, 70, 978–990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Dewey, C. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [Green Version]
- Audic, S.; Claverie, J.M. The significance of digital gene expression profiles. Genome Res. 1997, 7, 986–995. [Google Scholar] [CrossRef]
- Ye, J.; Fang, L.; Zheng, H.; Zhang, Y.; Chen, J.; Zhang, Z. WEGO: A web tool for plotting GO annotations. Nucleic. Acids Res. 2006, 34, W293–W297. [Google Scholar] [CrossRef]

| Traits | B73 | lpa1 |
|---|---|---|
| Leaf length/cm | 84.68 ± 6.86 a | 81.43 ± 4.89 a |
| Leaf width/cm | 14.16 ± 0.29 ab | 10.20 ± 0.32 b |
| Leaf angle/° | 33.19 ± 0.44 b | 38.94 ± 3.53 a |
| High point length/cm | 62.70 ± 4.70 b | 55.49 ± 2.29 a |
| Leaf space/cm | 12.32 ± 0.16 b | 17.06 ± 0.58 a |
| Leaf orientation value | 42.11 ± 1.64 b | 34.78 ± 1.44 a |
| Plant height/cm | 235.78 ± 0.92 a | 222.66 ± 0.66 b |
| Traits | B73 | lpa1 |
|---|---|---|
| Plant height/cm | 235.78 ± 0.92 a | 222.66 ± 0.66 b |
| Ear height/cm | 131.18 ± 0.43 a | 106.26 ± 0.32 b |
| Number of tassel branches | 11.50 ± 0.41 a | 5.50 ± 0.41 b |
| Total tassel length/cm | 38.55 ± 0.04 a | 29.20 ± 0.24 b |
| Tassel branch length/cm | 27.95 ± 0.37 a | 16.60 ± 0.08 b |
| Panicle length/cm | 20.90 ± 0.33 a | 15.43 ± 0.67 b |
| Per-panicle weight/g | 213.83 ± 2.43 a | 98.08 ± 1.27 b |
| Kernel weight/g | 186.87 ± 1.28 a | 74.35 ± 0.80 b |
| Seed setting rate/% | 87.40 ± 0.39 a | 75.81 ± 0.17 b |
| Hundred-grain weight/g | 43.99 ± 0.73 a | 28.95 ± 0.65 b |
| Ear coarse/cm | 5.72 ± 0.02 a | 4.38 ± 0.06 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ji, X.; Gao, Q.; Chen, F.; Bai, M.; Zhuang, Z.; Peng, Y. Mutant lpa1 Analysis of ZmLPA1 Gene Regulates Maize Leaf-Angle Development through the Auxin Pathway. Int. J. Mol. Sci. 2022, 23, 4886. https://doi.org/10.3390/ijms23094886
Ji X, Gao Q, Chen F, Bai M, Zhuang Z, Peng Y. Mutant lpa1 Analysis of ZmLPA1 Gene Regulates Maize Leaf-Angle Development through the Auxin Pathway. International Journal of Molecular Sciences. 2022; 23(9):4886. https://doi.org/10.3390/ijms23094886
Chicago/Turabian StyleJi, Xiangzhuo, Qiaohong Gao, Fenqi Chen, Mingxing Bai, Zelong Zhuang, and Yunling Peng. 2022. "Mutant lpa1 Analysis of ZmLPA1 Gene Regulates Maize Leaf-Angle Development through the Auxin Pathway" International Journal of Molecular Sciences 23, no. 9: 4886. https://doi.org/10.3390/ijms23094886
APA StyleJi, X., Gao, Q., Chen, F., Bai, M., Zhuang, Z., & Peng, Y. (2022). Mutant lpa1 Analysis of ZmLPA1 Gene Regulates Maize Leaf-Angle Development through the Auxin Pathway. International Journal of Molecular Sciences, 23(9), 4886. https://doi.org/10.3390/ijms23094886





