A Single-Cell Raman Spectroscopy Analysis of Bone Marrow Mesenchymal Stem/Stromal Cells to Identify Inter-Individual Diversity
Abstract
1. Introduction
2. Results
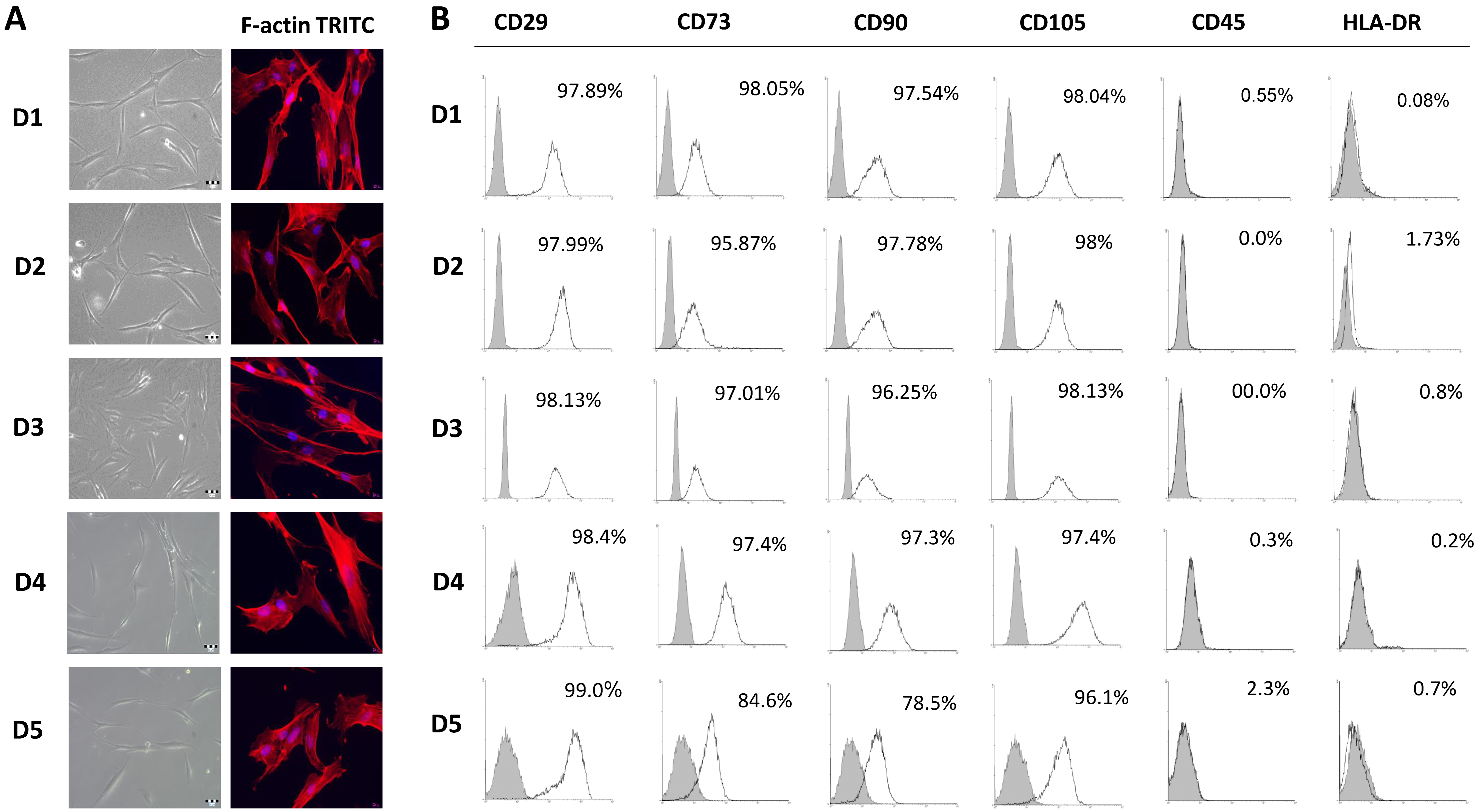
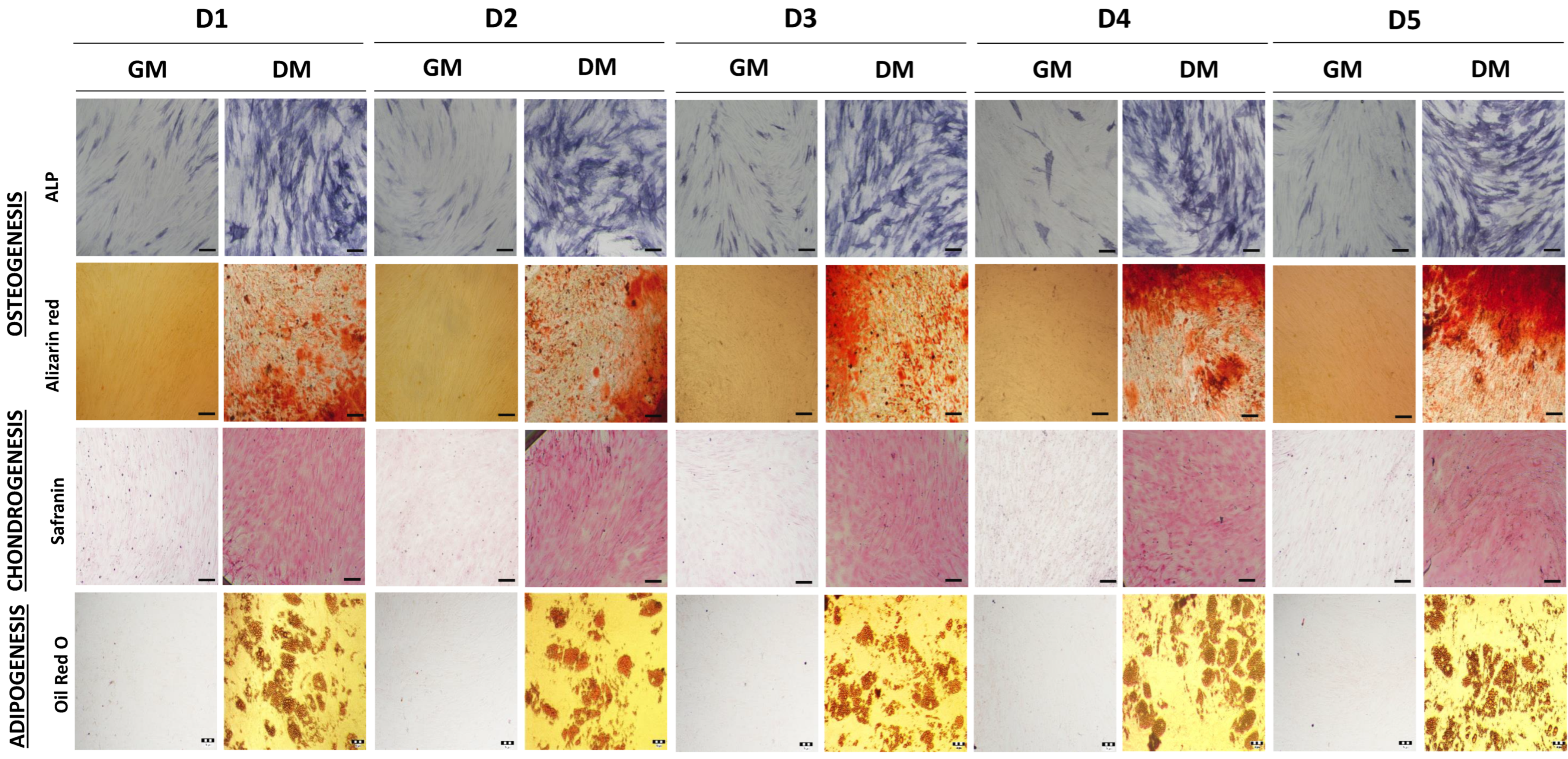
2.1. Comparison of Bone Marrow Mesenchymal Stem/Stromal Cell Features Isolated from Five Pediatric Donors
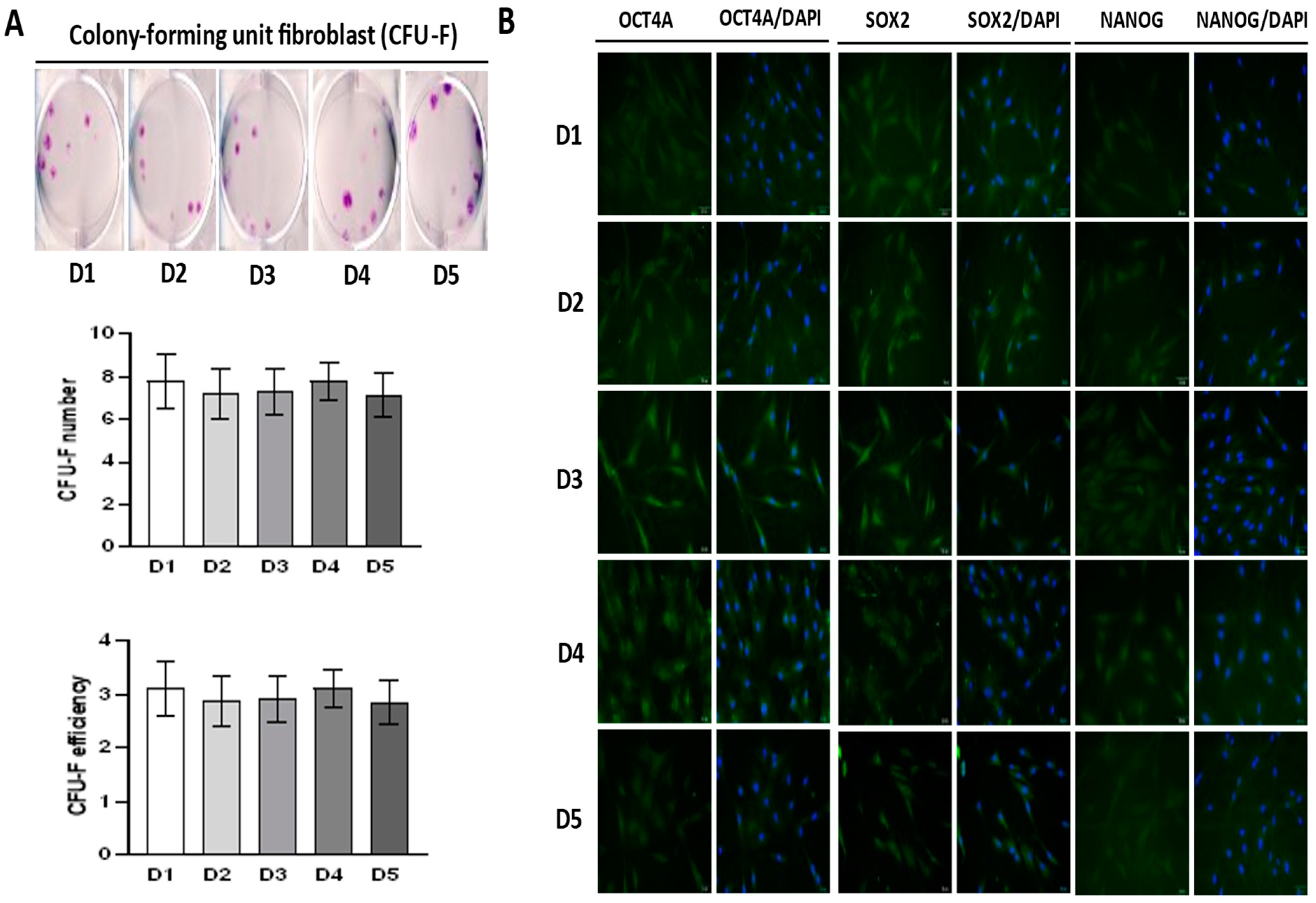
2.2. Self-Renewal of BM-MSCs and Expression of Markers Associated with Pluripotency
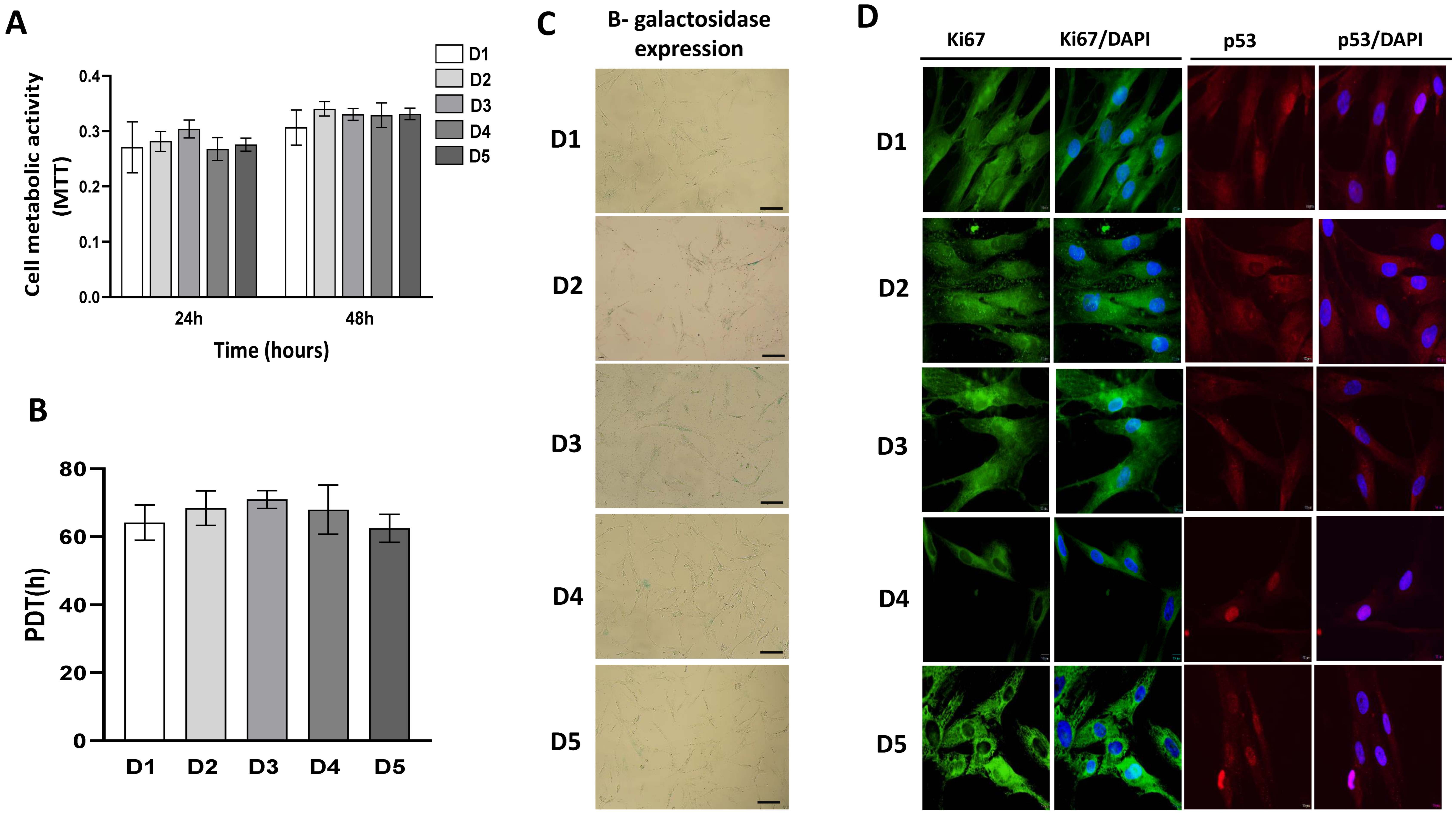
2.3. Growth Characteristics of BM-MSCs
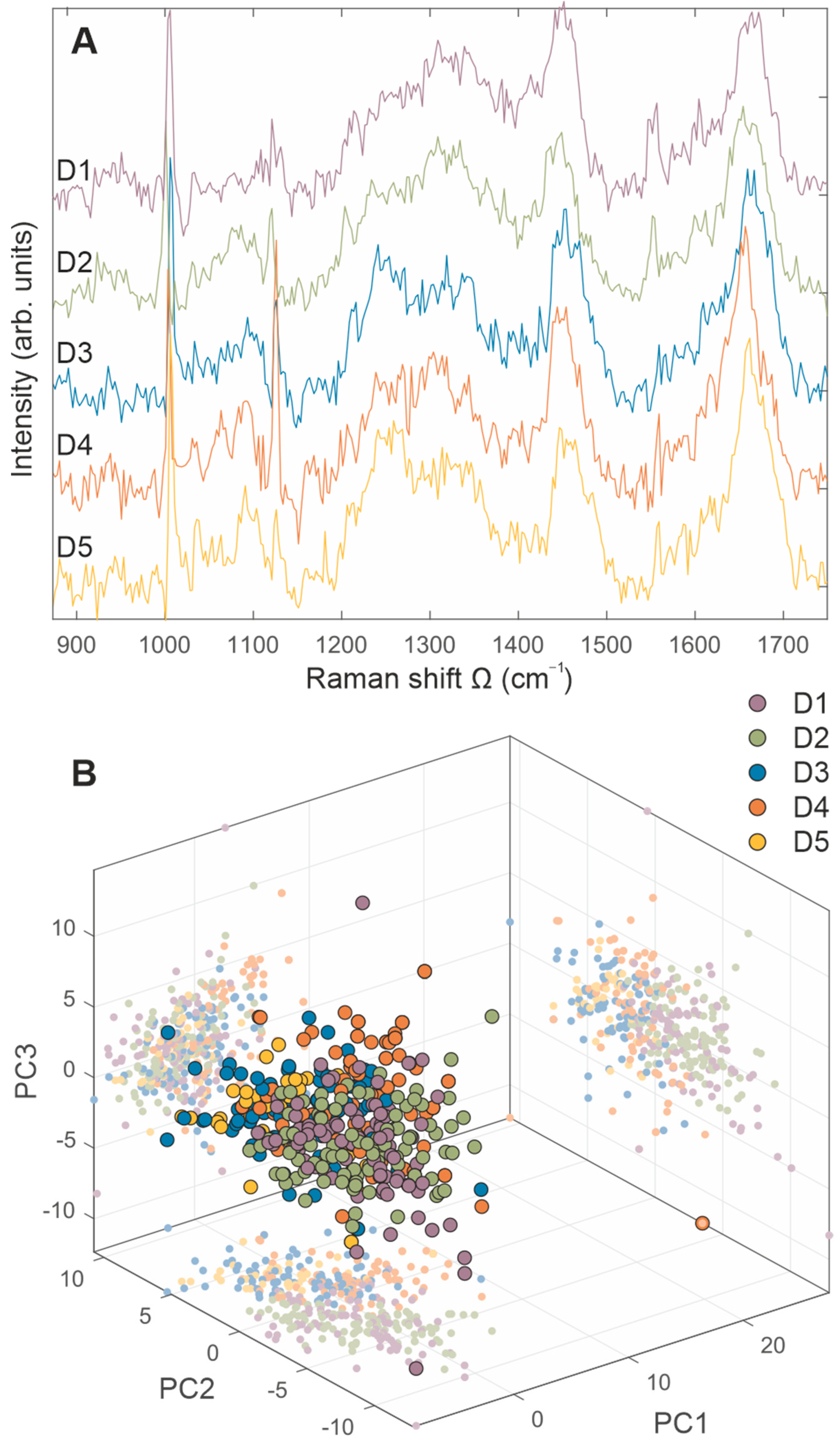
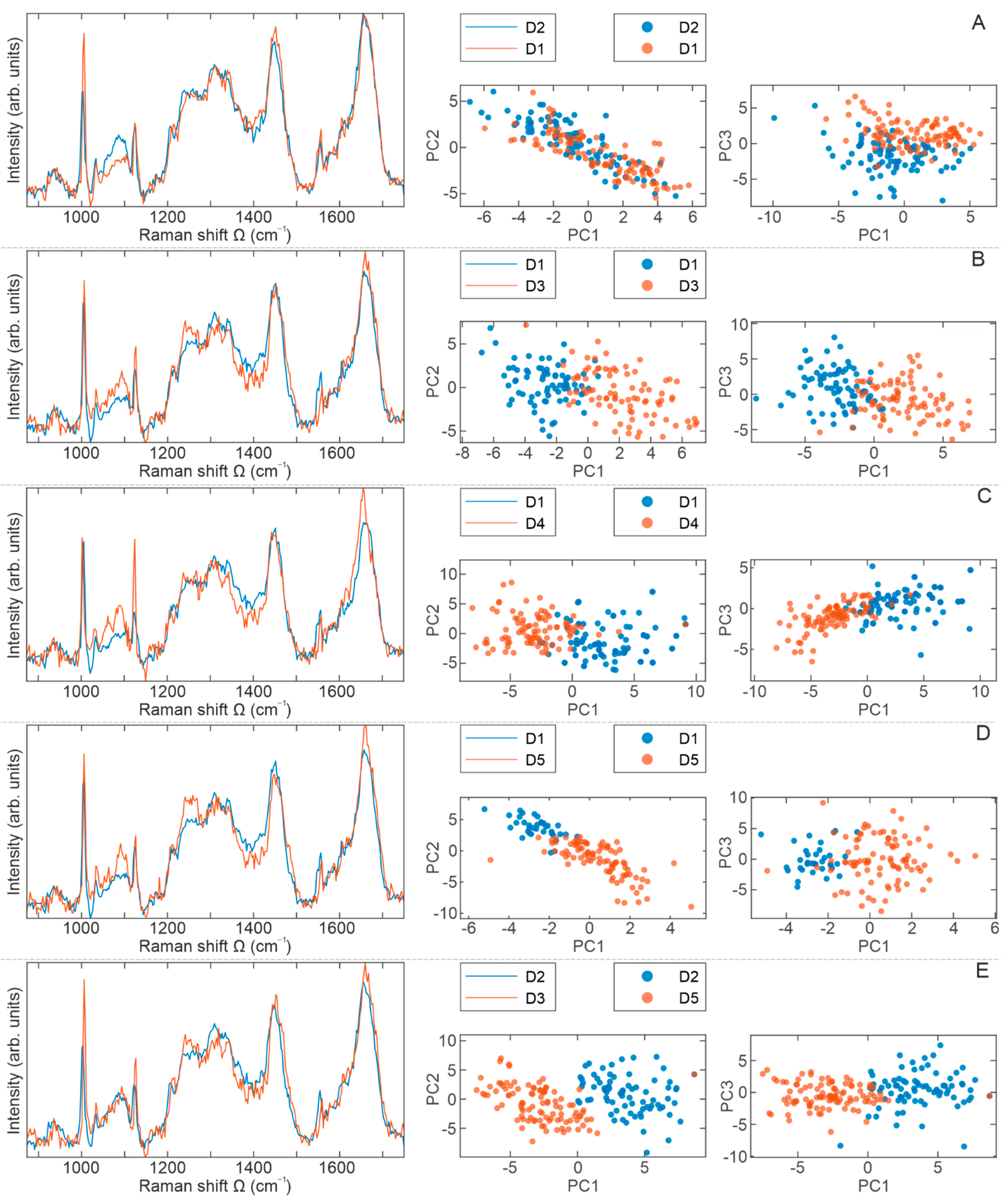
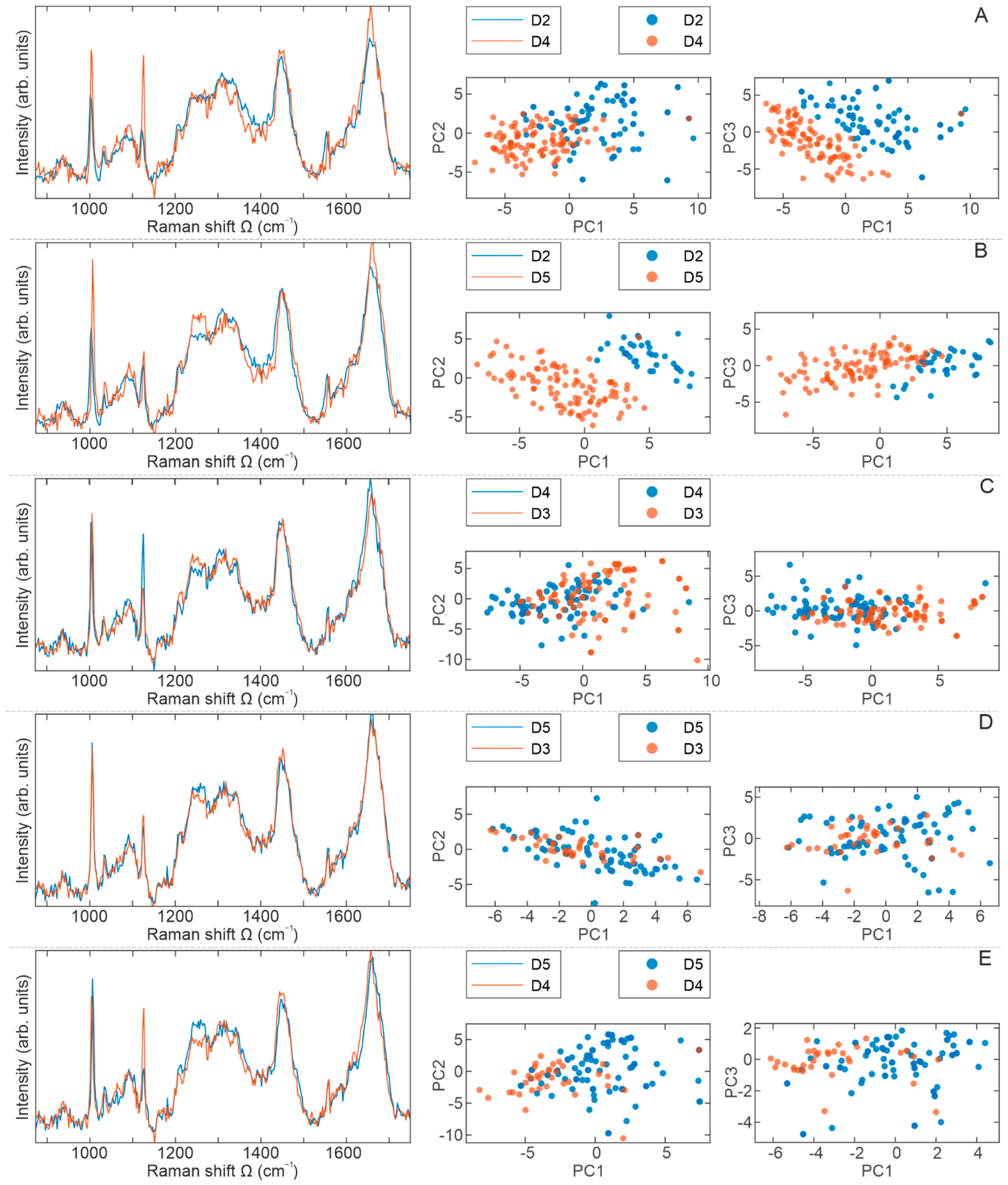
2.4. Raman Spectra Analyses
3. Discussion
4. Materials and Methods
4.1. Collection, Isolation, Expansion, and Cultivation of MSCs
4.2. Immunophenotyping
4.3. Multipotent Differentiation
4.4. CFU-F (Colony-Forming Units-Fibroblastic) Assay
4.5. Cellular Proliferation, Viability and Senescence
4.6. Immunofluorescence
4.7. Sample Preparation for Raman Experiment
4.8. µ-Raman Spectroscopy
4.9. Data Processing and Analysis
- (i)
- Removal of the irregular spectra from the datasets;
- (ii)
- Removal of the background noise from every spectrum in the data set;
- (iii)
- Spectra normalization.
4.10. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Teixeira, F.G.; Salgado, A.J. Mesenchymal Stem Cells Secretome: Current Trends and Future Challenges. Neural Regen. Res. 2020, 15, 75–77. [Google Scholar] [CrossRef]
- Zakrzewski, W.; Dobrzyński, M.; Szymonowicz, M.; Rybak, Z. Stem Cells: Past, Present, and Future. Stem Cell Res. Ther. 2019, 10, 68. [Google Scholar] [CrossRef]
- Nava, M.M.; Raimondi, M.T.; Pietrabissa, R. Controlling Self-Renewal and Differentiation of Stem Cells via Mechanical Cues. J. Biomed. Biotechnol. 2012, 2012, 797410. [Google Scholar] [CrossRef]
- Wei, X.; Yang, X.; Han, Z.; Qu, F.; Shao, L.; Shi, Y. Mesenchymal Stem Cells: A New Trend for Cell Therapy. Acta Pharmacol. Sin. 2013, 34, 747–754. [Google Scholar] [CrossRef]
- Caplan, A.I. Mesenchymal Stem Cells: Time to Change the Name! Stem Cells Transl. Med. 2017, 6, 1445–1451. [Google Scholar] [CrossRef]
- Harrell, C.R.; Fellabaum, C.; Jovicic, N.; Djonov, V.; Arsenijevic, N.; Volarevic, V. Molecular Mechanisms Responsible for Therapeutic Potential of Mesenchymal Stem Cell-Derived Secretome. Cells 2019, 8, 467. [Google Scholar] [CrossRef]
- Pittenger, M.F.; Discher, D.E.; Péault, B.M.; Phinney, D.G.; Hare, J.M.; Caplan, A.I. Mesenchymal Stem Cell Perspective: Cell Biology to Clinical Progress. NPJ Regen. Med. 2019, 4, 22. [Google Scholar] [CrossRef]
- Musiał-Wysocka, A.; Kot, M.; Majka, M. The Pros and Cons of Mesenchymal Stem Cell-Based Therapies. Cell Transpl. 2019, 28, 801–812. [Google Scholar] [CrossRef]
- Yin, P.T.; Han, E.; Lee, K.-B. Engineering Stem Cells for Biomedical Applications. Adv. Health. Mater. 2016, 5, 10–55. [Google Scholar] [CrossRef]
- Wilson, A.; Hodgson-Garms, M.; Frith, J.E.; Genever, P. Multiplicity of Mesenchymal Stromal Cells: Finding the Right Route to Therapy. Front. Immunol. 2019, 10, 1112. [Google Scholar] [CrossRef]
- Lv, F.-J.; Tuan, R.S.; Cheung, K.M.C.; Leung, V.Y.L. Concise Review: The Surface Markers and Identity of Human Mesenchymal Stem Cells. Stem Cells 2014, 32, 1408–1419. [Google Scholar] [CrossRef]
- Mo, M.; Wang, S.; Zhou, Y.; Li, H.; Wu, Y. Mesenchymal Stem Cell Subpopulations: Phenotype, Property and Therapeutic Potential. Cell. Mol. Life Sci. 2016, 73, 3311–3321. [Google Scholar] [CrossRef]
- Phinney, D.G. Functional Heterogeneity of Mesenchymal Stem Cells: Implications for Cell Therapy. J. Cell. Biochem. 2012, 113, 2806–2812. [Google Scholar] [CrossRef]
- Dominici, M.; Le Blanc, K.; Mueller, I.; Slaper-Cortenbach, I.; Marini, F.; Krause, D.; Deans, R.; Keating, A.; Prockop, D.; Horwitz, E. Minimal Criteria for Defining Multipotent Mesenchymal Stromal Cells. The International Society for Cellular Therapy Position Statement. Cytotherapy 2006, 8, 315–317. [Google Scholar] [CrossRef]
- Ullah, I.; Subbarao, R.B.; Rho, G.J. Human Mesenchymal Stem Cells—Current Trends and Future Prospective. Biosci. Rep. 2015, 35, e00191. [Google Scholar] [CrossRef]
- Chen, J.-Y.; Mou, X.-Z.; Du, X.-C.; Xiang, C. Comparative Analysis of Biological Characteristics of Adult Mesenchymal Stem Cells with Different Tissue Origins. Asian Pac. J. Trop. Med. 2015, 8, 739–746. [Google Scholar] [CrossRef]
- Guan, Y.-T.; Xie, Y.; Li, D.-S.; Zhu, Y.-Y.; Zhang, X.-L.; Feng, Y.-L.; Chen, Y.-P.; Xu, L.-J.; Liao, P.-F.; Wang, G. Comparison of Biological Characteristics of Mesenchymal Stem Cells Derived from the Human Umbilical Cord and Decidua Parietalis. Mol. Med. Rep. 2019, 20, 633–639. [Google Scholar] [CrossRef]
- Sangeetha, K.N.; Vennila, R.; Secunda, R.; Sakthivel, S.; Pathak, S.; Jeswanth, S.; Surendran, R. Functional Variations between Mesenchymal Stem Cells of Different Tissue Origins: A Comparative Gene Expression Profiling. Biotechnol. Lett. 2020, 42, 1287–1304. [Google Scholar] [CrossRef]
- Xu, L.; Liu, Y.; Sun, Y.; Wang, B.; Xiong, Y.; Lin, W.; Wei, Q.; Wang, H.; He, W.; Wang, B.; et al. Tissue Source Determines the Differentiation Potentials of Mesenchymal Stem Cells: A Comparative Study of Human Mesenchymal Stem Cells from Bone Marrow and Adipose Tissue. Stem Cell Res. Ther. 2017, 8, 275. [Google Scholar] [CrossRef]
- Costa, L.A.; Eiro, N.; Fraile, M.; Gonzalez, L.O.; Saá, J.; Garcia-Portabella, P.; Vega, B.; Schneider, J.; Vizoso, F.J. Functional Heterogeneity of Mesenchymal Stem Cells from Natural Niches to Culture Conditions: Implications for Further Clinical Uses. Cell. Mol. Life Sci. 2021, 78, 447–467. [Google Scholar] [CrossRef]
- Lähnemann, D.; Köster, J.; Szczurek, E.; McCarthy, D.J.; Hicks, S.C.; Robinson, M.D.; Vallejos, C.A.; Campbell, K.R.; Beerenwinkel, N.; Mahfouz, A.; et al. Eleven Grand Challenges in Single-Cell Data Science. Genome Biol. 2020, 21, 31. [Google Scholar] [CrossRef]
- Yuan, G.-C.; Cai, L.; Elowitz, M.; Enver, T.; Fan, G.; Guo, G.; Irizarry, R.; Kharchenko, P.; Kim, J.; Orkin, S.; et al. Challenges and Emerging Directions in Single-Cell Analysis. Genome Biol. 2017, 18, 84. [Google Scholar] [CrossRef]
- Zheng, G.; Xie, Z.-Y.; Wang, P.; Wu, Y.-F.; Shen, H.-Y. Recent Advances of Single-Cell RNA Sequencing Technology in Mesenchymal Stem Cell Research. World J. Stem Cells 2020, 12, 438–447. [Google Scholar] [CrossRef]
- Ember, K.J.I.; Hoeve, M.A.; McAughtrie, S.L.; Bergholt, M.S.; Dwyer, B.J.; Stevens, M.M.; Faulds, K.; Forbes, S.J.; Campbell, C.J. Raman Spectroscopy and Regenerative Medicine: A Review. NPJ Regen. Med. 2017, 2, 12. [Google Scholar] [CrossRef]
- García-Timermans, C.; Props, R.; Zacchetti, B.; Sakarika, M.; Delvigne, F.; Boon, N. Raman Spectroscopy-Based Measurements of Single-Cell Phenotypic Diversity in Microbial Populations. mSphere 2020, 5, e00806-20. [Google Scholar] [CrossRef]
- Downes, A.; Mouras, R.; Bagnaninchi, P.; Elfick, A. Raman Spectroscopy and CARS Microscopy of Stem Cells and Their Derivatives. J. Raman Spectrosc. 2011, 42, 1864–1870. [Google Scholar] [CrossRef]
- Bai, H.; Chen, P.; Fang, H.; Lin, L.; Tang, G.Q.; Mu, G.G.; Gong, W.; Liu, Z.P.; Wu, H.; Zhao, H.; et al. Detecting Viability Transitions of Umbilical Cord Mesenchymal Stem Cells by Raman Micro-Spectroscopy. Laser Phys. Lett. 2010, 8, 78. [Google Scholar] [CrossRef]
- Azrad, E.; Zahor, D.; Vago, R.; Nevo, Z.; Doron, R.; Robinson, D.; Gheber, L.A.; Rosenwaks, S.; Bar, I. Probing the Effect of an Extract of Elk Velvet Antler Powder on Mesenchymal Stem Cells Using Raman Microspectroscopy: Enhanced Differentiation toward Osteogenic Fate. J. Raman Spectrosc. 2006, 37, 480–486. [Google Scholar] [CrossRef]
- Molony, C.; McIntyre, J.; Maguire, A.; Hakimjavadi, R.; Burtenshaw, D.; Casey, G.; di Luca, M.; Hennelly, B.; Byrne, H.J.; Cahill, P.A. Label-Free Discrimination Analysis of de-Differentiated Vascular Smooth Muscle Cells, Mesenchymal Stem Cells and Their Vascular and Osteogenic Progeny Using Vibrational Spectroscopy. Biochim. Biophys. Acta Mol. Cell Res. 2018, 1865, 343–353. [Google Scholar] [CrossRef]
- Butler, H.J.; Ashton, L.; Bird, B.; Cinque, G.; Curtis, K.; Dorney, J.; Esmonde-White, K.; Fullwood, N.J.; Gardner, B.; Martin-Hirsch, P.L.; et al. Using Raman Spectroscopy to Characterize Biological Materials. Nat. Protoc. 2016, 11, 664–687. [Google Scholar] [CrossRef]
- Rangan, S.; Schulze, H.G.; Vardaki, M.Z.; Blades, M.W.; Piret, J.M.; Turner, R.F.B. Applications of Raman Spectroscopy in the Development of Cell Therapies: State of the Art and Future Perspectives. Analyst 2020, 145, 2070–2105. [Google Scholar] [CrossRef]
- Geng, J.; Zhang, W.; Chen, C.; Zhang, H.; Zhou, A.; Huang, Y. Tracking the Differentiation Status of Human Neural Stem Cells through Label-Free Raman Spectroscopy and Machine Learning-Based Analysis. Anal. Chem. 2021, 93, 10453–10461. [Google Scholar] [CrossRef]
- Wang, J.; Qi, G.; Qu, X.; Ling, X.; Zhang, Z.; Jin, Y. Molecular Profiling of Dental Pulp Stem Cells during Cell Differentiation by Surface Enhanced Raman Spectroscopy. Anal. Chem. 2020, 92, 3735–3741. [Google Scholar] [CrossRef]
- Hsu, C.-C.; Xu, J.; Brinkhof, B.; Wang, H.; Cui, Z.; Huang, W.E.; Ye, H. A Single-Cell Raman-Based Platform to Identify Developmental Stages of Human Pluripotent Stem Cell-Derived Neurons. Proc. Natl. Acad. Sci. USA 2020, 117, 18412–18423. [Google Scholar] [CrossRef]
- Lazarević, J.J.; Kukolj, T.; Bugarski, D.; Lazarević, N.; Bugarski, B.; Popović, Z.V. Probing Primary Mesenchymal Stem Cells Differentiation Status by Micro-Raman Spectroscopy. Spectrochim. Acta. A Mol. Biomol. Spectrosc. 2019, 213, 384–390. [Google Scholar] [CrossRef]
- Notingher, I.; Bisson, I.; Bishop, A.E.; Randle, W.L.; Polak, J.M.P.; Hench, L.L. In Situ Spectral Monitoring of MRNA Translation in Embryonic Stem Cells during Differentiation In Vitro. Anal. Chem. 2004, 76, 3185–3193. [Google Scholar] [CrossRef]
- Ghita, A.; Pascut, F.C.; Sottile, V.; Denning, C.; Notingher, I. Applications of Raman Micro-Spectroscopy to Stem Cell Technology: Label-Free Molecular Discrimination and Monitoring Cell Differentiation. EPJ Tech. Instrum. 2015, 2, 6. [Google Scholar] [CrossRef]
- Ravera, F.; Efeoglu, E.; Byrne, H.J. Vibrational Spectroscopy for In Vitro Monitoring Stem Cell Differentiation. Mol. Basel Switz. 2020, 25, 5554. [Google Scholar] [CrossRef]
- Carvalho, M.S.; Silva, J.C.; Hoff, C.M.; Cabral, J.M.S.; Linhardt, R.J.; da Silva, C.L.; Vashishth, D. Loss and Rescue of Osteocalcin and Osteopontin Modulate Osteogenic and Angiogenic Features of Mesenchymal Stem/Stromal Cells. J. Cell. Physiol. 2020, 235, 7496–7515. [Google Scholar] [CrossRef]
- Wiercigroch, E.; Szafraniec, E.; Czamara, K.; Pacia, M.Z.; Majzner, K.; Kochan, K.; Kaczor, A.; Baranska, M.; Malek, K. Raman and Infrared Spectroscopy of Carbohydrates: A Review. Spectrochim. Acta. A Mol. Biomol. Spectrosc. 2017, 185, 317–335. [Google Scholar] [CrossRef]
- Clemens, G.; Hands, J.R.; Dorling, K.M.; Baker, M.J. Vibrational Spectroscopic Methods for Cytology and Cellular Research. Analyst 2014, 139, 4411–4444. [Google Scholar] [CrossRef]
- Czamara, K.; Majzner, K.; Pacia, M.Z.; Kochan, K.; Kaczor, A.; Baranska, M. Raman Spectroscopy of Lipids: A Review. J. Raman Spectrosc. 2015, 46, 4–20. [Google Scholar] [CrossRef]
- de Gelder, J.; de Gussem, K.; Vandenabeele, P.; Moens, L. Reference Database of Raman Spectra of Biological Molecules. J. Raman Spectrosc. 2007, 38, 1133–1147. [Google Scholar] [CrossRef]
- Pezzotti, G.; Zhu, W.; Adachi, T.; Horiguchi, S.; Marin, E.; Boschetto, F.; Ogitani, E.; Mazda, O. Metabolic Machinery Encrypted in the Raman Spectrum of Influenza A Virus-Inoculated Mammalian Cells. J. Cell. Physiol. 2020, 235, 5146–5170. [Google Scholar] [CrossRef]
- Baker, M.J.; Byrne, H.J.; Chalmers, J.; Gardner, P.; Goodacre, R.; Henderson, A.; Kazarian, S.G.; Martin, F.L.; Moger, J.; Stone, N.; et al. Clinical Applications of Infrared and Raman Spectroscopy: State of Play and Future Challenges. Analyst 2018, 143, 1735–1757. [Google Scholar] [CrossRef]
- Tanwar, S.; Paidi, S.K.; Prasad, R.; Pandey, R.; Barman, I. Advancing Raman Spectroscopy from Research to Clinic: Translational Potential and Challenges. Spectrochim. Acta. A Mol. Biomol. Spectrosc. 2021, 260, 119957. [Google Scholar] [CrossRef]
- Crow, P.; Barrass, B.; Kendall, C.; Hart-Prieto, M.; Wright, M.; Persad, R.; Stone, N. The Use of Raman Spectroscopy to Differentiate between Different Prostatic Adenocarcinoma Cell Lines. Br. J. Cancer 2005, 92, 2166–2170. [Google Scholar] [CrossRef]
- Ilin, Y.; Kraft, M.L. Identifying the Lineages of Individual Cells in Cocultures by Multivariate Analysis of Raman Spectra. Analyst 2014, 139, 2177–2185. [Google Scholar] [CrossRef]
- Surmacki, J.M.; Woodhams, B.J.; Haslehurst, A.; Ponder, B.A.J.; Bohndiek, S.E. Raman Micro-Spectroscopy for Accurate Identification of Primary Human Bronchial Epithelial Cells. Sci. Rep. 2018, 8, 12604. [Google Scholar] [CrossRef]
- Pijanka, J.K.; Kumar, D.; Dale, T.; Yousef, I.; Parkes, G.; Untereiner, V.; Yang, Y.; Dumas, P.; Collins, D.; Manfait, M.; et al. Vibrational Spectroscopy Differentiates between Multipotent and Pluripotent Stem Cells. Analyst 2010, 135, 3126–3132. [Google Scholar] [CrossRef]
- Pudlas, M.; Carvajal Berrio, D.; Linneweh, M.; Koch, S.; Thude, S.; Walles, H.; Schenke-Layland, K. Non-Contact Discrimination of Human Bone Marrow-Derived Mesenchymal Stem Cells and Fibroblasts Using Raman Spectroscopy. Med. Laser Appl. 2011, 26, 119–125. [Google Scholar] [CrossRef]
- Simonović, J.; Toljić, B.; Rašković, B.; Jovanović, V.; Lazarević, M.; Milošević, M.; Nikolić, N.; Panajotović, R.; Milašin, J. Raman Microspectroscopy: Toward a Better Distinction and Profiling of Different Populations of Dental Stem Cells. Croat. Med. J. 2019, 60, 78–86. [Google Scholar] [CrossRef] [PubMed]
- Baghaei, K.; Hashemi, S.M.; Tokhanbigli, S.; Asadi Rad, A.; Assadzadeh-Aghdaei, H.; Sharifian, A.; Zali, M.R. Isolation, Differentiation, and Characterization of Mesenchymal Stem Cells from Human Bone Marrow. Gastroenterol. Hepatol. Bed Bench 2017, 10, 208–213. [Google Scholar] [PubMed]
- Mennan, C.; Garcia, J.; Roberts, S.; Hulme, C.; Wright, K. A Comprehensive Characterisation of Large-Scale Expanded Human Bone Marrow and Umbilical Cord Mesenchymal Stem Cells. Stem Cell Res. Ther. 2019, 10, 99. [Google Scholar] [CrossRef]
- Bhat, S.; Viswanathan, P.; Chandanala, S.; Prasanna, S.J.; Seetharam, R.N. Expansion and Characterization of Bone Marrow Derived Human Mesenchymal Stromal Cells in Serum-Free Conditions. Sci. Rep. 2021, 11, 3403. [Google Scholar] [CrossRef]
- Yang, Y.-H.K.; Ogando, C.R.; Wang See, C.; Chang, T.-Y.; Barabino, G.A. Changes in Phenotype and Differentiation Potential of Human Mesenchymal Stem Cells Aging in Vitro. Stem Cell Res. Ther. 2018, 9, 131. [Google Scholar] [CrossRef]
- Song, Y.; Zhao, H.-Y.; Lyu, Z.-S.; Cao, X.-N.; Shi, M.-M.; Wen, Q.; Tang, F.-F.; Wang, Y.; Xu, L.-P.; Zhang, X.-H.; et al. Dysfunctional Bone Marrow Mesenchymal Stem Cells in Patients with Poor Graft Function after Allogeneic Hematopoietic Stem Cell Transplantation. Biol. Blood Marrow Transplant. 2018, 24, 1981–1989. [Google Scholar] [CrossRef]
- Zhou, Y.; Chen, H.; Li, H.; Wu, Y. 3D Culture Increases Pluripotent Gene Expression in Mesenchymal Stem Cells through Relaxation of Cytoskeleton Tension. J. Cell. Mol. Med. 2017, 21, 1073–1084. [Google Scholar] [CrossRef]
- Petrenko, Y.; Vackova, I.; Kekulova, K.; Chudickova, M.; Koci, Z.; Turnovcova, K.; Kupcova Skalnikova, H.; Vodicka, P.; Kubinova, S. A Comparative Analysis of Multipotent Mesenchymal Stromal Cells Derived from Different Sources, with a Focus on Neuroregenerative Potential. Sci. Rep. 2020, 10, 4290. [Google Scholar] [CrossRef]
- Mareschi, K.; Ferrero, I.; Rustichelli, D.; Aschero, S.; Gammaitoni, L.; Aglietta, M.; Madon, E.; Fagioli, F. Expansion of Mesenchymal Stem Cells Isolated from Pediatric and Adult Donor Bone Marrow. J. Cell. Biochem. 2006, 97, 744–754. [Google Scholar] [CrossRef]
- Kern, S.; Eichler, H.; Stoeve, J.; Klüter, H.; Bieback, K. Comparative Analysis of Mesenchymal Stem Cells from Bone Marrow, Umbilical Cord Blood, or Adipose Tissue. Stem Cells Dayt. Ohio 2006, 24, 1294–1301. [Google Scholar] [CrossRef] [PubMed]
- Araújo, A.B.; Salton, G.D.; Furlan, J.M.; Schneider, N.; Angeli, M.H.; Laureano, Á.M.; Silla, L.; Passos, E.P.; Paz, A.H. Comparison of Human Mesenchymal Stromal Cells from Four Neonatal Tissues: Amniotic Membrane, Chorionic Membrane, Placental Decidua and Umbilical Cord. Cytotherapy 2017, 19, 577–585. [Google Scholar] [CrossRef]
- Ghaneialvar, H.; Soltani, L.; Rahmani, H.R.; Lotfi, A.S.; Soleimani, M. Characterization and Classification of Mesenchymal Stem Cells in Several Species Using Surface Markers for Cell Therapy Purposes. Indian J. Clin. Biochem. 2018, 33, 46–52. [Google Scholar] [CrossRef]
- Pittenger, M.F.; Mackay, A.M.; Beck, S.C.; Jaiswal, R.K.; Douglas, R.; Mosca, J.D.; Moorman, M.A.; Simonetti, D.W.; Craig, S.; Marshak, D.R. Multilineage Potential of Adult Human Mesenchymal Stem Cells. Science 1999, 284, 143–147. [Google Scholar] [CrossRef] [PubMed]
- Meppelink, A.M.; Wang, X.-H.; Bradica, G.; Barron, K.; Hiltz, K.; Liu, X.-H.; Goldman, S.M.; Vacanti, J.P.; Keating, A.; Hoganson, D.M. Rapid Isolation of Bone Marrow Mesenchymal Stromal Cells Using Integrated Centrifuge-Based Technology. Cytotherapy 2016, 18, 729–739. [Google Scholar] [CrossRef] [PubMed]
- Meuleman, N.; Tondreau, T.; Delforge, A.; Dejeneffe, M.; Massy, M.; Libertalis, M.; Bron, D.; Lagneaux, L. Human Marrow Mesenchymal Stem Cell Culture: Serum-Free Medium Allows Better Expansion than Classical Alpha-MEM Medium. Eur. J. Haematol. 2006, 76, 309–316. [Google Scholar] [CrossRef] [PubMed]
- Dmitrieva, R.I.; Minullina, I.R.; Bilibina, A.A.; Tarasova, O.V.; Anisimov, S.V.; Zaritskey, A.Y. Bone Marrow- and Subcutaneous Adipose Tissue-Derived Mesenchymal Stem Cells: Differences and Similarities. Cell Cycle Georget. Tex. 2012, 11, 377–383. [Google Scholar] [CrossRef]
- Vishnubalaji, R.; Al-Nbaheen, M.; Kadalmani, B.; Aldahmash, A.; Ramesh, T. Comparative Investigation of the Differentiation Capability of Bone-Marrow- and Adipose-Derived Mesenchymal Stem Cells by Qualitative and Quantitative Analysis. Cell Tissue Res. 2012, 347, 419–427. [Google Scholar] [CrossRef]
- Alcayaga-Miranda, F.; Cuenca, J.; Luz-Crawford, P.; Aguila-Díaz, C.; Fernandez, A.; Figueroa, F.E.; Khoury, M. Characterization of Menstrual Stem Cells: Angiogenic Effect, Migration and Hematopoietic Stem Cell Support in Comparison with Bone Marrow Mesenchymal Stem Cells. Stem Cell Res. Ther. 2015, 6, 32. [Google Scholar] [CrossRef]
- Kolf, C.M.; Cho, E.; Tuan, R.S. Mesenchymal Stromal Cells. Biology of Adult Mesenchymal Stem Cells: Regulation of Niche, Self-Renewal and Differentiation. Arthritis Res. Ther. 2007, 9, 204. [Google Scholar] [CrossRef][Green Version]
- Boyer, L.A.; Lee, T.I.; Cole, M.F.; Johnstone, S.E.; Levine, S.S.; Zucker, J.P.; Guenther, M.G.; Kumar, R.M.; Murray, H.L.; Jenner, R.G.; et al. Core Transcriptional Regulatory Circuitry in Human Embryonic Stem Cells. Cell 2005, 122, 947–956. [Google Scholar] [CrossRef] [PubMed]
- Riekstina, U.; Cakstina, I.; Parfejevs, V.; Hoogduijn, M.; Jankovskis, G.; Muiznieks, I.; Muceniece, R.; Ancans, J. Embryonic Stem Cell Marker Expression Pattern in Human Mesenchymal Stem Cells Derived from Bone Marrow, Adipose Tissue, Heart and Dermis. Stem Cell Rev. Rep. 2009, 5, 378–386. [Google Scholar] [CrossRef] [PubMed]
- Miletić, M.; Mojsilović, S.; Okić-Đorđević, I.; Kukolj, T.; Jauković, A.; Santibacez, J.F.; Jovčić, G.; Bugarski, D. Mesenchymal Stem Cells Isolated from Human Periodontal Ligament. Arch. Biol. Sci. 2014, 66, 261–271. [Google Scholar] [CrossRef]
- Tantrawatpan, C.; Manochantr, S.; Kheolamai, P.; U-Pratya, Y.; Supokawej, A.; Issaragrisil, S. Pluripotent Gene Expression in Mesenchymal Stem Cells from Human Umbilical Cord Wharton’s Jelly and Their Differentiation Potential to Neural-like Cells. J. Med. Assoc. Thail. Chotmaihet Thangphaet 2013, 96, 1208–1217. [Google Scholar]
- Park, E.; Patel, A.N. Changes in the Expression Pattern of Mesenchymal and Pluripotent Markers in Human Adipose-Derived Stem Cells. Cell Biol. Int. 2010, 34, 979–984. [Google Scholar] [CrossRef]
- Jaramillo-Ferrada, P.A.; Wolvetang, E.J.; Cooper-White, J.J. Differential Mesengenic Potential and Expression of Stem Cell-Fate Modulators in Mesenchymal Stromal Cells from Human-Term Placenta and Bone Marrow. J. Cell. Physiol. 2012, 227, 3234–3242. [Google Scholar] [CrossRef]
- Gao, L.R.; Zhang, N.K.; Ding, Q.A.; Chen, H.Y.; Hu, X.; Jiang, S.; Li, T.C.; Chen, Y.; Wang, Z.G.; Ye, Y.; et al. Common Expression of Stemness Molecular Markers and Early Cardiac Transcription Factors in Human Wharton’s Jelly-Derived Mesenchymal Stem Cells and Embryonic Stem Cells. Cell Transplant. 2013, 22, 1883–1900. [Google Scholar] [CrossRef]
- Kukolj, T.; Trivanović, D.; Mojsilović, S.; Okić Djordjević, I.; Obradović, H.; Krstić, J.; Jauković, A.; Bugarski, D. IL-33 Guides Osteogenesis and Increases Proliferation and Pluripotency Marker Expression in Dental Stem Cells. Cell Prolif. 2019, 52, e12533. [Google Scholar] [CrossRef]
- Jauković, A.; Kukolj, T.; Trivanović, D.; Okić-Đorđević, I.; Obradović, H.; Miletić, M.; Petrović, V.; Mojsilović, S.; Bugarski, D. Modulating Stemness of Mesenchymal Stem Cells from Exfoliated Deciduous and Permanent Teeth by IL-17 and BFGF. J. Cell. Physiol. 2021, 236, 7322–7341. [Google Scholar] [CrossRef]
- Han, S.-M.; Han, S.-H.; Coh, Y.-R.; Jang, G.; Chan Ra, J.; Kang, S.-K.; Lee, H.-W.; Youn, H.-Y. Enhanced Proliferation and Differentiation of Oct4- and Sox2-Overexpressing Human Adipose Tissue Mesenchymal Stem Cells. Exp. Mol. Med. 2014, 46, e101. [Google Scholar] [CrossRef]
- Piccinato, C.A.; Sertie, A.L.; Torres, N.; Ferretti, M.; Antonioli, E. High OCT4 and Low P16(INK4A) Expressions Determine In Vitro Lifespan of Mesenchymal Stem Cells. Stem Cells Int. 2015, 2015, 369828. [Google Scholar] [CrossRef] [PubMed]
- Pitrone, M.; Pizzolanti, G.; Tomasello, L.; Coppola, A.; Morini, L.; Pantuso, G.; Ficarella, R.; Guarnotta, V.; Perrini, S.; Giorgino, F.; et al. NANOG Plays a Hierarchical Role in the Transcription Network Regulating the Pluripotency and Plasticity of Adipose Tissue-Derived Stem Cells. Int. J. Mol. Sci. 2017, 18, 1107. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Qu, H.; Qi, D.; Xu, W.; Liu, S.; Jin, X.; Song, P.; Guo, Y.; Jia, Y.; Wang, X.; et al. OCT4 Maintains Self-Renewal and Reverses Senescence in Human Hair Follicle Mesenchymal Stem Cells through the Downregulation of P21 by DNA Methyltransferases. Stem Cell Res. Ther. 2019, 10, 28. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Bai, J.; Ji, X.; Li, R.; Xuan, Y.; Wang, Y. Comprehensive Characterization of Four Different Populations of Human Mesenchymal Stem Cells as Regards Their Immune Properties, Proliferation and Differentiation. Int. J. Mol. Med. 2014, 34, 695–704. [Google Scholar] [CrossRef] [PubMed]
- Kundrotas, G.; Gasperskaja, E.; Slapsyte, G.; Gudleviciene, Z.; Krasko, J.; Stumbryte, A.; Liudkeviciene, R. Identity, Proliferation Capacity, Genomic Stability and Novel Senescence Markers of Mesenchymal Stem Cells Isolated from Low Volume of Human Bone Marrow. Oncotarget 2016, 7, 10788–10802. [Google Scholar] [CrossRef] [PubMed]
- Nawrocka, D.; Kornicka, K.; Szydlarska, J.; Marycz, K. Basic Fibroblast Growth Factor Inhibits Apoptosis and Promotes Proliferation of Adipose-Derived Mesenchymal Stromal Cells Isolated from Patients with Type 2 Diabetes by Reducing Cellular Oxidative Stress. Oxid. Med. Cell. Longev. 2017, 2017, 3027109. [Google Scholar] [CrossRef]
- Remnant, L.; Kochanova, N.Y.; Reid, C.; Cisneros-Soberanis, F.; Earnshaw, W.C. The Intrinsically Disorderly Story of Ki-67. Open Biol. 2021, 11, 210120. [Google Scholar] [CrossRef]
- Sun, X.; Kaufman, P.D. Ki-67: More than a Proliferation Marker. Chromosoma 2018, 127, 175–186. [Google Scholar] [CrossRef]
- Sales Gil, R.; Vagnarelli, P. Ki-67: More Hidden behind a “Classic Proliferation Marker”. Trends Biochem. Sci. 2018, 43, 747–748. [Google Scholar] [CrossRef]
- Chierico, L.; Rizzello, L.; Guan, L.; Joseph, A.S.; Lewis, A.; Battaglia, G. The Role of the Two Splice Variants and Extranuclear Pathway on Ki-67 Regulation in Non-Cancer and Cancer Cells. PLoS ONE 2017, 12, e0171815. [Google Scholar] [CrossRef]
- Hafner, A.; Bulyk, M.L.; Jambhekar, A.; Lahav, G. The Multiple Mechanisms That Regulate P53 Activity and Cell Fate. Nat. Rev. Mol. Cell Biol. 2019, 20, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Xia, C.; Jiang, T.; Wang, Y.; Chen, X.; Hu, Y.; Gao, Y. The P53/MiR-145a Axis Promotes Cellular Senescence and Inhibits Osteogenic Differentiation by Targeting Cbfb in Mesenchymal Stem Cells. Front. Endocrinol. 2020, 11, 609186. [Google Scholar] [CrossRef] [PubMed]
- Velletri, T.; Xie, N.; Wang, Y.; Huang, Y.; Yang, Q.; Chen, X.; Chen, Q.; Shou, P.; Gan, Y.; Cao, G.; et al. P53 Functional Abnormality in Mesenchymal Stem Cells Promotes Osteosarcoma Development. Cell Death Dis. 2016, 7, e2015. [Google Scholar] [CrossRef] [PubMed]
- Boregowda, S.V.; Krishnappa, V.; Strivelli, J.; Haga, C.L.; Booker, C.N.; Phinney, D.G. Basal P53 Expression Is Indispensable for Mesenchymal Stem Cell Integrity. Cell Death Differ. 2018, 25, 679–692. [Google Scholar] [CrossRef] [PubMed]
- O’Brate, A.; Giannakakou, P. The Importance of P53 Location: Nuclear or Cytoplasmic Zip Code? Drug Resist. Updat. Rev. Comment. Antimicrob. Anticancer Chemother. 2003, 6, 313–322. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Jiang, J.; Tan, W.; Xia, Y.; Cao, H.; Meng, Y.; Da, Z.; Liu, H.; Cheng, C. P53/P21 Pathway Involved in Mediating Cellular Senescence of Bone Marrow-Derived Mesenchymal Stem Cells from Systemic Lupus Erythematosus Patients. Clin. Dev. Immunol. 2013, 2013, 134243. [Google Scholar] [CrossRef]
- Solozobova, V.; Blattner, C. P53 in Stem Cells. World J. Biol. Chem. 2011, 2, 202–214. [Google Scholar] [CrossRef]
- Solozobova, V.; Rolletschek, A.; Blattner, C. Nuclear Accumulation and Activation of P53 in Embryonic Stem Cells after DNA Damage. BMC Cell Biol. 2009, 10, 46. [Google Scholar] [CrossRef]
- Ferretti, C.; Lucarini, G.; Andreoni, C.; Salvolini, E.; Bianchi, N.; Vozzi, G.; Gigante, A.; Mattioli-Belmonte, M. Human Periosteal Derived Stem Cell Potential: The Impact of Age. Stem Cell Rev. Rep. 2015, 11, 487–500. [Google Scholar] [CrossRef]
- Sullivan, K.D.; Galbraith, M.D.; Andrysik, Z.; Espinosa, J.M. Mechanisms of Transcriptional Regulation by P53. Cell Death Differ. 2018, 25, 133–143. [Google Scholar] [CrossRef]
- Aksoy, C.; Severcan, F. Role of Vibrational Spectroscopy in Stem Cell Research. Spectrosc. Int. J. 2012, 27, 167–184. [Google Scholar] [CrossRef]
- Matthews, Q.; Jirasek, A.; Lum, J.; Duan, X.; Brolo, A.G. Variability in Raman Spectra of Single Human Tumor Cells Cultured in Vitro: Correlation with Cell Cycle and Culture Confluency. Appl. Spectrosc. 2010, 64, 871–887. [Google Scholar] [CrossRef] [PubMed]
- Movasaghi, Z.; Rehman, S.; Rehman, I.U. Raman Spectroscopy of Biological Tissues. Appl. Spectrosc. Rev. 2007, 42, 493–541. [Google Scholar] [CrossRef]
- Brauchle, E.; Noor, S.; Holtorf, E.; Garbe, C.; Schenke-Layland, K.; Busch, C. Raman Spectroscopy as an Analytical Tool for Melanoma Research. Clin. Exp. Dermatol. 2014, 39, 636–645. [Google Scholar] [CrossRef] [PubMed]
- Hoppe, A. What MRNA Abundances Can Tell Us about Metabolism. Metabolites 2012, 2, 614–631. [Google Scholar] [CrossRef] [PubMed]
- Verrier, S.; Notingher, I.; Polak, J.M.; Hench, L.L. In Situ Monitoring of Cell Death Using Raman Microspectroscopy. Biopolymers 2004, 74, 157–162. [Google Scholar] [CrossRef]
- Parrotta, E.; De Angelis, M.T.; Scalise, S.; Candeloro, P.; Santamaria, G.; Paonessa, M.; Coluccio, M.L.; Perozziello, G.; De Vitis, S.; Sgura, A.; et al. Two Sides of the Same Coin? Unraveling Subtle Differences between Human Embryonic and Induced Pluripotent Stem Cells by Raman Spectroscopy. Stem Cell Res. Ther. 2017, 8, 271. [Google Scholar] [CrossRef]
- Bogdan, P.; Deasy, B.M.; Gharaibeh, B.; Roehrs, T.; Marculescu, R. Heterogeneous Structure of Stem Cells Dynamics: Statistical Models and Quantitative Predictions. Sci. Rep. 2014, 4, 4826. [Google Scholar] [CrossRef]
- McLeod, C.M.; Mauck, R.L. On the Origin and Impact of Mesenchymal Stem Cell Heterogeneity: New Insights and Emerging Tools for Single Cell Analysis. Eur. Cell. Mater. 2017, 34, 217–231. [Google Scholar] [CrossRef]
- Tan, Y.; Konorov, S.O.; Schulze, H.G.; Piret, J.M.; Blades, M.W.; Turner, R.F.B. Comparative Study Using Raman Microspectroscopy Reveals Spectral Signatures of Human Induced Pluripotent Cells More Closely Resemble Those from Human Embryonic Stem Cells than Those from Differentiated Cells. Analyst 2012, 137, 4509–4515. [Google Scholar] [CrossRef]
- Germond, A.; Panina, Y.; Shiga, M.; Niioka, H.; Watanabe, T.M. Following Embryonic Stem Cells, Their Differentiated Progeny, and Cell-State Changes During IPS Reprogramming by Raman Spectroscopy. Anal. Chem. 2020, 92, 14915–14923. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, T.; Wang, L.; Cai, Y.; Zhong, X.; He, X.; Hu, L.; Tian, S.; Wu, M.; Hui, L.; et al. Fatty Acid Synthesis Is Critical for Stem Cell Pluripotency via Promoting Mitochondrial Fission. EMBO J. 2017, 36, 1330–1347. [Google Scholar] [CrossRef] [PubMed]
- Ow, Y.-L.P.; Green, D.R.; Hao, Z.; Mak, T.W. Cytochrome c: Functions beyond Respiration. Nat. Rev. Mol. Cell Biol. 2008, 9, 532–542. [Google Scholar] [CrossRef] [PubMed]
- Zaidi, S.; Hassan, M.I.; Islam, A.; Ahmad, F. The Role of Key Residues in Structure, Function, and Stability of Cytochrome-c. Cell. Mol. Life Sci. 2014, 71, 229–255. [Google Scholar] [CrossRef] [PubMed]
- Kühlbrandt, W. Structure and Function of Mitochondrial Membrane Protein Complexes. BMC Biol. 2015, 13, 89. [Google Scholar] [CrossRef] [PubMed]
- Eleftheriadis, T.; Pissas, G.; Liakopoulos, V.; Stefanidis, I. Cytochrome c as a Potentially Clinical Useful Marker of Mitochondrial and Cellular Damage. Front. Immunol. 2016, 7, 279. [Google Scholar] [CrossRef]
- Cai, J.; Yang, J.; Jones, D.P. Mitochondrial Control of Apoptosis: The Role of Cytochrome c. Biochim. Biophys. Acta 1998, 1366, 139–149. [Google Scholar] [CrossRef]
- Martínez-Fábregas, J.; Díaz-Moreno, I.; González-Arzola, K.; Díaz-Quintana, A.; De la Rosa, M.A. A Common Signalosome for Programmed Cell Death in Humans and Plants. Cell Death Dis. 2014, 5, e1314. [Google Scholar] [CrossRef]
- Okada, M.; Smith, N.I.; Palonpon, A.F.; Endo, H.; Kawata, S.; Sodeoka, M.; Fujita, K. Label-Free Raman Observation of Cytochrome c Dynamics during Apoptosis. Proc. Natl. Acad. Sci. USA 2012, 109, 28–32. [Google Scholar] [CrossRef]
- Salehi, H.; Middendorp, E.; Panayotov, I.; Collart Dutilleul, P.-Y.; Dutilleul, P.-Y.C.; Vegh, A.-G.; Ramakrishnan, S.; Gergely, C.; Cuisinier, F. Confocal Raman Data Analysis Enables Identifying Apoptosis of MCF-7 Cells Caused by Anticancer Drug Paclitaxel. J. Biomed. Opt. 2013, 18, 56010. [Google Scholar] [CrossRef]
- Ichimura, T.; Chiu, L.; Fujita, K.; Kawata, S.; Watanabe, T.M.; Yanagida, T.; Fujita, H. Visualizing Cell State Transition Using Raman Spectroscopy. PLoS ONE 2014, 9, e84478. [Google Scholar] [CrossRef] [PubMed]
| Energy (cm−1) | Biomolecule Assignment |
|---|---|
| 940 | Skeletal modes in polysaccharides |
| 957 | O-P-O symmetric stretch in adenosine-monophosphate |
| 1003 | Symmetric ring breathing mode in phenylalanine (Phe) |
| 1010 | Ring breathing in benzene ring of tryptophan (Trp) |
| 1033 | C-H in plane bend (Phe) |
| 1050 | C-O and C-N stretch in proteins |
| 1080 | O-P-O symmetric stretch |
| 1100 | PO2- symmetric stretching in RNA and phosphatidylinositol |
| 1123 | Cytochrome C; C-C asymmetric stretch in fatty acids |
| 1155 | C-C and C-N stretch in proteins |
| 1165 | C-O stretch, C-OH bending, C=C stretch in lipids, C-C stretch in proteins |
| 1173 | G-ring stretch, C-C-H bending in phenol ring (DNA) |
| 1206 | C-C stretch in phenol ring of tyrosine (Tyr) |
| 1245 | NH2 bending in Amide IIIβ |
| 1266 | Amide IIIα |
| 1310 | C-H deformation (saturated. lipids) |
| 1335 | DNA purine bases (CH3CH2 wagging mode of polynucleotide chain) |
| 1450 | CH2 scissoring in lipids |
| 1554 | Amide II |
| 1604 | Phe, Tyr |
| 1655 | Amide Iα |
| 1669 | Amide Iβ |
| Donor | Sex | Blood Type | Karyotype | Age |
|---|---|---|---|---|
| D1 | male | AB+ | 46, XY, 20 | 8 years and 10 months |
| D2 | female | B+ | 46, XX, 20 | 12 years and 3 months |
| D3 | female | O+ | 46, XX, 20 | 2 years and 5 months |
| D4 | male | AB+ | 46, XY, 20 | 12 years and 4 months |
| D5 | female | O+ | 46, XX, 20 | 12 years and 2 months |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kukolj, T.; Lazarević, J.; Borojević, A.; Ralević, U.; Vujić, D.; Jauković, A.; Lazarević, N.; Bugarski, D. A Single-Cell Raman Spectroscopy Analysis of Bone Marrow Mesenchymal Stem/Stromal Cells to Identify Inter-Individual Diversity. Int. J. Mol. Sci. 2022, 23, 4915. https://doi.org/10.3390/ijms23094915
Kukolj T, Lazarević J, Borojević A, Ralević U, Vujić D, Jauković A, Lazarević N, Bugarski D. A Single-Cell Raman Spectroscopy Analysis of Bone Marrow Mesenchymal Stem/Stromal Cells to Identify Inter-Individual Diversity. International Journal of Molecular Sciences. 2022; 23(9):4915. https://doi.org/10.3390/ijms23094915
Chicago/Turabian StyleKukolj, Tamara, Jasmina Lazarević, Ana Borojević, Uroš Ralević, Dragana Vujić, Aleksandra Jauković, Nenad Lazarević, and Diana Bugarski. 2022. "A Single-Cell Raman Spectroscopy Analysis of Bone Marrow Mesenchymal Stem/Stromal Cells to Identify Inter-Individual Diversity" International Journal of Molecular Sciences 23, no. 9: 4915. https://doi.org/10.3390/ijms23094915
APA StyleKukolj, T., Lazarević, J., Borojević, A., Ralević, U., Vujić, D., Jauković, A., Lazarević, N., & Bugarski, D. (2022). A Single-Cell Raman Spectroscopy Analysis of Bone Marrow Mesenchymal Stem/Stromal Cells to Identify Inter-Individual Diversity. International Journal of Molecular Sciences, 23(9), 4915. https://doi.org/10.3390/ijms23094915






