Human Serine Racemase Weakly Binds the Third PDZ Domain of PSD-95
Abstract
1. Introduction
2. Results
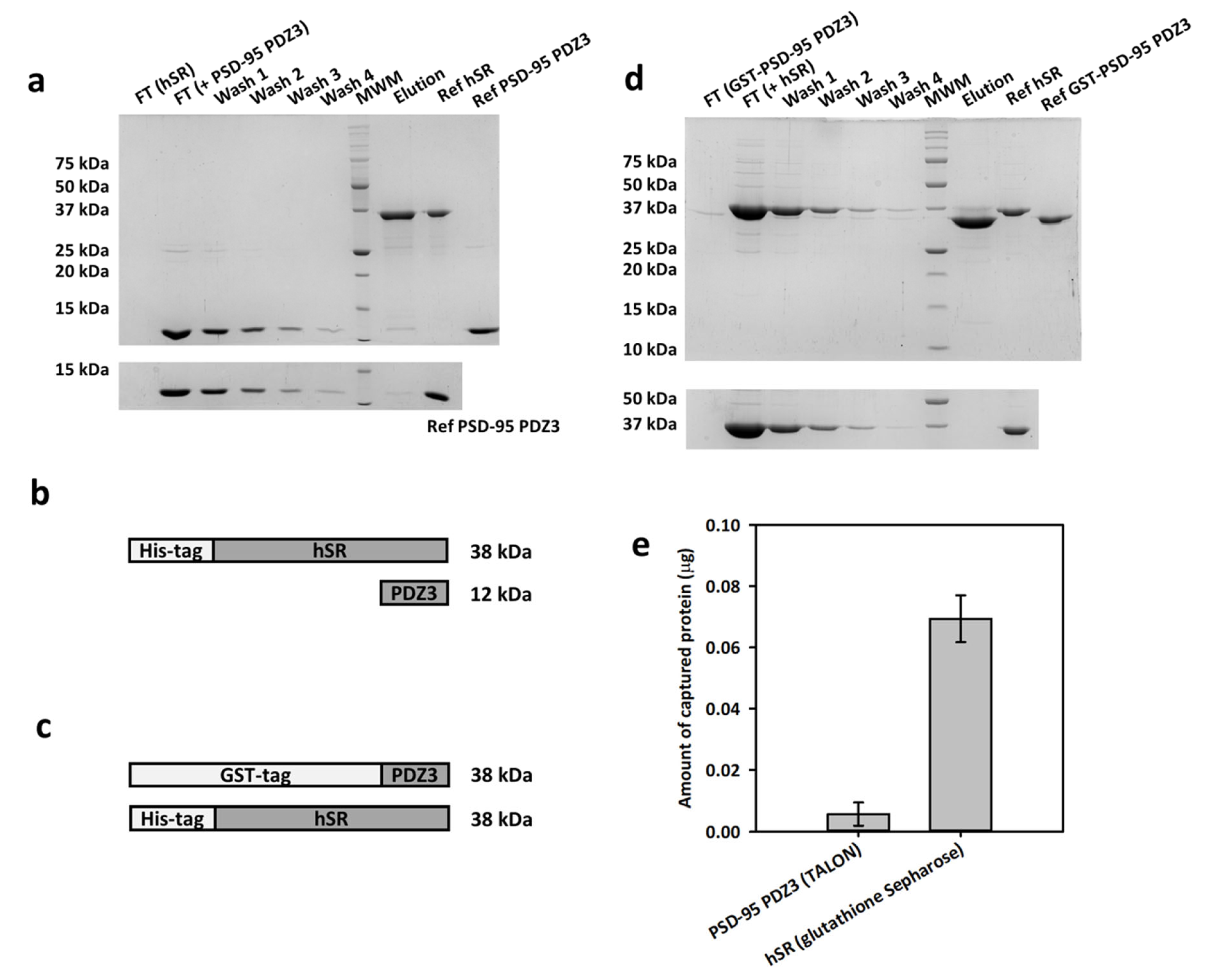
2.1. Pull-Down Assays exclude the Presence of a Strong Binding between hSR and PSD-95 PDZ3
2.2. NMR Titration Confirms That the Binding between the Two Proteins Is Weak
2.3. Cross-Linking Experiments Suggest That hSR Does Bind to PSD-95 PDZ3
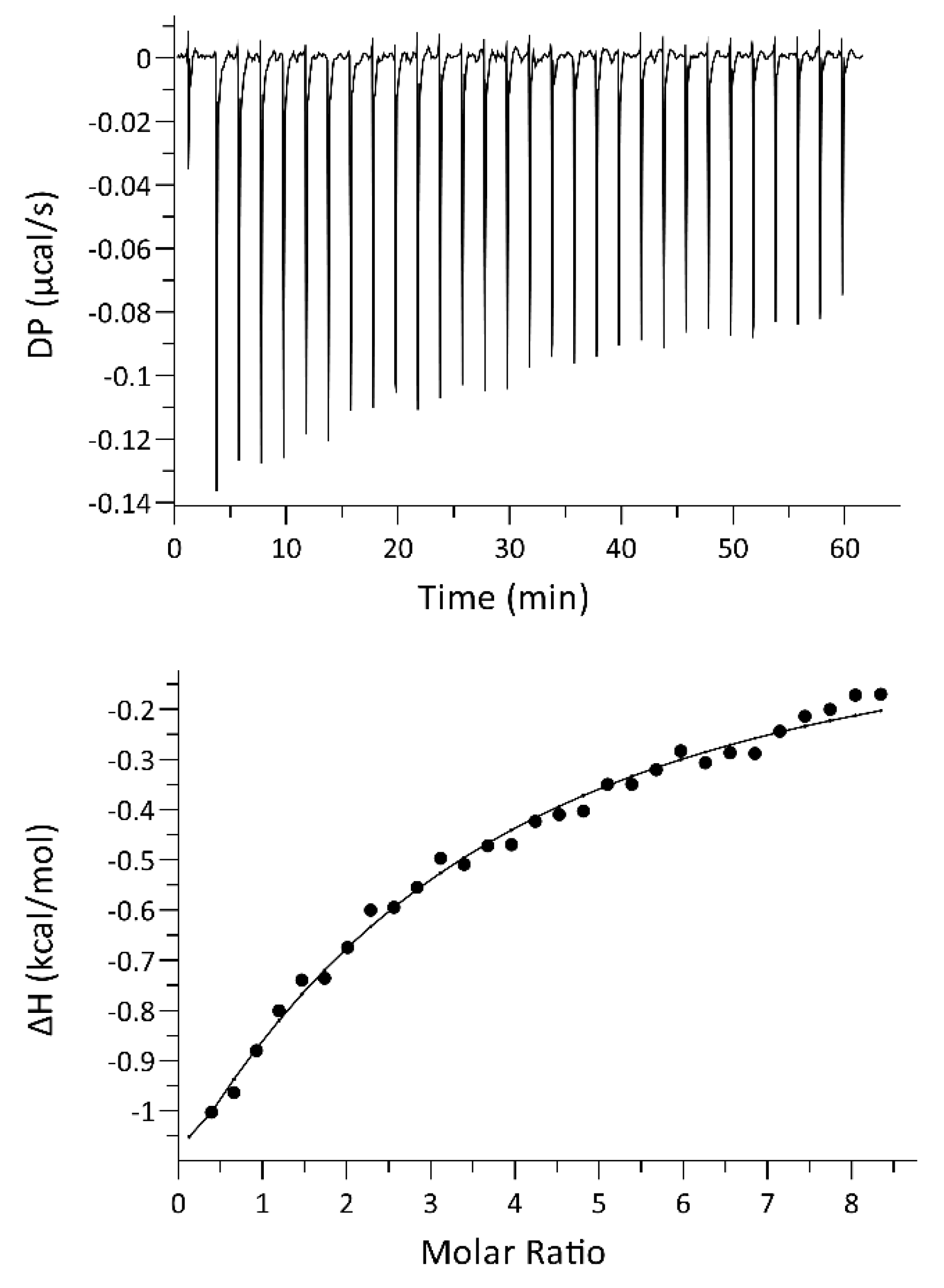
2.4. ITC Titrations Confirm That hSR Binds with Low Affinity to PSD-95 PDZ3
2.5. The Enzymatic Activity of hSR Is Moderately Increased by the Presence of PSD-95 PDZ3
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Purification of hSR and PSD-95 PDZ3
4.3. Pull-Down Assays
4.4. NMR Spectroscopy
4.5. Cross-Linking
4.6. Isothermal Titration Calorimetry (ITC)
4.7. Enzymatic Assays
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Campanini, B.; Spyrakis, F.; Peracchi, A.; Mozzarelli, A. Serine racemase: A key player in neuron activity and in neuropathologies. Front. Biosci. 2013, 18, 1112–1128. [Google Scholar]
- Raboni, S.; Marchetti, M.; Faggiano, S.; Campanini, B.; Bruno, S.; Marchesani, F.; Margiotta, M.; Mozzarelli, A. The Energy Landscape of Human Serine Racemase. Front. Mol. Biosci. 2018, 5, 112. [Google Scholar] [CrossRef] [PubMed]
- Wolosker, H.; Sheth, K.N.; Takahashi, M.; Mothet, J.P.; Brady, R.O., Jr.; Ferris, C.D.; Snyder, S.H. Purification of serine racemase: Biosynthesis of the neuromodulator D-serine. Proc. Natl. Acad. Sci. USA 1999, 96, 721–725. [Google Scholar] [CrossRef] [PubMed]
- Pollegioni, L.; Sacchi, S. Metabolism of the neuromodulator D-serine. Cell. Mol. Life Sci. 2010, 67, 2387–2404. [Google Scholar] [CrossRef] [PubMed]
- Wolosker, H.; Mori, H. Serine racemase: An unconventional enzyme for an unconventional transmitter. Amino Acids 2012, 43, 1895–1904. [Google Scholar] [CrossRef]
- De Miranda, J.; Santoro, A.; Engelender, S.; Wolosker, H. Human serine racemase: Molecular cloning, genomic organization and functional analysis. Gene 2000, 256, 183–188. [Google Scholar] [CrossRef]
- Graham, D.L.; Beio, M.L.; Nelson, D.L.; Berkowitz, D.B. Human Serine Racemase: Key Residues/Active Site Motifs and Their Relation to Enzyme Function. Front. Mol. Biosci. 2019, 6, 8. [Google Scholar] [CrossRef]
- Coyle, J.T.; Balu, D.T. The Role of Serine Racemase in the Pathophysiology of Brain Disorders. Adv. Pharmacol. 2018, 82, 35–56. [Google Scholar]
- Dellafiora, L.; Marchetti, M.; Spyrakis, F.; Orlandi, V.; Campanini, B.; Cruciani, G.; Cozzini, P.; Mozzarelli, A. Expanding the chemical space of human serine racemase inhibitors. Bioorganic Med. Chem. Lett. 2015, 25, 4297–4303. [Google Scholar] [CrossRef]
- Jiraskova-Vanickova, J.; Ettrich, R.; Vorlova, B.; Hoffman, H.E.; Lepsik, M.; Jansa, P.; Konvalinka, J. Inhibition of human serine racemase, an emerging target for medicinal chemistry. Curr. Drug Targets 2011, 12, 1037–1055. [Google Scholar] [CrossRef]
- Takahara, S.; Nakagawa, K.; Uchiyama, T.; Yoshida, T.; Matsumoto, K.; Kawasumi, Y.; Mizuguchi, M.; Obita, T.; Watanabe, Y.; Hayakawa, D.; et al. Design, synthesis, and evaluation of novel inhibitors for wild-type human serine racemase. Bioorganic Med. Chem. Lett. 2018, 28, 441–445. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, H.E.; Jiraskova, J.; Ingr, M.; Zvelebil, M.; Konvalinka, J. Recombinant human serine racemase: Enzymologic characterization and comparison with its mouse ortholog. Protein Expr. Purif. 2009, 63, 62–67. [Google Scholar] [CrossRef] [PubMed]
- Bruno, S.; Marchesani, F.; Dellafiora, L.; Margiotta, M.; Faggiano, S.; Campanini, B.; Mozzarelli, A. Human serine racemase is allosterically modulated by NADH and reduced nicotinamide derivatives. Biochem. J. 2016, 473, 3505–3516. [Google Scholar] [CrossRef] [PubMed]
- Bruno, S.; Margiotta, M.; Marchesani, F.; Paredi, G.; Orlandi, V.; Faggiano, S.; Ronda, L.; Campanini, B.; Mozzarelli, A. Magnesium and calcium ions differentially affect human serine racemase activity and modulate its quaternary equilibrium toward a tetrameric form. Biochim. Biophys. Acta Proteins Proteom. 2017, 1865, 381–387. [Google Scholar] [CrossRef]
- Marchetti, M.; Bruno, S.; Campanini, B.; Bettati, S.; Peracchi, A.; Mozzarelli, A. Regulation of human serine racemase activity and dynamics by halides, ATP and malonate. Amino Acids 2015, 47, 163–173. [Google Scholar] [CrossRef]
- Marchetti, M.; Bruno, S.; Campanini, B.; Peracchi, A.; Mai, N.; Mozzarelli, A. ATP binding to human serine racemase is cooperative and modulated by glycine. FEBS J. 2013, 280, 5853–5863. [Google Scholar] [CrossRef]
- Michielon, A.; Marchesani, F.; Faggiano, S.; Giaccari, R.; Campanini, B.; Bettati, S.; Mozzarelli, A.; Bruno, S. Human serine racemase is inhibited by glyceraldehyde 3-phosphate, but not by glyceraldehyde 3-phosphate dehydrogenase. Biochim. Biophys. Acta Proteins Proteom. 2021, 1869, 140544. [Google Scholar] [CrossRef]
- Baumgart, F.; Rodriguez-Crespo, I. D-amino acids in the brain: The biochemistry of brain serine racemase. FEBS J. 2008, 275, 3538–3545. [Google Scholar] [CrossRef]
- Fujii, K.; Maeda, K.; Hikida, T.; Mustafa, A.K.; Balkissoon, R.; Xia, J.; Yamada, T.; Ozeki, Y.; Kawahara, R.; Okawa, M.; et al. Serine racemase binds to PICK1: Potential relevance to schizophrenia. Mol. Psychiatry 2006, 11, 150–157. [Google Scholar] [CrossRef]
- Ma, T.M.; Abazyan, S.; Abazyan, B.; Nomura, J.; Yang, C.; Seshadri, S.; Sawa, A.; Snyder, S.H.; Pletnikov, M.V. Pathogenic disruption of DISC1-serine racemase binding elicits schizophrenia-like behavior via D-serine depletion. Mol. Psychiatry 2013, 18, 557–567. [Google Scholar] [CrossRef]
- Zhuang, Z.; Yang, B.; Theus, M.H.; Sick, J.T.; Bethea, J.R.; Sick, T.J.; Liebl, D.J. EphrinBs regulate D-serine synthesis and release in astrocytes. J. Neurosci. Off. J. Soc. Neurosci. 2010, 30, 16015–16024. [Google Scholar] [CrossRef] [PubMed]
- Hung, A.Y.; Sheng, M. PDZ domains: Structural modules for protein complex assembly. J. Biol. Chem. 2002, 277, 5699–5702. [Google Scholar] [CrossRef] [PubMed]
- Tonikian, R.; Zhang, Y.; Sazinsky, S.L.; Currell, B.; Yeh, J.H.; Reva, B.; Held, H.A.; Appleton, B.A.; Evangelista, M.; Wu, Y.; et al. A specificity map for the PDZ domain family. PLoS Biol. 2008, 6, e239. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Zheng, J.J. PDZ domains and their binding partners: Structure, specificity, and modification. Cell Commun. Signal. CCS 2010, 8, 8. [Google Scholar] [CrossRef]
- Ye, F.; Zhang, M. Structures and target recognition modes of PDZ domains: Recurring themes and emerging pictures. Biochem. J. 2013, 455, 1–14. [Google Scholar] [CrossRef]
- Chi, C.N.; Bach, A.; Stromgaard, K.; Gianni, S.; Jemth, P. Ligand binding by PDZ domains. BioFactors 2012, 38, 338–348. [Google Scholar] [CrossRef]
- Xu, W. PSD-95-like membrane associated guanylate kinases (PSD-MAGUKs) and synaptic plasticity. Curr. Opin. Neurobiol. 2011, 21, 306–312. [Google Scholar] [CrossRef]
- Ma, T.M.; Paul, B.D.; Fu, C.; Hu, S.; Zhu, H.; Blackshaw, S.; Wolosker, H.; Snyder, S.H. Serine racemase regulated by binding to stargazin and PSD-95: Potential N-methyl-D-aspartate-alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (NMDA-AMPA) glutamate neurotransmission cross-talk. J. Biol. Chem. 2014, 289, 29631–29641. [Google Scholar] [CrossRef]
- Lin, H.; Jacobi, A.A.; Anderson, S.A.; Lynch, D.R. D-Serine and Serine Racemase Are Associated with PSD-95 and Glutamatergic Synapse Stability. Front. Cell. Neurosci. 2016, 10, 34. [Google Scholar] [CrossRef]
- Brymora, A.; Valova, V.A.; Robinson, P.J. Protein-protein interactions identified by pull-down experiments and mass spectrometry. Curr. Protoc. Cell Biol. 2004, 17, 17.5.1–17.5.51. [Google Scholar] [CrossRef]
- Chi, C.N.; Bach, A.; Engstrom, A.; Stromgaard, K.; Lundstrom, P.; Ferguson, N.; Jemth, P. Biophysical characterization of the complex between human papillomavirus E6 protein and synapse-associated protein 97. J. Biol. Chem. 2011, 286, 3597–3606. [Google Scholar] [CrossRef] [PubMed]
- Merino-Gracia, J.; Costas-Insua, C.; Canales, M.A.; Rodriguez-Crespo, I. Insights into the C-terminal Peptide Binding Specificity of the PDZ Domain of Neuronal Nitric-oxide Synthase: Characterization of the interaction with the Tight Junction Protein Claudin-3. J. Biol. Chem. 2016, 291, 11581–11595. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, A.; Konuma, T.; Yanaka, S.; Sugase, K. Quantitative analysis of protein-ligand interactions by NMR. Prog. Nucl. Magn. Reson. Spectrosc. 2016, 96, 47–57. [Google Scholar] [CrossRef] [PubMed]
- Williamson, M.P. Using chemical shift perturbation to characterise ligand binding. Prog. Nucl. Magn. Reson. Spectrosc. 2013, 73, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Bruce, J.E. Chemical cross-linking for protein-protein interaction studies. Methods Mol. Biol. 2009, 492, 283–293. [Google Scholar]
- Amacher, J.F.; Brooks, L.; Hampton, T.H.; Madden, D.R. Specificity in PDZ-peptide interaction networks: Computational analysis and review. J. Struct. Biol. X 2020, 4, 100022. [Google Scholar] [CrossRef]
- Cobos, E.S.; Sanchez, I.E.; Chemes, L.B.; Martinez, J.C.; Murciano-Calles, J. A Thermodynamic Analysis of the Binding Specificity between Four Human PDZ Domains and Eight Host, Viral and Designed Ligands. Biomolecules 2021, 11, 1071. [Google Scholar] [CrossRef]
- Saro, D.; Li, T.; Rupasinghe, C.; Paredes, A.; Caspers, N.; Spaller, M.R. A thermodynamic ligand binding study of the third PDZ domain (PDZ3) from the mammalian neuronal protein PSD-95. Biochemistry 2007, 46, 6340–6352. [Google Scholar] [CrossRef][Green Version]
- Songyang, Z.; Fanning, A.S.; Fu, C.; Xu, J.; Marfatia, S.M.; Chishti, A.H.; Crompton, A.; Chan, A.C.; Anderson, J.M.; Cantley, L.C. Recognition of unique carboxyl-terminal motifs by distinct PDZ domains. Science 1997, 275, 73–77. [Google Scholar] [CrossRef]
- Chi, C.N.; Haq, S.R.; Rinaldo, S.; Dogan, J.; Cutruzzola, F.; Engstrom, A.; Gianni, S.; Lundstrom, P.; Jemth, P. Interactions outside the boundaries of the canonical binding groove of a PDZ domain influence ligand binding. Biochemistry 2012, 51, 8971–8979. [Google Scholar] [CrossRef]
- Toto, A.; Pedersen, S.W.; Karlsson, O.A.; Moran, G.E.; Andersson, E.; Chi, C.N.; Stromgaard, K.; Gianni, S.; Jemth, P. Ligand binding to the PDZ domains of postsynaptic density protein 95. Protein Eng. Des. Sel. 2016, 29, 169–175. [Google Scholar] [CrossRef] [PubMed]
- Murciano-Calles, J.; Corbi-Verge, C.; Candel, A.M.; Luque, I.; Martinez, J.C. Post-translational modifications modulate ligand recognition by the third PDZ domain of the MAGUK protein PSD-95. PLoS ONE 2014, 9, e90030. [Google Scholar] [CrossRef] [PubMed]
- Doyle, D.A.; Lee, A.; Lewis, J.; Kim, E.; Sheng, M.; MacKinnon, R. Crystal structures of a complexed and peptide-free membrane protein-binding domain: Molecular basis of peptide recognition by PDZ. Cell 1996, 85, 1067–1076. [Google Scholar] [CrossRef]
- Gianni, S.; Engstrom, A.; Larsson, M.; Calosci, N.; Malatesta, F.; Eklund, L.; Ngang, C.C.; Travaglini-Allocatelli, C.; Jemth, P. The kinetics of PDZ domain-ligand interactions and implications for the binding mechanism. J. Biol. Chem. 2005, 280, 34805–34812. [Google Scholar] [CrossRef] [PubMed]
- Haq, S.R.; Chi, C.N.; Bach, A.; Dogan, J.; Engstrom, A.; Hultqvist, G.; Karlsson, O.A.; Lundstrom, P.; Montemiglio, L.C.; Stromgaard, K.; et al. Side-chain interactions form late and cooperatively in the binding reaction between disordered peptides and PDZ domains. J. Am. Chem. Soc. 2012, 134, 599–605. [Google Scholar] [CrossRef]
- Ivarsson, Y. Plasticity of PDZ domains in ligand recognition and signaling. FEBS Lett. 2012, 586, 2638–2647. [Google Scholar] [CrossRef]
- Cushing, P.R.; Fellows, A.; Villone, D.; Boisguerin, P.; Madden, D.R. The relative binding affinities of PDZ partners for CFTR: A biochemical basis for efficient endocytic recycling. Biochemistry 2008, 47, 10084–10098. [Google Scholar] [CrossRef]
- Mustafa, A.K.; Kumar, M.; Selvakumar, B.; Ho, G.P.; Ehmsen, J.T.; Barrow, R.K.; Amzel, L.M.; Snyder, S.H. Nitric oxide S-nitrosylates serine racemase, mediating feedback inhibition of D-serine formation. Proc. Natl. Acad. Sci. USA 2007, 104, 2950–2955. [Google Scholar] [CrossRef]
- Marchesani, F.; Bruno, S.; Paredi, G.; Raboni, S.; Campanini, B.; Mozzarelli, A. Human serine racemase is nitrosylated at multiple sites. Biochim. Biophys. Acta Proteins Proteom. 2018, 1866, 813–821. [Google Scholar] [CrossRef]
- Marchesani, F.; Gianquinto, E.; Autiero, I.; Michielon, A.; Campanini, B.; Faggiano, S.; Bettati, S.; Mozzarelli, A.; Spyrakis, F.; Bruno, S. The allosteric interplay between S-nitrosylation and glycine binding controls the activity of human serine racemase. FEBS J. 2021, 288, 3034–3054. [Google Scholar] [CrossRef]
- Pedersen, S.W.; Albertsen, L.; Moran, G.E.; Levesque, B.; Pedersen, S.B.; Bartels, L.; Wapenaar, H.; Ye, F.; Zhang, M.; Bowen, M.E.; et al. Site-Specific Phosphorylation of PSD-95 PDZ Domains Reveals Fine-Tuned Regulation of Protein-Protein Interactions. ACS Chem. Biol. 2017, 12, 2313–2323. [Google Scholar] [CrossRef] [PubMed]
- Petit, C.M.; Zhang, J.; Sapienza, P.J.; Fuentes, E.J.; Lee, A.L. Hidden dynamic allostery in a PDZ domain. Proc. Natl. Acad. Sci. USA 2009, 106, 18249–18254. [Google Scholar] [CrossRef] [PubMed]
- Stevens, A.O.; He, Y. Allosterism in the PDZ Family. Int. J. Mol. Sci. 2022, 23, 1454. [Google Scholar] [CrossRef] [PubMed]
- Dixon, S.M.; Li, P.; Liu, R.; Wolosker, H.; Lam, K.S.; Kurth, M.J.; Toney, M.D. Slow-binding human serine racemase inhibitors from high-throughput screening of combinatorial libraries. J. Med. Chem. 2006, 49, 2388–2397. [Google Scholar] [CrossRef] [PubMed]
- Canosa, A.V.; Faggiano, S.; Marchetti, M.; Armao, S.; Bettati, S.; Bruno, S.; Percudani, R.; Campanini, B.; Mozzarelli, A. Glutamine 89 is a key residue in the allosteric modulation of human serine racemase activity by ATP. Sci. Rep. 2018, 8, 9016. [Google Scholar] [CrossRef]
- The UniProt Consortium. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Research 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Gouet, P.; Courcelle, E.; Stuart, D.I.; Metoz, F. ESPript: Analysis of multiple sequence alignments in PostScript. Bioinformatics 1999, 15, 305–308. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Giaccari, R.; Marchesani, F.; Compari, C.; Fisicaro, E.; Mozzarelli, A.; Campanini, B.; Bettati, S.; Bruno, S.; Faggiano, S. Human Serine Racemase Weakly Binds the Third PDZ Domain of PSD-95. Int. J. Mol. Sci. 2022, 23, 4959. https://doi.org/10.3390/ijms23094959
Giaccari R, Marchesani F, Compari C, Fisicaro E, Mozzarelli A, Campanini B, Bettati S, Bruno S, Faggiano S. Human Serine Racemase Weakly Binds the Third PDZ Domain of PSD-95. International Journal of Molecular Sciences. 2022; 23(9):4959. https://doi.org/10.3390/ijms23094959
Chicago/Turabian StyleGiaccari, Roberta, Francesco Marchesani, Carlotta Compari, Emilia Fisicaro, Andrea Mozzarelli, Barbara Campanini, Stefano Bettati, Stefano Bruno, and Serena Faggiano. 2022. "Human Serine Racemase Weakly Binds the Third PDZ Domain of PSD-95" International Journal of Molecular Sciences 23, no. 9: 4959. https://doi.org/10.3390/ijms23094959
APA StyleGiaccari, R., Marchesani, F., Compari, C., Fisicaro, E., Mozzarelli, A., Campanini, B., Bettati, S., Bruno, S., & Faggiano, S. (2022). Human Serine Racemase Weakly Binds the Third PDZ Domain of PSD-95. International Journal of Molecular Sciences, 23(9), 4959. https://doi.org/10.3390/ijms23094959








